Dissection of leucine-rich repeat receptor-like protein kinases: insight into resistance to Fusarium wilt in tung tree
- Published
- Accepted
- Received
- Academic Editor
- Lin Zhang
- Subject Areas
- Agricultural Science, Bioinformatics, Biotechnology, Plant Science
- Keywords
- LRR-RLK, Tung tree, Expression patterns, Evolution analysis
- Copyright
- © 2022 Cao et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. Dissection of leucine-rich repeat receptor-like protein kinases: insight into resistance to Fusarium wilt in tung tree. PeerJ 10:e14416 https://doi.org/10.7717/peerj.14416
Abstract
The tung tree is a woody oil plant native to China and widely distributed in the subtropics. The three main species commonly known as Vernicia are V. fordii, V. montana, and V. cordata. The growth and development of V. fordii are affected by a large number of plant pathogens, such as Fusarium wilt caused by Fusarium sp. In contrast, V. montana shows significant resistance to Fusarium wilt. The leucine-rich repeat receptor-like protein kinase (LRR-RLK) is the largest class of receptor-like kinases associated with plant resistance to Fusarium wilt. Here, we identified 239 VmLRR-RLKs in V. montana, and found that there were characteristic domains of resistance to Fusarium wilt in them. Phylogenetic analysis suggested that the VmLRR-RLKs are divided into 14 subfamilies, indicating that homologous genes in the same group may have similar functions. Chromosomal localization analysis showed that VmLRR-RLKs were unevenly distributed on chromosomes, and segment duplications were the main reason for the expansion of VmLRR-RLK family members. The transcriptome data showed that six orthologous pairs were up-regulated in V. montana in response to Fusarium wilt, while the corresponding orthologous genes showed low or no expression in V. fordii in resistance Fusarium wilt, further indicating the important role of LRR-RLKs in V. montana’s resistance to infection by Fusarium spp. Our study provides important reference genes for the future use of molecular breeding to improve oil yield and control of Fusarium wilt in tung tree.
Introduction
The tung tree is an important industrial oil tree species in the world. The three most important tung tree species in the world are Vernicia fordii, V. montana and V. cordata (Haw, 2017; Stuppy et al., 1999). Where V. fordii is widely planted because of its high oil production (Cui et al., 2018). However, compared with the other two tree species, the growth and development of V. fordii are more susceptible to Fusarium wilt (Cao et al., 2021; Chen et al., 2016). On the contrary, V. montana showed significant resistance to Fusarium wilt (Chen et al., 2016; Jiang et al., 2022a; Zhang et al., 2016).
In the process of plant growth and development, cell-environment signals and cell-cell interaction signals can stimulate and induce plants to produce a variety of different signal transduction pathways. The reversible phosphorylation regulation mechanism of protein kinases (PK) is involved in the process of cell signal transduction (Cao et al., 2021). Receptor-like kinase (RLK) family members can sense and process external or internal signals in living cells of plants (Gou et al., 2010; Shiu & Bleecker, 2001). The first plant leucine-rich repeat receptor-like kinase (LRR-RLK) gene was cloned and found in maize (Walker & Zhang, 1990). The researchers then found that LRR-RLKs are widely distributed in plant genomes and have expanded to hundreds of members per genome, such as 309 members in rice, 303 in Brassica rapa, and 379 in poplar (Hwang, Kim & Jang, 2011; Rameneni et al., 2015; Zan et al., 2013). The typical plant LRR-RLKs proteins contain three characteristic domains: extracellular domain (ECD), transmembrane domain (TM), and intracellular kinase domain (KD) (Gou et al., 2010; Shiu & Bleecker, 2001; Song et al., 2017).
At present, the functions of LRR-RLKs in many plants, especially model plants, have been fully studied (Ariza-Suarez et al., 2022; Gottin et al., 2021). For instance, brassinolide-insensitive 1 (BRI1) plays a key role in a variety of plant growth and development processes by sensing the steroid hormone brassinolide (BRs) (Nolan, Chen & Yin, 2017). PXC1 from Arabidopsis, an LRR-RLK protein, is essential to regulate secondary wall formation (Wang et al., 2013). The Arabidopsis LRR-RLK, HSL3, is a regulator of the drought stress response and stomatal closure correlated with hydrogen peroxide homeostasis (Liu et al., 2020). COE1, also known as LRR-RLK, plays a critical role in the formation of commissural patterns in rice (Sakaguchi et al., 2010). In addition, studies have confirmed that the RECEPTOR-LIKE PROTEIN KINASE 1 (RPK1) acts as a defense-related receptor in Oryza rufipogon, while the Arabidopsis homolog of RPK1, AtRPK1, has also reported to play key roles in leaf senescence and drought stress responses (Law et al., 2012; Lee et al., 2011; Osakabe et al., 2005).
Recently, systematic identification of LRR-RLKs has been carried out in five Rosaceae species (Sun et al., 2017), two citrus species (Magalhães et al., 2016), Solanum lycopersicum (Wei et al., 2015), Amborella trichopoda (Liu et al., 2016), and other plant species (Liu et al., 2017; Sun et al., 2018; Wang et al., 2019). However, it is still excluded whether the function of LRR-RLKs as alarm genes in tung tree in response to Fusarium wilt infection. In our study, we used the Fusarium wilt-susceptible V. fordii and Fusarium wilt-resistant V. montana as materials to study the genetic mechanisms of LRR-RLKs in resistance to Fusarium wilt infection in tung tree. Our data might provide important candidate genes for future molecular-assisted breeding in tung tree.
Materials and Methods
Database search
The proteins, CDSs, and GFF files of V. montana and V. fordii were obtained from NCBI database, as described by Cui et al. (2018), Li et al. (2022) and Cao et al. (2022). The proteins and CDSs of LRR-RLKs in V. fordii were obtained from Cao et al. (2021) and Cao et al. (2020). Subsequently, we used InterProScan (Jones et al., 2014) to identify distinct protein signatures in these predicted datasets and developed a local database to analyze the datasets in each included plant genome. Pkinase (PF00069) and Pkinase_Tyr (PF07714) are considered to be KD domains specific to LRR-RLK proteins. LRR domains containing LRRNT_2 (PF08263), LRRCT (PF01463), LRV (PF01816), LRRNT (PF01462), LRR_1 (PF00560), LRR_2 (PF07723), LRR_3 (PF07725), LRR_4 (two copies; PF12799), LRR_5 (six copies; PF13306), LRR_8 (PF13855) and LRR_9 (PF14580) are LRR-RLK-specific R domains. To detect the candidate LRR-RLKs in V. montana, we first downloaded and obtained their HMM models from the Pfa, website (El-Gebali et al., 2019). The local database was then retrieved using the HMM model using the HMMER 3.0 software (Mistry et al., 2013). Each amino acid sequence of V. fordii VfLRR-RLKs was also used as a query to determine LRR-RLKs in the V. montana local genome database using BLASTp with an E-value less than 1e−5. Finally, if identified candidate genes contained complete KD and R domains, we considered them to be candidate LRR-RLKs.
Phylogenetic analysis
All full-length VmLRR-RLK proteins were subjected to multiple sequence alignment using MAFFT (version 7) software with default parameters (Katoh & Standley, 2013). MEGA (version 5) software was used to construct the neighbor-joining tree (Tamura et al., 2011). The bootstrap value is an important method to analyze the reliability of the phylogenetic tree, so to confirm the reliability of the phylogenetic relationship, this study used 1,000 bootstrap values to test the reliability of this tree. The iTOL website and FigTree software were used to edit and visualize the phylogenetic tree (Letunic & Bork, 2019).
Gene structure and collinear analysis
The information about the starting position of each VmLRR-RLK on the chromosome was obtained from the GFF file of the V. montana genome, and then TBtools was used to visualize the location of the VmLRR-RLK gene on the chromosomes (Chen et al., 2018). The DNA and CDS sequence of each VmLRR-RLK gene were obtained from the V. montana genome and compared using Tbtools (version 1.098769) software to obtain the gene structure information of VmLRR-RLK. The collinear analysis was identified among genome regions by MCScanX software with an E-value of 1e−10 (Wang et al., 2012), as described by Jiang et al. (2022b). The TBtools (version 1.098769) software was carried out to visualize the collinear relationships (Chen et al., 2018).
Transcriptome analysis
Transcriptome data (PRJNA445068, PRJNA483508, and PRJNA318350) were collected and retrieved from NCBI databases to analyze expression patterns. SRA toolkit was used to decompress raw data into fastq format. Then, each dataset was mapped into the corresponding reference genome using HISAT2 using default parameters (Kim et al., 2019; Pertea et al., 2016). The expression levels of genes were calculated using StringTie using default parameters and normalized using FPKM (Pertea et al., 2016). In this study, we normalized and visualized all expression data using TBtools (version 1.098769) software (Chen et al., 2018).
Results and Discussion
Identified of VmLRR-RLKs in V. montana
V. fordii and V. montana are the two most important main varieties in China. V. fordii has high oil content but is susceptible to Fusarium wilt, while V. montana is resistant to Fusarium wilt (Cui et al., 2018). More studies have confirmed that LRR-RLKs play important roles in plant stress resistance (Ariza-Suarez et al., 2022; Cao et al., 2021; Geng et al., 2021; Gottin et al., 2021). In the previous study, we only identified and analyzed the VfLRR-RLKs in the V. fordii due to the lack of the V. montana genome (Cao et al., 2021). Identification of VmLRR-RLKs from Fusarium wilt-resistant V. montana and comparison with corresponding members in Fusarium wilt-susceptible V. fordii will help to further clarify the roles of LRR-RLKs in Fusarium wilt resistance.
To identify LRR-RLK gene family members, we first obtained the Hidden Markov Model to identify LRR-RLK members from the Pfam (El-Gebali et al., 2019). At the same time, we also used the LRR-RLK sequence of V. fordii as a template and used BlastP software to search the genome of V. montana. Subsequently, 243 candidate VmLRR-RLKs were found in V. montana genome. Three tools, including Pfam (El-Gebali et al., 2019), InterProScan (Jones et al., 2014), and SMART (Letunic & Bork, 2018), were used to determine whether the identified LRR-RLK proteins contained the PK and R domains. Two members were found to lack or not contain the complete LRR-RLK domain. Therefore, this study finally identified 239 VmLRR-RLKs in the V. montana genome for further analysis (Table S1).
The LRR-RLK gene family contains many members, such as 226 members in rice and 236 members in Arabidopsis (Hwang, Kim & Jang, 2011). Further studies determined that the number of LRR-RLK family members was not necessarily related to genome size. For example, V. fordii and V. montana have almost the same genome size (Cui et al., 2018; Zhang et al., 2019), but V. fordii only had 167 members (Cao et al., 2021), while V. montana did contain 239 members. Interestingly, the number of LRR-RLK members in cassava and rubber trees was about twice as high as that in V. montana (Cao et al., 2021). This may be due to the fact that cassava and rubber trees have additionally experienced recent whole-genome duplication events in addition to those shared by Euphorbiaceae (Cui et al., 2018; Mansfeld et al., 2021). These results suggested that the number of LRR-RLKs is closely related to duplication events in addition to whether the species is resistant to disease.
Gene structure analysis of VmLRR-RLKs in V. montana
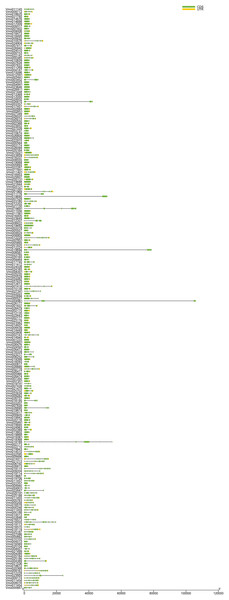
The gene structures are not only closely related to their functions but also may reflect the evolutionary history of gene family members (Cao et al., 2016; Song et al., 2017). In general, ancient genes are generally intronless, and then introns appeared in the long evolutionary process, resulting in more and more complex gene structures (Chen et al., 2020; Liu et al., 2021; Zou, Guo & He, 2011). To gain insight into the evolutionary history of VmLRR-RLKs in V. montana, we analyzed their exon-intron structures. As shown in Fig. 1, the results showed that the VmLRR-RLKs presented a complex gene structure. Four genes, including Vmo023646, Vmo000564, Vmo025607, and Vmo001233, were intronless genes, suggesting that these genes might be the VmLRR-RLKs ancestral genes. Vmo023859 contained the most introns (28), followed by Vmo012229 (27) and Vmo014511 (27), indicating these genes might be young genes. Taken together, our results indicated that each member of VmLRR-RLK exhibits a complex genetic structure during the long evolutionary process, which might contribute to the resistance to Fusarium wilt in V. montana.
Figure 1: Gene structure of VmLRR-RLKs in V. montana.
The exon-intron organizations of all VmLRR-RLKs were visualized by Tbtools software. Introns and exons were plotted by lines and boxes, respectively.Phylogenetic analysis of VmLRR-RLKs in V. montana
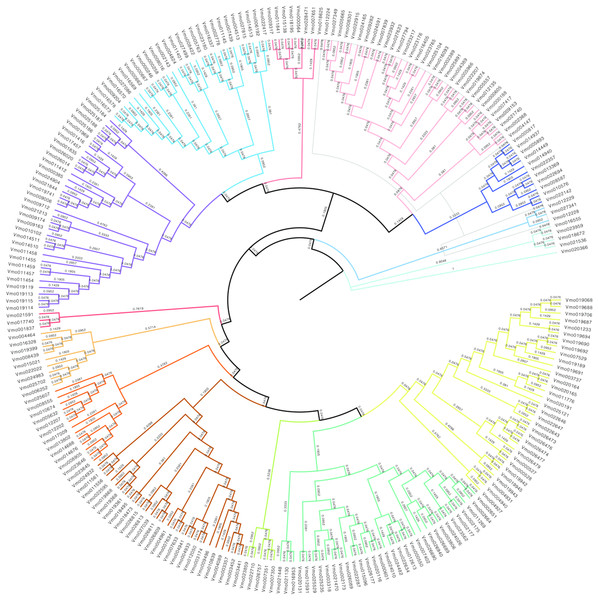
In order to further obtain the evolutionary relationships of VfLRR-RLKs in V. montana, we first used MAFFT software to conduct multiple sequence alignment analysis of all VmLRR-RLK protein sequences and then used MEGA (version 5) software to construct a phylogenetic tree using the neighbor-joining method. The bootstrap value was used to check whether the constructed evolutionary tree was reliable, as described by Li et al. (2022). As shown in Fig. 2, all VmLRR-RLKs were divided into 14 major subgroups, which were named C1 to C14 and distinguished by different colors. This result was basically consistent with the exon-intron distributions, that were, members of the same subgroup contained similar exon-intron structures. Gene duplication and loss events may play important roles in the expansion and contraction of LRR-RLK family members (Liu et al., 2016; Magalhães et al., 2016; Zou, Guo & He, 2011). In this study, at the branch tip, the shorter branch length demonstrated the strong amino acid conservation of the cluster genes, suggesting that these members may share the conserved evolutionary relationships, leading them to possibly have similar functions with functional redundancy. In each branch, the two genes from the tip might have undergone a gene duplication event during evolution, or a gene loss event might have occurred.
Figure 2: The phylogenetic tree was generated from the alignment result of the full-length amino acid sequences by the neighbor-joining (NJ) method.
The chromosome localization and gene duplication analysis of VmLRR-RLKs in V. montana
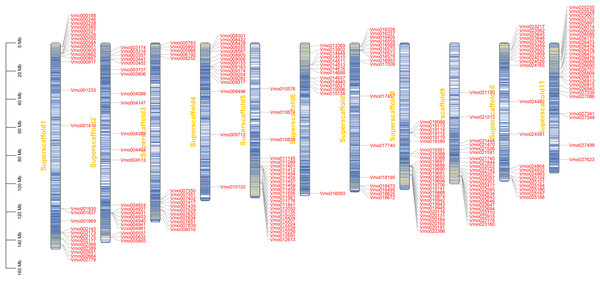
Gene location analysis can determine the distribution of genes on chromosomes (Mishra et al., 2021). The chromosome location of each of VmLRR-RLK was determined on V. montana chromosomes. As shown in Fig. 3, 238 of the 239 VmLRR-RLKs were unevenly assigned to chromosomes, while the remaining three VmLRR-RLKs were localized on scaffolds in V. montana genome. The Vm8 and Vm11 chromosomes each contained 28 VmLRR-RLKs, the Vm3 and Vm4 chromosomes each had 16 VmLRR-RLKs, the Vm1 chromosome contained 27 VmLRR-RLKs, the Vm2 chromosome located 22 VmLRR-RLKs, and the Vm5 chromosome contained 25 VmLRR-RLKs. There were 14 VmLRR-RLKs in Vm6 chromosome, 17 VmLRR-RLKs in Vm7 chromosome, 24 VmLRR-RLKs in Vm9 chromosome, and 21 VmLRR-RLKs in Vm10 chromosome.
Figure 3: VmLRR-RLK genes were mapped to all V. montana chromosomes.
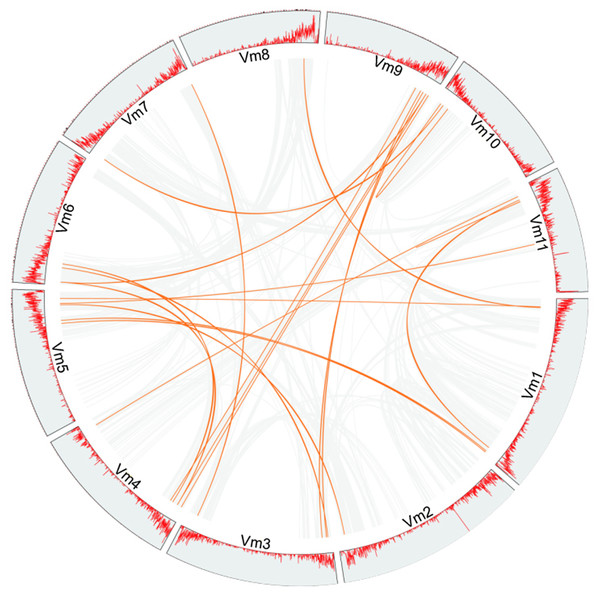
Phylogenetic analysis suggested that gene duplication events might be the main cause of the expansion of members of the VmLRR-RLK gene family in V. montana. For example, a total of 15 tandem duplications and 31 segmental duplications were found in cucumber genome (Yu et al., 2022). In Thinopyrum elongatum genome, Mishra et al. (2021) identified 191 segmental duplications and 145 tandem duplications, respectively (Mishra et al., 2021). In the potato genome, 16 and 20 genes were predicted to be the results of tandem duplications and segmental duplications, respectively (Li et al., 2018). To further elucidate the expansion mechanism of members of the VmLRR-RLK gene family, we analyzed the gene duplication events in V. montana (Fig. 4). The results indicated that segmental duplications were the main reason for the expansion of VmLRR-RLK gene family members. It is worth noting that this study did not find that members of the VmLRR-RLK gene family have undergone tandem duplications in V. montana, which was different from the previous results in other plants, such as cucumber, potato, and T. elongatum (Li et al., 2018; Mishra et al., 2021; Yu et al., 2022). These data suggested that the evolution mechanisms of LRR-RLKs were different in different plant genomes.
Figure 4: The duplication analysis of VmLRR-RLKs in V. montana.
All putative segmental duplications are linked by the colored lines respectively.Expression pattern analysis of LRR-RLK family genes
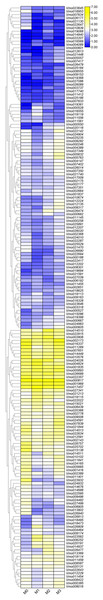
More LRR-RLKs have been reported involved in disease resistance in plants (Cao et al., 2020; Geng et al., 2021). For example, an LRR receptor-like kinase protein, ERECTA can affect resistance to bacterial wilt controlling development pleiotropically (Godiard et al., 2003). The LRR-RLK/malectin-like IOS1 play important role in BAK1-independent and BAK1-dependent pattern-triggered immunity in Arabidopsis (Yeh et al., 2016). XIK1 from Oryzae pv. oryzae, an LRR receptor-like kinase gene is involved in Xa21-mediated disease resistance (Hu et al., 2015). In this study, to further determine the functions of the LRR-RLKs, we selected the Fusarium wilt-resistant V. montana and Fusarium wilt-susceptible V. fordii as the research objects. Among them, the expression data of the V. fordii VfLRR-RLKs were derived from a previously published manuscript (Cao et al., 2020, 2021). As shown in Fig. 5, we found that the FPKM values of 26.6% (64/239) VmLRR-RLKs genes were <1 response to Fusarium wilt infection, proposing that these genes might be expressed in other tissues, such as flowers, roots, and leaves. Remarkably, 22 VmLRR-RLKs showed down-regulated expression, while 124 VmLRR-RLKs were up-regulated during Fusarium wilt infection (Fig. 5).
Figure 5: Expression of VmLRR-RLK genes during Fusarium wilt infection.
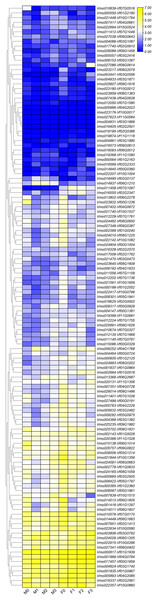
M0, M1, M2 and M3 indicated the expression of VmLRR-RLKs in V. montana during the infection stage (0, uninfected stage; 1, 2 days after Fusarium wilt infection (dpi); 8 dpi; 3, 13 dpi) by the pathogen Fusarium wilt.Due to the lack of the genome of V. montana, in the previous study, we aligned the transcriptome data of V. montana infected with Fusarium wilt and V. fordii infected with Fusarium wilt to the genome of V. fordii for determine the potential roles of LRR-RLKs in resistance to Fusarium wilt (Cao et al., 2021). In this study, one-to-one LRR-RLK orthologous gene pairs were identified using reciprocal Blast between V. fordii and V. montana (Cao et al., 2022). Subsequently, we analyzed the expression patterns of these orthologous genes in Fusarium wilt-susceptible V. fordii and in Fusarium wilt-resistant V. montana during Fusarium wilt infection (Table 1). As shown in Fig. 6, most of the orthologous genes showed opposite expression patterns, of which 55 (44.7%) LRR-RLKs pairs were all up-regulated in V. montana, while their corresponding orthologous genes were all down-regulated in V. fordii; 16 (13%) LRR-RLKs pairs were all down-regulated in V. montana, while their corresponding orthologous genes were all up-regulated in V. fordii. Notably, six orthologous LRR-RLK gene pairs (Vf01G2180-Vmo000550, Vf02G0137-Vmo018495, Vf06G0012-Vmo016093, Vf06G1210-Vmo014937, Vf07G1580-Vmo012050, and Vf11G1090-Vmo019368) were up-regulated in V. montana, but were not expressed or expressed relatively low in V. fordii, suggesting indicated that these LRR-RLKs might play important roles in the resistance of tung tree species to the infection of Fusarium spp.
| Gene1 | Gene2 | Gene1 | Gene2 |
|---|---|---|---|
| Vmo000188 | Vf01G2552 | Vmo014940 | Vf00G0360 |
| Vmo000385 | Vf01G2360 | Vmo015096 | Vf00G0329 |
| Vmo000550 | Vf01G2180 | Vmo015138 | Vf06G1014 |
| Vmo000564 | Vf01G2163 | Vmo016093 | Vf06G0012 |
| Vmo000605 | Vf01G2125 | Vmo016328 | Vf02G2570 |
| Vmo000817 | Vf01G1939 | Vmo016337 | Vf02G2561 |
| Vmo001416 | Vf01G1297 | Vmo016405 | Vf02G2501 |
| Vmo001835 | Vf01G0967 | Vmo016555 | Vf02G2347 |
| Vmo001837 | Vf01G0964 | Vmo016569 | Vf02G2333 |
| Vmo001969 | Vf04G0784 | Vmo016573 | Vf09G0613 |
| Vmo002143 | Vf01G0028 | Vmo017009 | Vf02G1762 |
| Vmo002368 | Vf01G0240 | Vmo017457 | Vf00G1959 |
| Vmo002664 | Vf01G0518 | Vmo017740 | Vf02G0910 |
| Vmo002778 | Vf01G0633 | Vmo018195 | Vf02G0386 |
| Vmo003441 | Vf03G0556 | Vmo018495 | Vf02G0137 |
| Vmo003806 | Vf03G0792 | Vmo018625 | Vf02G0002 |
| Vmo004089 | Vf03G1087 | Vmo019368 | Vf11G1090 |
| Vmo004147 | Vf00G1381 | Vmo019399 | Vf11G0681 |
| Vmo004389 | Vf03G1382 | Vmo019874 | Vf11G1118 |
| Vmo004464 | Vf00G0724 | Vmo020131 | Vf11G1306 |
| Vmo004824 | Vf03G1740 | Vmo020366 | Vf11G1528 |
| Vmo004931 | Vf03G1869 | Vmo021448 | Vf10G1794 |
| Vmo004933 | Vf03G1871 | Vmo021470 | Vf03G0473 |
| Vmo004961 | Vf03G1880 | Vmo021591 | Vf10G1659 |
| Vmo005557 | Vf03G2416 | Vmo021740 | Vf10G1537 |
| Vmo005632 | Vf03G2482 | Vmo021844 | Vf10G1359 |
| Vmo005665 | Vf03G2505 | Vmo022142 | Vf10G1082 |
| Vmo005763 | Vf04G2229 | Vmo022207 | Vf10G1004 |
| Vmo005893 | Vf04G2085 | Vmo022357 | Vf10G0860 |
| Vmo005966 | Vf04G2023 | Vmo022417 | Vf10G0796 |
| Vmo006192 | Vf04G1833 | Vmo022634 | Vf10G0580 |
| Vmo006252 | Vf04G1768 | Vmo022694 | Vf10G0524 |
| Vmo007351 | Vf04G0728 | Vmo022915 | Vf10G0296 |
| Vmo007402 | Vf04G0672 | Vmo023176 | Vf10G0017 |
| Vmo007417 | Vf04G0661 | Vmo023180 | Vf10G0012 |
| Vmo007439 | Vf04G0638 | Vmo023217 | Vf08G2079 |
| Vmo007651 | Vf02G1413 | Vmo023645 | Vf08G1609 |
| Vmo007839 | Vf10G1515 | Vmo023859 | Vf08G1383 |
| Vmo008301 | Vf05G1941 | Vmo023932 | Vf00G1236 |
| Vmo008422 | Vf05G1797 | Vmo024010 | Vf08G1225 |
| Vmo008587 | Vf05G1661 | Vmo024026 | Vf08G1205 |
| Vmo009006 | Vf05G1214 | Vmo024165 | Vf08G1059 |
| Vmo009153 | Vf05G1097 | Vmo024482 | Vf08G0650 |
| Vmo009282 | Vf05G0975 | Vmo024591 | Vf08G0663 |
| Vmo009317 | Vf05G0929 | Vmo024904 | Vf00G1004 |
| Vmo010576 | Vf07G0170 | Vmo024983 | Vf08G0202 |
| Vmo010674 | Vf07G0327 | Vmo025235 | Vf09G1982 |
| Vmo010839 | Vf07G0516 | Vmo025607 | Vf09G1916 |
| Vmo011145 | Vf07G0781 | Vmo025702 | Vf09G1831 |
| Vmo011401 | Vf07G1038 | Vmo025891 | Vf09G1626 |
| Vmo011412 | Vf07G1049 | Vmo026014 | Vf09G1496 |
| Vmo011458 | Vf07G1087 | Vmo026089 | Vf09G1458 |
| Vmo011556 | Vf07G1158 | Vmo026757 | Vf09G0822 |
| Vmo012050 | Vf07G1580 | Vmo027039 | Vf09G0643 |
| Vmo012135 | Vf07G1668 | Vmo027086 | Vf09G0614 |
| Vmo012202 | Vf07G1736 | Vmo027341 | Vf09G0402 |
| Vmo012224 | Vf07G1755 | Vmo027349 | Vf09G0387 |
| Vmo012229 | Vf07G1761 | Vmo027499 | Vf00G0181 |
| Vmo013369 | Vf06G2687 | Vmo027623 | Vf11G0584 |
| Vmo013802 | Vf06G2278 | Vmo014513 | Vf06G1605 |
| Vmo014449 | Vf06G1663 | Vmo014937 | Vf06G1210 |
| Vmo014511 | Vf06G1607 |
Figure 6: Expression profiles of LRR-RLK genes under Fusarium wilt infection between Fusarium wilt-susceptible V. fordii and Fusarium wilt-resistant V. montana.
M0–M3 suggested the expressions of LRR-RLK genes in V. montana during the infection stage (0, 1, 2, 3) by the pathogen Fusarium wilt, and F0–F3 suggested the expression of LRR-RLK genes in V. fordii during the infection stage (0, 1, 2, 3) by the pathogen Fusarium wilt.Conclusions
The tung tree LRR-RLK family members were investigated through gene structure, gene duplication, chromosomal distribution, phylogeny, and expression patterns analysis, which help us to further understand the evolutionary history of this gene family. Our study analyzed the LRR-RLKs in Fusarium wilt-susceptible V. fordii and Fusarium wilt-resistant V. montana to reveal their expression patterns in response to Fusarium wilt infection. Taken together, these data revealed the genetic mechanisms of resistance to Fusarium wilt infection and provided important candidate genes for future molecular-assisted breeding in tung tree.