Incidence, genetic diversity, and antimicrobial resistance profiles of Vibrio parahaemolyticus in seafood in Bangkok and eastern Thailand
- Published
- Accepted
- Received
- Academic Editor
- Michael LaMontagne
- Subject Areas
- Aquaculture, Fisheries and Fish Science, Food Science and Technology, Microbiology, Molecular Biology
- Keywords
- V. parahaemolyticus, Seafood, Antimicrobial resistance, Genetic diversity, Multilocus sequence typing, Thailand
- Copyright
- © 2023 Changsen et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Incidence, genetic diversity, and antimicrobial resistance profiles of Vibrio parahaemolyticus in seafood in Bangkok and eastern Thailand. PeerJ 11:e15283 https://doi.org/10.7717/peerj.15283
Abstract
Background
Emergence of Vibrio parahaemolyticus pandemic strain O3:K6 was first documented in 1996. Since then it has been accounted for large outbreaks of diarrhea globally. In Thailand, prior studies on pandemic and non-pandemic V. parahaemolyticus had mostly been done in the south. The incidence and molecular characterization of pandemic and non-pandemic strains in other parts of Thailand have not been fully characterized. This study examined the incidence of V. parahaemolyticus in seafood samples purchased in Bangkok and collected in eastern Thailand and characterized V. parahaemolyticus isolates. Potential virulence genes, VPaI-7, T3SS2, and biofilm were examined. Antimicrobial resistance (AMR) profiles and AMR genes (ARGs) were determined.
Methods
V. parahaemolyticus was isolated from 190 marketed and farmed seafood samples by a culture method and confirmed by polymerase chain reaction (PCR). The incidence of pandemic and non-pandemic V. parahaemolyticus and VPaI-7, T3SS2, and biofilm genes was examined by PCR. AMR profiles were verified by a broth microdilution technique. The presence of ARGs was verified by genome analysis. V. parahaemolyticus characterization was done by multilocus sequence typing (MLST). A phylogenomic tree was built from nucleotide sequences by UBCG2.0 and RAxML softwares.
Results
All 50 V. parahaemolyticus isolates including 21 pathogenic and 29 non-pathogenic strains from 190 samples had the toxRS/old sequence, indicating non-pandemic strains. All isolates had biofilm genes (VP0950, VP0952, and VP0962). None carried T3SS2 genes (VP1346 and VP1367), while VPaI-7 gene (VP1321) was seen in two isolates. Antimicrobial susceptibility profiles obtained from 36 V. parahaemolyticus isolates revealed high frequency of resistance to colistin (100%, 36/36) and ampicillin (83%, 30/36), but susceptibility to amoxicillin/clavulanic acid and piperacillin/tazobactam (100%, 36/36). Multidrug resistance (MDR) was seen in 11 isolates (31%, 11/36). Genome analysis revealed ARGs including blaCARB (100%, 36/36), tet(34) (83%, 30/36), tet(35) (42%, 15/36), qnrC (6%, 2/36), dfrA6 (3%, 1/36), and blaCTX-M-55 (3%, 1/36). Phylogenomic and MLST analyses classified 36 V. parahaemolyticus isolates into 5 clades, with 12 known and 13 novel sequence types (STs), suggesting high genetic variation among the isolates.
Conclusions
Although none V. parahaemolyticus strains isolated from seafood samples purchased in Bangkok and collected in eastern Thailand were pandemic strains, around one third of isolates were MDR V. parahaemolyticus strains. The presence of resistance genes of the first-line antibiotics for V. parahaemolyticus infection raises a major concern for clinical treatment outcome since these resistance genes could be highly expressed under suitable circumstances.
Introduction
Seafood is generally a good source of high-quality proteins, essential amino acids, and healthy fats for a lower calorie intake per portion compared to other animal meats. Due to the increase in global consumption of seafood products, oversight of seafood quality is a priority to prohibit contamination from seafood-borne pathogens (FAO, 2020; Choudhury et al., 2022). Vibrio parahaemolyticus is a Gram-negative bacterium commonly found in seawater, seafood (particularly oysters), and aquatic products (Nakaguchi, 2013; Odeyemi, 2016). It is a leading cause of human gastroenteritis following consumption of raw or improperly cooked contaminated seafood (Martinez-Urtaza & Baker-Austin, 2020). Pathogenicity of V. parahaemolyticus infection has been attributed to the expression of virulent determinant genes including tdh (thermostable direct hemolysin, TDH), trh (TDH-related hemolysin, TRH), pathogenicity islands (PAIs), type III secretion system (T3SS1 and T3SS2), and biofilm formation genes (Raghunath, 2015; Klein, Pipes & Lovell, 2018; Li et al., 2019; Sharan et al., 2022). V. parahaemolyticus strains are classified into pathogenic, non-pathogenic, pandemic, and non-pandemic strains (Okura et al., 2003; Meador et al., 2007; Chao et al., 2009; Lopez-Joven et al., 2015). Pathogenic strains harbor tdh and/or trh genes while non-pathogenic strains lack both tdh and trh genes. Pandemic strains harbor both tdh gene and toxRS/new gene (the pandemic marker gene containing the unique base changes in the regulatory gene, toxRS). Non-pandemic strains lack both tdh and toxRS/new genes genes (Okura et al., 2003; Meador et al., 2007; Chao et al., 2009; Lopez-Joven et al., 2015).
The first pandemic strain of V. parahaemolyticus O3:K6 was documented in February 1996 in Kolkata, India (Okuda et al., 1997). Infections caused by the O3:K6 strain and related serovariants, collectively called pandemic strains, were responsible for several large gastroenteritis outbreaks in humans (Nair et al., 2007). V. parahaemolyticus new/pandemic O3:K6 strains isolated since 1996 produced only TDH while the old O3:K6 strains isolated before 1996 produced only TRH (Okuda et al., 1997; Honda, Ni & Miwatani, 1988). Several studies have detected pandemic V. parahaemolyticus serovariants not only in clinical samples (Li et al., 2014; Pazhani et al., 2014; Ueno et al., 2016), but also in seafood and other environmental samples (Arakawa et al., 1999; Deepanjali et al., 2005; Quilici et al., 2005; Caburlotto et al., 2010; Haley et al., 2014; Jingjit et al., 2021), indicating that the pandemic strains have established ecological niches around the world. Although tdh and trh are associated with pathogenic strains, several studies have reported about 10% of clinical strains do not contain tdh and trh suggesting that pathogenicity could be caused by additional virulence factor(s) (Jones et al., 2012; Li et al., 2014; Pazhani et al., 2014).
The pathogenic potentials of V. parahaemolyticus are also associated with increased resistance to antimicrobials because of excessive use and misuse of antimicrobials in humans, agriculture, and aquaculture systems (Mazel & Davies, 1999; Cabello, 2006; Igbinosa, 2016). In recent reports, V. parahaemolyticus strains isolated from seafood, clinical, and environmental samples were highly resistant to multiple antibiotics including amoxicillin, ampicillin, ciprofloxacin, cefazolin, ceftazidime, cefotaxime, cefuroxime sodium, colistin, gentamicin, penicillin, spectinomycin, tetracycline and doxycycline (Tan et al., 2020; Ashrafudoulla et al., 2021; Dutta et al., 2021). Among antibiotic-resistant profiles, tetracycline, doxycycline, a 3rd-generation cephalosporin, and quinolone including ciprofloxacin are recommended for the treatment of severe or prolonged V. parahaemolyticus infection (Ashrafudoulla et al., 2021; Dutta et al., 2021; Rezny & Evans, 2022). Several studies demonstrated that seafood is a potential reservoir for the dissemination of multidrug resistant (MDR) V. parahaemolyticus (Letchumanan et al., 2015; Jeamsripong, Khant & Chuanchuen, 2020; Li et al., 2020).
In Thailand, V. parahaemolyticus is the major causative agent of human gastroenteritis, occurring in about 50–60% of gastroenteritis cases (DDC, 2020). Incidence of human gastroenteritis in Thailand has increased every year and has been linked to consumption and mishandled of seafood contaminated with V. parahaemolyticus (DDC, 2020). The first report of the O3:K6 pandemic strain was obtained from patients at Hat Yai Hospital located in southern Thailand in 1988 (Matsumoto et al., 2000). Since then the studies of pandemic and non-pandemic V. parahaemolytics strains have been mainly conducted in southern parts of the country (Vuddhakul et al., 2000; Laohaprertthisan et al., 2003; Vuddhakul et al., 2006; Wootipoom et al., 2007; Bhoopong et al., 2007; Thongjun et al., 2013; Han et al., 2016; Jingjit et al., 2021). Molecular fingerprinting methods have suggested possible epidemiological linkage between the clinical and the environmental strains in southern Thailand (Vuddhakul et al., 2006). Very limited information on occurrence and molecular characterization of pandemic and non-pandemic strains is available in other areas of Thailand with high concentration of seafood sources. This study aimed to investigate the occurrence of V. parahaemolyticus pandemic and non-pandemic strains, the characterization of V. parahaemolyticus isolates, the presence of pathogenic potential genes including VPaI-7, T3SS2 and biofilm formation genes, and AMR profiles of V. parahaemolyticus isolated from raw seafood in Bangkok and eastern Thailand (highly populated areas). These findings will add to the body of knowledge and surveillance information of V. parahaemolyticus that would be useful for relevant stakeholders to make informed-decision in the aquaculture management, food safety, and public health strategies.
Materials and Methods
Sample collection and V. parahaemolyticus isolation
Sample size estimation was based on the assumption of previous prevalence of 59% of V. parahaemolyticus contamination in seafood collected in Bangkok, Thailand (Atwill & Jeamsripong, 2021). The formula (n = Z2P(1−P)/d2), 95% confidence level and 7% margin of error were used in this study to obtain the representative sample with a less expensive survey (Daniel, 1999; Pourhoseingholi, Vahedi & Rahimzadeh, 2013). In this formula, n is the sample size, Z is the statistic for a level of confidence, P is expected prevalence, and d is precision or margin of error. The required samples were 190 samples which were collected from different sources, time, and locations. Of 190 samples, 143 and 31 samples were purchased in Bangkok, Thailand in 2018 and 2021 to 2022, respectively and 16 samples were collected from different shrimp farms in eastern Thailand in 2013. Among 174 of samples purchased in Bangkok, Thailand, 84 and 90 samples were purchased from fresh markets and supermarkets, respectively. Of 90 samples purchased from supermarkets, 40 samples were frozen seafood. One hundred and ninety samples comprised of crabs (n = 20; blue swimming crabs, red swimming crabs, and mud crabs), fish (n = 50; groupers, mackerels, ornate threadfin bream, and giant seaperch), mollusc shellfish (n = 50; blood cockles, green mussels, oysters, short-necked clams, and spiral babylon snails), shrimp (n = 50; giant tiger prawns and Pacific white shrimp) and squids (n = 20; splendid squids, Dollfus’ octopuses, and giant squid tentacle).
For sample purchasing, markets were chosen based on geography to ensure complete coverage of Bangkok and each market was visited only once. After purchasing from markets or collecting from farms, samples were individually packed in a sterilized plastic bag and transported on ice to the laboratory and processed within 2 h. For V. parahaemolyticus isolation from marketed and farmed samples, a 2.5-g portion of seafood sample was aseptically transferred into a stomacher bag containing 22.5 mL of tryptic soy broth (TSB) supplemented with 2% NaCl, mixed thoroughly by hand for 1 min and kept at room temperature for 30 min. Thereafter, the sample was removed from the broth which was incubated at 37 °C for 16 h before culturing on selective media of thiosulfate citrate bile salts sucrose agar (TCBSA, Difco Laboratories, Franklin Lakes, NJ, USA). V. parahaemolyticus colonies were opaque and blue-green color with 2–3 mm in diameter on the TCBS agar plates. V. parahaemolyticus colonies were further confirmed using CHROMagar™ Vibrio (CHROMagar, Paris, France) of which the positive colonies gave mauve color and were collected for further characterization (Ahmmed et al., 2019).
Polymerase chain reaction (PCR)
The isolates obtained from CHROMagar™ Vibrio were further confirmed for V. parahaemolyticus by PCR using species-specific PCR primers targeting toxR gene (Kim et al., 1999). To identify the pandemic O3:K6 strain and its serovariants, PCR primers targeting tdh and toxRS/new genes were used (Tada et al., 1992; Matsumoto et al., 2000). PCR primers targeting toxRS/old were used for determining the O3:K6 strains isolated before 1996 (Okura et al., 2003). PCR primers targeting one VPaI-7 open reading frame (ORF), two TSSS2 genes, and three biofilm genes, were used to identify genes involved in pathogenesis (Chao et al., 2009; Chao et al., 2010). PCR primers and conditions used in this study were summarized in Table 1. Briefly, the PCR reaction was performed in a 25-μL reaction volume, containing 12.5 μL GoTaq® Green Master Mix solution (Promega, Madison, WI, USA), 2 μL of DNA template (50 to 70 ng) and milli-Q water for adjusting final volume up to 25 μL. Gene-specific primer sets were added to corresponding PCR reactions: 0.1 μM of each primer for tdh gene; 0.6 μM of each primer for three biofilm genes; 0.8 μM of each primer for toxR, toxRS/old, and toxRS/new genes; and 1 μM of each primer for VPaI-7 and T3SS2 genes. PCR amplifications were performed in triplicate for each sample. PCR products were analyzed by electrophoresis at 75 V for 40 min in 1.5% agarose (Vivantis Technologies, Malaysia) with a 1X TAE (Tris-Acetate + EDTA) buffer. Agarose gels were stained in SYBRTM Safe DNA Gel Stain (Thermo Fisher Scientific, Waltham, MA, USA). Amplicon bands were observed under UV light.
| Primer name | Primer sequence (5′ to 3′) | Target gene | Amplicon size (bp) | Reference | PCR condition |
|---|---|---|---|---|---|
|
toxR-F toxR-R |
GTCTTCTGACGCAATCGTTG ATACGAGTGGTTGCTGTCATG |
toxR | 368 | Kim et al. (1999) | 95 °C-30 s; 63 °C-30 s; 72 °C-30 s |
|
toxRS/old-F toxRS/old-R |
TAATGAGGTAGAAACG ACGTAACGGGCCTACG |
toxRS of the old O3:K6 clone |
651 | Matsumoto et al. (2000) | 96 °C-1 min; 47.1 °C-2 min; 72 °C-3 min |
|
toxRS/new-F toxRS/new-R |
TAATGAGGTAGAAACA ACGTAACGGGCCTACA |
toxRS of the new O3:K6 clone |
651 | Matsumoto et al. (2000) | 94 °C-30 s; 52.3 °C-30 s; 72 °C-1 min |
|
tdh-F tdh-R |
CCACTACCACTCTCATATGC GGTACTAAATGGCTGACATC |
tdh | 251 | Tada et al. (1992) | 94 °C-1 min; 55 °C-1 min; 72 °C-1 min |
| VP1321-F VP1321-R |
CCTTGGAAGACAAATGTGGAT ATGGCTTACCAATGTCAAACTAT |
VPaI-7 | 261 | Chao et al. (2009) | 94 °C-40 s; 54.3 °C-40 s; 72 °C-30 s |
| VP1346-F VP1346-R |
TACCATCAGAGGATACAACC ACAATGAGAACATCAAACA |
VPaI-7 (T3SS2) | 262 | Chao et al. (2009) | 94 °C-40 s; 51.2 °C-40 s; 72 °C-30 s |
| VP1367-F VP1367-R |
CTATGGCGTGCTGGTAGAC TCACTCGTAAGATGTTGGG |
VPaI-7 (T3SS2) | 209 | Chao et al. (2009) | 94 °C-40 s; 56.6 °C-40 s; 72 °C-30 s |
| VP0950-F VP0950-R |
GCCAAACTTCTCAAACAACA ATGAAACGCAATTTACCATC |
Biofilm | 298 | Chao et al. (2010) | 94 °C-55 s; 50 °C-50 s; 72 °C-2 min |
| VP0952-F VP0952-R |
TATGATGGTGTTTGGTGC TGTTTTTCTGAGCGTTTC |
Biofilm | 276 | Chao et al. (2010) | 94 °C-55 s; 50 °C-50 s; 72 °C-2 min |
| VP0962-F VP0962-R |
GACCAAGACCCAGTGAGA GGTAAAGCCAGCAAAGTT |
Biofilm | 358 | Chao et al. (2010) | 94 °C-55 s; 50 °C-50 s; 72 °C-2 min |
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs), the lowest drug concentration inhibiting visible growth, of different antimicrobial drugs were determined by the broth microdilution technique utilizing a semi-automatic procedure (Sensititre, Trek Diagnostic Systems Ltd., West Sussex, UK) according to Clinical and Laboratory Standards Institute (CLSI) recommendations (CLSI, 2021). Two sets of dehydrated 96-well microtiter plates, including THAN2F and CMV4AGNF were used. In all, 27 antibiotics of different drug classes and action mechanisms were tested in varied concentration as summarized in Table S1. Most of the tested antimicrobial agents in this study are recommended by Centers for Disease Control and Prevention (CDC) for the therapeutics of Vibrio spp. infections including fluoroquinolones (ciprofloxacin and levofloxacin), 3rd-generation cephalosporins (cefotaxime, ceftazidime, and ceftriaxone), aminoglycosides (gentamicin and amikacin), tetracycline, folate synthesis inhibitors (trimethoprim/sulfamethoxazole) (Daniels & Shafaie, 2000; Shaw et al., 2014). The MIC analysis conditions were done as specified by manufacturer’s guidelines with slight adaptation. Briefly, isolates were cultured overnight on tryptic soy agar (TSA) with 2% NaCl at 37 °C in 5% CO2 incubator (Ahmmed et al., 2019). Selected colonies were suspended in Sensititre cation-adjusted Mueller-Hinton broth (CAMHBT) and adjusted to a 0.5 McFarland standard. Thereafter, a 10-μL aliquot of suspension was transferred into a tube of CAMHBT to get an inoculum density of 5 × 105 CFU/mL. THAN2F and CMV4AGNF panels were reconstituted by adding 50 μL/well and were enclosed with an adhesive seal and incubated at 35 ± 2 °C in Sensititre ARISTM 2X for 20–24 h. The MIC value was determined automatically on the Sensititre ARISTM 2X and visualized using a manual viewbox in accordance with instructions in Sensititre SWIN software. Escherichia coli ATCC 25922 was used as an antimicrobial-susceptible control strain. Results were interpreted according to CLSI, National Antimicrobial Resistance Monitoring System (NARMS), and European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (CLSI, 2021; NARMS, 2021; EUCAST, 2022). Strains were classified as “non-susceptible” when they were resistant to at least one antimicrobial. Multidrug resistance (MDR) was defined as non-susceptible to at least one agent in three or more antimicrobial classes according to the definition proposed for other bacterial groups (Magiorakos et al., 2012). The Multiple Antibiotic Resistance (MAR) index was determined for each isolate using the formula MAR = a/b, where “a” is the number of antibiotics to which the test isolate is resistant, and “b” is the total number of antibiotics tested. MAR index values greater than 0.2 indicated that the isolates were obtained from a high-risk source of contamination where antibiotics are often used (Krumperman, 1983).
Whole-genome sequencing (WGS), genome assembly and annotation
To determine the presence of antimicrobial resistance gene (ARG), a pure culture of V. parahaemolyticus was grown overnight on TSA with 2% NaCl at 37 °C in 5% CO2. Subsequently, colonies were selected and suspended in 5 mL TSB with 2% NaCl. The broth culture was incubated at 37 °C in 5% CO2 for 16 h. Thereafter, bacterial cells were collected by centrifugation and used for genomic DNA (gDNA) preparation. V. parahaemolyticus gDNA was extracted with phenol-chloroform DNA extraction (Green & Sambrook, 2017). The purity and quantity of gDNA was determined by measuring OD260/280 and OD260/230 values with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The integrity of intact gDNA was verified with agarose gel electrophoresis. Whole-genome sequencing analysis of 36 high quality gDNA samples with OD260/OD280 of ≥1.8 and OD260/OD230 of ≥2.0 were performed using Illumina® DNA sequencing with HiSeq 2X150 paired-end (PE) configuration (serviced by GeneWiz Inc., Chelmsford, MA, USA). Raw sequencing reads were trimmed prior to genome assembly using the JGI bbduk tool (k = 27, ktrim = 1, hdist = 1, minlength = 50). The genome assembly and scaffolding were performed using SPAdes version 3.15 (Prjibelski et al., 2020). The genome annotation was performed using Prokka version 1.13 (Seemann, 2014). V. parahaemolyticus RIMD 2210633 (accession no. GCA_000196095.1) was used as the reference genome. Sequence data of 36 V. parahaemolyticus genomes were deposited in DDBJ/EMBL/GenBank. The GenBank accession numbers are shown in Table S2.
Detection of antimicrobial resistance genes (ARGs)
To detect ARGs in each annotated genome, nucleotide sequences of CDS genes of each annotated genomes were analyzed using ResFinder version 4.1 with minimum sequence alignment coverage = 0.6 and minimum threshold for sequence identity = 0.8 (Florensa et al., 2022).
Multi-locus sequence typing (MLST) analysis
Molecular typing using MLST was done with 7 conserved housekeeping genes dnaE, gyrB, recA, dtdS, pntA, pyrC, and tnaA. Briefly, ORF and scaffold sequences of each isolate were BLASTN searched against the V. parahaemolyticus typing allele sequence database downloaded from PubMLST.org (2022-05-09) (Jolley, Bray & Maiden, 2018) to obtain sequence types (STs). The genome sequences of new ST profiles identified in this study were submitted to PubMLST.org databases.
Phylogenomic tree construction
A maximum-likelihood phylogenomic tree was inferred using UBCG2.0 (version Feb, 2021) (Kim et al., 2021) and RAxML (version 8.2.12) (Stamatakis, 2014) softwares with a default parameter setting, a GTR + CAT substitution model on a nucleotide alignment of 81 bacterial universal core genes from 36 V. parahaemolyticus genomes. These universal core genes previously identified by UBCG2 method as single-copy core genes covering 3,508 species of 43 bacterial phyla are suitable for phylogenomic tree analyses (Kim et al., 2021). The phylogenomic tree from the maximum likelihood analysis was visualized with MEGA X software.
Results
Detection of V. parahaemolyticus potential virulence genes
Among 174 samples purchased from fresh markets and supermarkets, 34 V. parahaemolyticus were isolated. A total 50 V. parahaemolyticus isolates, 34 isolates from markets and 16 isolates from shrimp farms, were used in this study. Of 50 isolates, 21 and 29 were pathogenic and non-pathogenic strains, respectively (A. Lamalee, 2023, unpublished data) (Table S3). All 50 V. parahaemolyticus isolates exhibited positive PCR amplification to toxR, toxRS/old (a non-pandemic gene marker), and biofilm formation genes (VP0950, VP0952, and VP0962) (Table 2). None of 50 isolates showed positive PCR amplification for both pandemic gene markers (both tdh positive and toxRS/new positive) and for two T3SS2 genes (VP1346 and VP1367) (Table 2). VPaI-7 (VP1321 ORF) was detected in two isolates (F2CK02 and F3CK01) accounting for 4% (2/50) (Tables 2 and S3). Altogether, the results indicated that all isolates were non-pandemic strains. Based on the presence or absence of genetic markers, 50 V. parahaemolyticus isolates could be classified as follows: 21 were non-pandemic and pathogenic strains (tdh− or toxRS/new−; toxRS/old+; tdh+ or trh +) of which two isolates carried VPaI-7 genes, and 29 were non-pandemic and non-pathogenic strains (tdh− or toxRS/new−; toxRS/old+; tdh− and trh−).
Antimicrobial susceptibilities
Of 50 V. parahaemolyticus isolates, 14 isolates were not examined for antimicrobial susceptibilities due to poor recovery from glycerol stocks. Thus, antimicrobial susceptibility tests were performed on 36 isolates against 27 agents from five antimicrobial categories (Table S1). Based on the results, the susceptibility rate of 36 V. parahaemolyticus strains was 100% (36/36) to fluoroquinolones, carbapenems, amikacin, gentamicin, netilmicin, tetracyclines chloramphenicol, azithromycin, amoxicillin/clavulanic acid, ampicillin/sulbactam, piperacillin/tazobactam, and trimethoprim/sulfamethoxazole (Tables 3 and S4). The susceptible rate was 97% (35/36) to cefoxitin, cefotaxime, ceftazidime, ceftriaxone, and cefepime, 92% (33/36) to sulfisoxazole, and 83% (30/36) to streptomycin. A high number of intermediate susceptibility was observed for cefuroxime (81%, 29/36). The low resistance rate was observed for cefuroxime and sulfisoxazole (8%, 3/36) and for cefotaxime, ceftazidime, ceftriaxone, and cefepime (3%, 1/36). In contrast, high resistance pattern was observed for ampicillin (83%, 30/36) and colistin (100%, 36/36) (Tables 3 and S4).
| Antibiotic drugs | MIC breakpoints, µg/mL | MIC values (µg/mL)a | MIC50 | MIC90 | S (%) | I (%) | R (%) | MIC ranges | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.50 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | |||||||
| Amikacin | ≤16 | 32 | ≥64 | 36 | 0| | 0∣ | ≤8 | ≤8 | 100.0 | 0.0 | 0.0 | ≤8 | |||||||||||||
| Amoxicillin-clavulanic acid | ≤8/4 | 16/8 | ≥32/16 | 1 | 31 | 4 | 0| | 0∣ | 0 | 2 | 4 | 100.0 | 0.0 | 0.0 | ≤1–4 | ||||||||||
| Ampicillin | ≤8 | 16 | ≥32 | 0 | 0 | 0 | 0| | 6∣ | 18 | 12 | 32 | >32 | 0.0 | 16.7 | 83.3 | 16–>32 | |||||||||
| Ampicillin-sulbactam | ≤8/4 | 16/8 | ≥32/16 | 35 | 1| | 0∣ | ≤4 | ≤4 | 100.0 | 0.0 | 0.0 | ≤4–8 | |||||||||||||
| Azithromycin | ≤4 | – | >4 | 26 | 10 | 0 | 0∣ | 0 | 0 | 0 | 0 | ≤0.25 | 0.5 | 100.0 | – | 0.0 | ≤0.25–0.5 | ||||||||
| Cefepime | ≤2 | 4-8 | ≥16 | 35 | 0| | 0 | 0∣ | 0 | 1 | ≤1 | ≤1 | 97.2 | 0.0 | 2.8 | ≤1–>32 | ||||||||||
| Cefotaxime | ≤1 | 2 | ≥4 | 35| | 0∣ | 0 | 0 | 0 | 1 | ≤1 | ≤1 | 97.2 | 0.0 | 2.8 | ≤1–>32 | ||||||||||
| Cefoxitin | ≤8 | 16 | ≥32 | 0 | 0 | 0 | 7 | 28| | 1∣ | 0 | 8 | 8 | 97.2 | 2.8 | 0.0 | 4–16 | |||||||||
| Ceftazidime | ≤4 | 8 | ≥16 | 35 | 0 | 0| | 0∣ | 1 | 0 | ≤1 | ≤1 | 97.2 | 0.0 | 2.8 | ≤1–16 | ||||||||||
| Ceftriaxone | ≤1 | 2 | ≥4 | 35 | 0 | 0| | 0∣ | 0 | 0 | 0 | 0 | 1 | ≤0.25 | ≤0.25 | 97.2 | 0.0 | 2.8 | ≤0.25–>64 | |||||||
| Cefuroxime (sodium) | ≤8 | 16 | ≥32 | 4| | 29∣ | 3 | 16 | 16 | 11.1 | 80.6 | 8.3 | ≤8–>16 | |||||||||||||
| Chloramphenicol | ≤8 | 16 | ≥32 | 36 | 0 | 0| | 0∣ | 0 | ≤2 | ≤2 | 100.0 | 0.0 | 0.0 | ≤2 | |||||||||||
| Ciprofloxacin | ≤1 | 2 | ≥4 | 4 | 24 | 8 | 0 | 0| | 0∣ | 0 | 0.12 | 0.25 | 100.0 | 0.0 | 0.0 | 0.06–0.25 | |||||||||
| Colistin | ≤2 | – | >2 | 0∣ | 0 | 2 | 2 | 32 | >8 | >8 | 0.0 | – | 100.0 | 4–>8 | |||||||||||
| Doripenem | ≤1 | 2 | ≥4 | 36 | 0| | 0∣ | 0 | 0 | 0 | ≤0.5 | ≤0.5 | 100.0 | 0.0 | 0.0 | ≤0.5 | ||||||||||
| Ertapenem | ≤0.5 | 1 | ≥2 | 36| | 0∣ | 0 | 0 | ≤0.5 | ≤0.5 | 100.0 | 0.0 | 0.0 | ≤0.5 | ||||||||||||
| Gentamicin | ≤4 | 8 | ≥16 | 0 | 0 | 7 | 26 | 3| | 0∣ | 0 | 2 | 2 | 100.0 | 0.0 | 0.0 | 1–4 | |||||||||
| Imipenem | ≤1 | 2 | ≥4 | 36 | 0| | 0∣ | 0 | 0 | 0 | ≤0.5 | ≤0.5 | 100.0 | 0.0 | 0.0 | ≤0.5 | ||||||||||
| Levofloxacin | ≤2 | 4 | ≥8 | 4 | 28 | 4 | 0 | 0 | 0| | 0∣ | 0 | 0.12 | 0.25 | 100.0 | 0.0 | 0.0 | ≤0.06–0.25 | ||||||||
| Meropenem | ≤1 | 2 | ≥4 | 36 | 0 | 0 | 0 | 0| | 0∣ | 0 | 0 | 0 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 | ≤0.06 | |||||||
| Nalidixic acid | ≤16 | – | ≥32 | 25 | 10 | 1 | 0 | 0 | 0| | 0∣ | ≤0.5 | 1 | 100.0 | – | 0.0 | ≤0.5–2 | |||||||||
| Netilmicin | ≤8 | 16 | ≥32 | 36| | 0∣ | ≤8 | ≤8 | 100.0 | 0.0 | 0.0 | ≤8 | ||||||||||||||
| Piperacillin-tazobactam | ≤16/4 | 32/4-64/4 | ≥128/4 | 36 | 0| | 0 | 0∣ | ≤8 | ≤8 | 100.0 | 0.0 | 0.0 | ≤8 | ||||||||||||
| Streptomycin | ≤16 | – | ≥32 | 0 | 0 | 0 | 30∣ | 6 | 0 | 16 | 32 | 83.3 | – | 16.7 | 16–32 | ||||||||||
| Sulfisoxazole | ≤256 | – | ≥512 | 2 | 9 | 15 | 5 | 2∣ | 3 | 64 | 256 | 91.7 | – | 8.3 | ≤16–>256 | ||||||||||
| Tetracycline | ≤4 | 8 | ≥16 | 36| | 0∣ | 0 | 0 | ≤4 | ≤4 | 100.0 | 0.0 | 0.0 | ≤4 | ||||||||||||
| Trimethoprim-sulfamethoxazole | ≤2/38 | – | ≥4/76 | 35 | 0 | 1 | 0 | 0∣ | 0 | ≤0.12 | ≤0.12 | 100.0 | – | 0.0 | ≤0.12–0.5 | ||||||||||
Note:
AMR patterns of 36 V. parahaemolyticus isolates could be classified into six patterns in which pattern No. 2 (AMP/COL) was predominant (53%, 19/36) (Table 4). MDR V. parahaemolyticus was observed in 11 isolates (31%,11/36) and all of them exhibited resistance to ampicillin and colistin. Interestingly, one MDR isolate, VP42 isolated from Pacific white shrimp, exhibited additional resistance to cefuroxime, cefotaxime, ceftazidime, ceftriaxone, and cefepime. All 36 V. parahaemolyticus isolates exhibited MAR index range of 0.04 to 0.3, indicating that V. parahaemolyticus isolates were resistant to 1–8 types of tested antibiotics (Table S5). VP42 showed resistant to 8/27 antimicrobial agents, which corresponded to the highest MAR index (0.3). The MAR index value greater than 0.2 suggested that VP42 was isolated from a high-risk source of antimicrobial contamination (Krumperman, 1983).
| Resistance pattern | Number of V. parahaemolyticus isolate |
Percentage (%) |
|---|---|---|
| 1. COL | 6 | 16.7 |
| 2. AMP/COL | 19 | 52.8 |
| 3. AMP/COL/S | 6 | 16.7 |
| 4. AMP/COL/SIX | 2 | 5.6 |
| 5. AMP/COL/FUR | 2 | 5.6 |
| 6. AMP/CPM/CTX/CAZ/CRO/FUR/COL/SIX | 1 | 2.8 |
Note:
AMP, Ampicillin; CPM, Cefepime; CTX, Cefotaxime; CAZ, Ceftazidime, CRO, Ceftriaxone; FUR, Cefuroxime (sodium); COL, Colistin; S, Streptomycin, and SIX, Sulfisoxazole.
Analysis of antimicrobial resistance genes (ARGs)
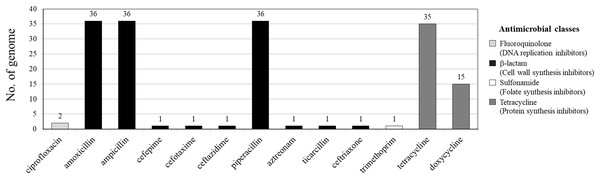
Among 36 V. parahaemolyticus genomes, 13 AMR genotypes and six ARGs were detected. Of six ARGs observed, blaCARB (100%, 36/36), tet(34) (83%, 30/36) and tet(35) (42%, 15/36) were the most common, followed by qnrC (6%, 2/36), dfrA6 (3%, 1/36), and blaCTX-M-55 (3%, 1/36) (Figs. 1 and 2). All 36 V. parahaemolyticus genomes carried at least one blaCARB gene (encoding for β-lactamase enzyme), causing resistance to amoxicillin, ampicillin, and piperacillin. Interestingly, VP42 carried blaCTX-M-55 gene encoding for extended spectrum β-lactamase (ESBL) conferring resistance to amoxicillin, ampicillin, piperacillin, cefepime, cefotaxime, ceftazidime, ceftriaxone, aztreonam, and ticarcillin.
Figure 1: Antimicrobial resistance gene (ARG) profiles of 36 Vibrio parahaemolyticus genomes detected with ResFinder.
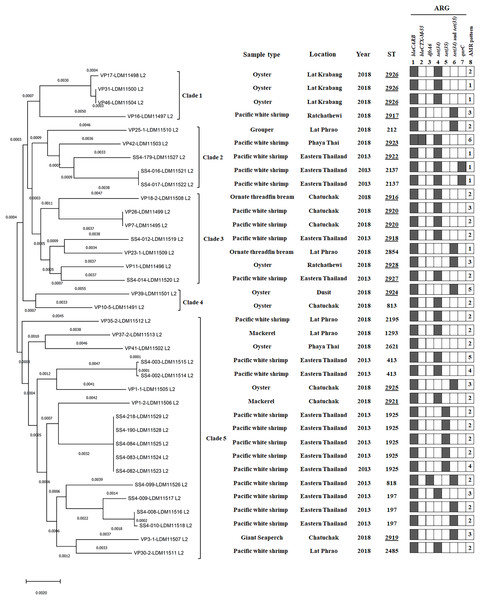
Figure 2: A phylogenomic tree from maximum likelihood analyzes of 81 bacterial core genes in Vibrio parahaemolyticus genomes and MLST types shows the relationship among 36 V. parahaemolyticus isolates.
Antimicrobial resistance gene (ARG) profiles: (1) blaCARB gene causing resistance to amoxicillin, ampicillin and piperacillin; (2) blaCTX-M-55 gene causing resistance to amoxicillin, ampicillin, piperacillin, cefepime, cefotaxime, ceftazidime, ceftriaxone, aztreonam, and ticarcillin; (3) dfrA6 gene giving resistance to trimethoprim; (4, 5 and 6) tet(34), tet(35), and tet(34) and tet(35) genes, respectively, causing resistance to tetracycline and doxycycline; (7) qnrC gene conferring ciprofloxacin resistance; (8) AMR patterns No. 1–6, as shown in Table 4. A filled box in column 1–7 specifies the identification of the particular gene. The sequence type (ST) with the underline represents the novel ST. The location column written eastern Thailand indicates the samples obtained from shrimp farms.Of 36 V. parahaemolyticus isolates, 35 and 15 isolates harbored ARG of tetracycline and doxycycline, respectively. Of 35 isolates with tetracycline ARG, 20 isolates harbored tet(34), 5 isolates harbored tet(35), and 10 isolates harbored tet(34) and tet(35). All 15 isolates with doxycycline ARG harbored tet(35). SS4-099 isolated from Pacific white shrimp carried dfrA6 gene giving resistance to trimethoprim. SS4-016-and SS4-017 isolated from Pacific white shrimp carried qnrC gene giving resistance to ciprofloxacin (Fig. 2).
Multilocus sequence type (MLST) analysis
MLST analysis revealed 25 different sequence types (STs) of 36 V. parahaemolyticus isolates (Table S6). Twelve STs (48%, 12/25) of 20 isolates were previously reported in pubMLST Database, whereas 13 novel STs (ST2916-ST2928, 52%, 13/25) of 16 isolates obtained in this study were submitted to PubMLST/V. parahaemolyticus database (http://pubmlst.org/vparahaemolyticus). GenBank accession numbers of 36 V. parahaemolyticus genomes are shown in Table S2. Among the novel STs (16 isolates), ST2926 was predominant (three isolates), followed by ST2920 (two isolates). Most of the novel STs were identified from marketed seafood (10 STs) and the rest three novel STs (ST2918, ST2922, and ST2927) were identified from shrimp farms. The most common STs detected in this study were ST1925 (20%, 5/25), ST197 (12%, 3/25), ST2926 (12%, 3/25), ST413 (8%, 2/25), ST2137 (8%, 2/25), and ST2920 (8%, 2/25) (Table S6).
Phylogenomic analysis
A phylogenomic tree from maximum likelihood analyzes of 81 bacterial core genes in 36 V. parahaemolyticus genomes exhibited five distinct clades (Fig. 2). All four isolates in clade 1 were identified as novel STs, ST2926 (VP17, VP31, and VP46) and ST2917 (VP16) and the other 11 novel STs were distributed to clade 2–5 (12 isolates). ST2926, ST2137, and ST2920 were identified in three isolates of clade 1, 2 isolates of clade 2 and 2 isolates of clade 3, respectively. ST197, ST413, ST1925, and ST818 were identified in 3, 2, 5, and 1 isolates of clade 5, respectively. These results exhibited a close relationship among V. parahaemolyticus strains which were identified from the same seafood types collected in the same year and from the same location. A close association among the strains isolated from the same seafood types collected in the same year but from different locations was also found in clade 4 (VP39 and VP10/5). Additionally, a close relationship among the strains isolated from different types of seafood in different years and different locations was exhibited in clade 3 (SS4-012 and VP23/1 as well as VP11 and SS4-014 isolates). SS4-016 and SS4-017 with the same ST2137 carrying qnrC isolated from farmed shrimp in eastern Thailand and VP42 (ST2923), carrying blaCTX-M-55 isolated from shrimp sold in Bangkok were in the same clade 2. These results indicated the association between these isolates from different geographical regions. Moreover, SS4-099 isolate carrying dfrA6 was in clade 5 and is distinct from SS4-016 and SS4-017 isolates carrying qnrC in clade 2. These three isolates were isolated from farmed shrimp in eastern Thailand indicating a distance relationship among these strains isolated from the same region.
Discussion
In this study, all V. parahaemolyticus strains isolated from seafood purchased in Bangkok and collected in eastern Thailand belonged to the old O3:K6 strains or non-pandemic strains. In contrast, 12 strains isolated from 302 seafood samples purchased in southern Thailand were pandemic strains (Vuddhakul et al., 2006). Jingjit et al. (2021) also identified 15 pandemic V. parahaemolyticus strains from 51 seafood-isolated V. parahaemolyticus strains obtained from southern Thailand (Jingjit et al., 2021). These pandemic strains were detected from hard clams, mussels, and cockles (Vuddhakul et al., 2006; Jingjit et al., 2021) Mussels, and cockles were also purchased from markets in this study. The discrepancies in pandemic strain occurrences are likely due to the differences in geographical regions for seafood sample collection. All seafood samples in this study were purchased from fresh markets and supermarkets in Bangkok and collected from farms in the upper Gulf of Thailand (central and eastern Thailand), while seafood samples in previous studies were collected from the lower Gulf of Thailand (southern Thailand). The O3:K6 pandemic strain had been firstly identified in 1998 at Hat Yai Hospital in southern Thailand (Matsumoto et al., 2000), later pandemic V. parahaemolyticus strains were reported from seafood samples in the same area (Vuddhakul et al., 2006; Jingjit et al., 2021) indicating that pandemic strains have established ecological niches in this region.
The incidence of V. parahaemolyticus in this study is somewhat low at 20%, compared to previous reports which seafood samples were also purchased in Bangkok, Thailand at 58% (Thitipetchrakul, Somyoonsup & Hatayananont, 2016) and at 59% (Atwill & Jeamsripong, 2021). This is likely due to the differences in seafood types, market types and sample storage condition between this study and previous studies. V. parahaemolyticus contamination was found more often in seafood purchased from fresh markets than those from supermarkets (Zhang et al., 2016; Martinez-Urtaza & Baker-Austin, 2020) and in refrigerated products more than frozen products (Caburlotto et al., 2016). In this study, more different seafood types were purchased from fresh markets and supermarkets of which some samples were frozen seafood whereas the study by Atwill & Jeamsripong (2021) used chilled seafood samples consisting of Pacific white shrimp, oysters, blood cockles, and Asian seabass purchased form open-air retail fresh markets (Atwill & Jeamsripong, 2021). Boiled crab meats from fresh and fair markets were used for V. parahaemolyticus surveillance (Thitipetchrakul, Somyoonsup & Hatayananont, 2016). Thus, the incidence of V. parahaemolyticus in this study was likely lower than those of previous reports.
Our results showed that none of 50 V. parahaemolyticus isolates harbored T3SS2 genes while VPaI-7 gene (VP1321) was present in two isolates. The absence of T3SS2 genes in two isolates positive for VPaI-7 is probably due to the partial loss of T3SS2 genes as previously reported (Chao et al., 2009). VPaI-7 on chromosome two typically encodes for two tdh (tdh1 and tdh2) genes and a set of T3SS2 genes which are virulence genes (Makino et al., 2003; Matsuda et al., 2020). V. parahaemolyticus harboring T3SS2 genes has been linked to clinical cases of inflammatory gastroenteritis (Makino et al., 2003; Hiyoshi et al., 2011). Although most previous studies reported the presence of T3SS2 only in highly virulent strains; however, T3SS2 has also been observed in a non-virulent strain (Meador et al., 2007). This study focused on VP1321 ORF of VPaI-7 and two T3SS2 genes (VP1346 and VP1367) as they were observed in all the pandemic strains reported previously (Chao et al., 2009). Although, none of tested seafood samples purchased in Bangkok and collected in eastern Thailand were pandemic strains, detection of VPaI-7 in non-pandemic-pathogenic V. parahaemolyticus strains provided the importance of seafood safety through consumer health awareness and practices. Strong biofilm formation ability has been linked to environmental survival ability, infectivity, and transmissibility of antibiotic-resistance microorganisms to humans (Elexson et al., 2014; Mizan, Jahid & Ha, 2015). Our results indicated that all 50 V. parahaemolyticus isolates consisting of 21 pathogenic and 29 non-pathogenic strains harbored three biofilm-associated genes (VP0950, VP0952, and VP0962). Under proper circumstances, these genes could be activated and expressed leading to biofilm formation in V. parahaemolyticus contaminated seafood that could cause a great threat to human health and economic values.
Our study demonstrated a high prevalence of AMR phenotypes among the 36 V. parahaemolyticus isolates for ampicillin (83%, 30/36) and colistin (100%, 36/36), which was in consistent with the frequent outbreaks of ampicillin- and colistin-resistant V. parahaemolyticus isolates reported in the past 5 years (Lopatek, Wieczorek & Osek, 2018; Dahanayake et al., 2020; Mok et al., 2021; Nishino et al., 2021; Vu et al., 2022). The blaCARB gene was present in all 36 V. parahaemolyticus isolates, confirming ampicillin-resistant phenotypes and susceptible phenotypes to amoxicillin/clavulanic acid, ampicillin/sulbactam, and piperacillin/tazobactam. The high distribution of blaCARB gene is similar to a previous study which found that all 30 V. parahaemolyticus isolates obtained from shrimp samples harbored blaCARB (Hossain et al., 2020). Consistently with our findings, a report by Chiou, Li & Chen (2015) suggested an intrinsic resistance of ampicillin in V. parahaemolyticus. The observed amoxicillin resistance phenotypes in all isolates and the higher prevalence of tet(34), conferring resistance to oxytetracycline (Nonaka & Suzuki, 2002), than that of tet(35) are in agreement with the excessive uses of amoxicillin and oxytetracycline in aquaculture. These two antimicrobial agents have been permitted for use in Asian aquaculture industries including Thailand (Yano et al., 2014; AAHRDD, 2022). Based on this finding, we recommend that antimicrobial use in aquaculture should follow guidelines recommended by Food and Drug Administration (FDA) of Thailand including using antimicrobials based on clinical diagnosis and using narrow-spectrum antimicrobials. Rotation of permitted antimicrobials should also be done to decrease the antimicrobial pressure due to the dominant use. The blaCTX-M-55 gene was identified in VP42 isolate confirming phenotypic resistance against cefuroxime, cefotaxime, ceftazidime, ceftriaxone, and cefepime. The low distribution of blaCTX-M-55 is in agreement with a study conducted in China which blaCTX-M-55 was found in 2% (2/116) of V. parahaemolyticus strains isolated from shrimp samples (Zheng et al., 2019). Detection of cephalosporins resistant isolates causes concern as cephalosporins are among the β-lactams currently used as the last line of antibiotics to treat V. parahaemolyticus infections (Ashrafudoulla et al., 2021; Dutta et al., 2021). Our results showed significant correlations between phenotypes and genotypes that conferred resistance to ampicillin (blaCARB) and cephalosporins (blaCTX-M-55). No genotypic resistance was observed for colistin, sulfisoxazole, and streptomycin. It is possible that ARGs are mobile genetic elements which may not be detected by a short-read sequencing method (Christaki, Marcou & Tofarides, 2020; Dutta et al., 2021). No phenotypic resistance was observed despite genotypic resistance for trimethoprim, ciprofloxacin, doxycycline, and tetracycline. It is plausible that these genes were silenced or expressed at low resistance level below the interpretive breakpoint utilized. Under suitable circumstances, these genes could be highly expressed and/or transferred horizontally among bacteria posing the risk of AMR and/or MDR phenotypes (Dutta et al., 2021). The discrepancy between phenotypic and genotypic resistance observed here was similar to the previous finding of pathogenic V. parahaemolyticus in seafood samples (Lou et al., 2016). Antibiotic misuse, warm temperature, acid-base and organic contamination can directly activate the expression of ARGs and virulence genes and also affect horizontal gene transfer (Guijarro et al., 2015; Deng et al., 2020). Thus, the occurrence of tetracycline, doxycycline and ciprofloxacin resistance genes is a major concern since they are the antibiotics of choice for treatment of severe or prolonged illnesses of V. parahaemolyticus infection (Tan et al., 2020; Ashrafudoulla et al., 2021; Dutta et al., 2021). The high resistance for ampicillin and colistin observed here suggests an alarming trend of widespread ampicillin and colistin resistance which compromise treatment efficacy for V. parahaemolyticus infection. It is important to note that due to limited number of isolates, specific locations, and samples per seafood species, years and market types in this study, it might be very challenging to conclude the current AMR situation of V. parahaemolyticus in Thailand. However, the high resistance to ampicillin and high susceptibility to cephalosporins found in seafood V. parahaemolyticus confirms previous findings from studies conducted in Thailand and other Asian countries (Elmahdi, DaSilva & Parveen, 2016; Palamae et al., 2022).
MLST analysis demonstrated a high number (25) of STs and particularly 13 novel STs among the 36 isolates suggesting a high genetic variation of Thai V. parahaemolyticus isolates. The relationship among the strains with specific ST found in the same seafood type collected from the same location in the same year revealed a close relationship among these strains suggesting cross-contamination during harvesting or handling seafood at the same processing site due to poor hygiene practices. Thus, local interventions should be addressed including using disinfected seawater or potable water to wash and process. seafood, wearing gloves during processing and keeping seafood at ≤10 °C during distribution and storage (Hara-Kudo & Kumagai, 2014). A close association among the V. parahaemolyticus strains isolated from the same type of seafood from different locations in the same year or different types of seafood from distinct locations and sampling years could indicated either the possibility of cross-contamination during food processing steps of supply chains or persistence of these strains in the environment. Overall, MLST analysis revealed that V. parahaemolyticus strains in Thailand were geographically distinct and genetically diverse. The strains isolated from marketed samples were more diverse than those from shrimp farms. These observations suggest the potential cross-contamination of V. parahaemolyticus during seafood processing steps and distribution chains including a contaminated container for transporting and improper handling. In contrast, V. parahaemolyticus in farmed shrimp is mainly derived from environmental contamination including estuarine water and water sediment at the cultivation site (Lovell, 2017). Several decontamination techniques are available to effectively reduce the number of V. parahaemolyticus without compromising the flavor, texture, and nutritional of seafood products. The decontamination techniques comprise chemical techniques (antibiotics, disinfectants and natural organic treatments), physical techniques (high-pressure processing, ozonation, irradiation, refrigeration and seafood suspension), and biological techniques (bacteriophage and probiotic treatments) (Ramos et al., 2012; Hara-Kudo et al., 2013; Wang et al., 2013; Ronholm, Lau & Banerjee, 2016; Zhang et al., 2018; Kontominas et al., 2021).
Conclusions
Our findings reveal that all V. parahaemolyticus isolates obtained from seafood in Bangkok and eastern Thailand are non-pandemic strains; some of them contain pathogenic potential genes. The presence of the first-line antimicrobial resistance genes of tetracyclines, doxycycline and ciprofloxacin raises a major concern of inducible gene expression and/or horizontal gene transfer among bacteria. The widespread of MDR V. parahaemolyticus confirms the emergence of AMR problems in seafood. Thus, monitoring of AMR of V. parahaemolyticus in seafood is highly recommended to tackle AMR problem and provide useful information for therapeutic treatment in humans. Based on our findings, antimicrobial use to treat V. parahaemolyticus infection in aquaculture should restrictedly follow guidelines recommended by FDA of Thailand to avoid AMR and MDR problems. Moreover, most of V. parahaemolyticus strains possess new STs suggesting high genetic diversity of the isolates. V. parahaemolyticus diversity in marketed samples suggests the cross-contamination possibility among samples in seafood production chains. Overall, our findings highlight the importance of a surveillance program to help strengthen safety guidelines of seafood production and promote public health awareness among health professional and consumers.
Supplemental Information
Antimicrobial categories, agents and concentrations used in this study.
GenBank accession numbers of 36 Vibrio parahaemolyticus genomes.
The genome sequences of 36 Vibrio parahaemolyticus isolates in this study are available at NCBI database: PRJNA859558 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA859558). Each assembled genome sequence is assigned to BioSample.
Distribution of pandemic and non-pandemic strains, and potential pathogenic genes of 50 Vibrio parahaemolyticus isolates.
Antimicrobial categories, agents, and susceptibility of 36 Vibrio parahaemolyticus isolated from seafood in Thailand.
Antimicrobial resistance (AMR) patterns and multiple antibiotic resistance (MAR) indexes of 36 Vibrio parahaemolyticusisolates.
Distribution of 25 sequence types (STs) with 12 known and 13 novel STs of 36 Vibrio parahaemolyticus isolated from seafood in Thailand.
The raw data for minimum inhibitory concentration (MIC) values of the 36 Vibrio parahaemolyticus isolates.
The raw data of the antimicrobial susceptibility test performed on the 36 Vibrio parahaemolyticus isolates against 27 agents from 5 antimicrobial categories.