An analytical study on the identification of N-linked glycosylation sites using machine learning model
- Published
- Accepted
- Received
- Academic Editor
- Giuseppe Agapito
- Subject Areas
- Bioinformatics, Artificial Intelligence, Data Mining and Machine Learning
- Keywords
- N-linked, Glycosylation, Machine learning, Deep learning, Artificial intelligence, Performance evaluation criteria
- Copyright
- © 2022 Akmal et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ Computer Science) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. An analytical study on the identification of N-linked glycosylation sites using machine learning model. PeerJ Computer Science 8:e1069 https://doi.org/10.7717/peerj-cs.1069
Abstract
N-linked is the most common type of glycosylation which plays a significant role in identifying various diseases such as type I diabetes and cancer and helps in drug development. Most of the proteins cannot perform their biological and psychological functionalities without undergoing such modification. Therefore, it is essential to identify such sites by computational techniques because of experimental limitations. This study aims to analyze and synthesize the progress to discover N-linked places using machine learning methods. It also explores the performance of currently available tools to predict such sites. Almost seventy research articles published in recognized journals of the N-linked glycosylation field have shortlisted after the rigorous filtering process. The findings of the studies have been reported based on multiple aspects: publication channel, feature set construction method, training algorithm, and performance evaluation. Moreover, a literature survey has developed a taxonomy of N-linked sequence identification. Our study focuses on the performance evaluation criteria, and the importance of N-linked glycosylation motivates us to discover resources that use computational methods instead of the experimental method due to its limitations.
Introduction
The process of glycosylation is considered to be one of the most complex type of post translation modification (PTM) in eukaryotes cells (Akmal, Rasool & Khan, 2017; Yang et al., 2019). The post translation modification occurs when protein, after synthesis, undergo different type of changes and without these modification proteins cannot perform their psychological functionalities properly (Yang et al., 2019). Nearly 200 different types of such post translation modification have been discovered and glycosylation is most important amongst them as it plays a vital role in different biological functions such as cell communication, protein folding, recognition of antigens and −50% of the human genomes are glycosylated (Akmal, Rasool & Khan, 2017; Akmal et al., 2020; Yang et al., 2019). The glycosylation sites are very relevant for cancer discovery as well as for further drug development (He, Wei & Zou, 2019; Hwang et al., 2020). Glycosylation sites are classified into five types: N-linked, O-linked, C-linked, glypiation and phospho glycosylation (Lei, Tang & Du, 2017). It is very much important to identify such sites.
There are various techniques to identify such sites, broadly it can be classified into experimental and computational method (Audagnotto & Dal Peraro, 2017). The experimental method requires the understanding of cell biology and the functions of cell structure (Hwang et al., 2020). The well-know techniques used for experimental identification are: radioactive label, chromatin immunoprecipitation (ChIP), mass spectrometry (MS) and liquid chromatography (LCG) (Akmal et al., 2020; Hwang et al., 2020; Naseer et al., 2020a). In computational method, researchers discover valuable information from the structure of protein sequences and apply some artificially intelligent algorithms to predict the relevant glycosylation or any other PTM sites (Hamby & Hirst, 2008; He, Wei & Zou, 2019; Shek, Kotidis & Betenbaugh, 2021; Naseer et al., 2021b; Murad et al., 2021).
The N-linked glycosylation is the primary glycosylation type, as 90% of glycosylated sites belong to the N-linked glycosylation (Akmal, Rasool & Khan, 2017). Usually, N-glycans are attached to glycoproteins on asparagine residues within the Asn-X-Ser/Thr sequon (except proline, X could be any amino residue) (Zhang et al., 2021b; Alkuhlani et al., 2021). N-linked glycans plays vital role in intrinsic and extrinsic (Alkuhlani et al., 2021). Apart from improving the protein’s stability, it provides a structural component to the cell surface. N-glycan also mediate cell-to-cell interaction and controls the glycoprotein in the cellular environment (Naseer et al., 2020b). N-linked glycan helps is identification of various diseases such as type I diabetes, cancer, rheumatoid arthritis, and Crohn’s disease (Alkuhlani et al., 2021; Naseer et al., 2020a; Khan et al., 2020b). Therefore, it is very much important to identify such sites, but the identification of such sites using experimental technique is time-consuming and expensive as well (Coff et al., 2020; Akmal et al., 2020; Qiu et al., 2018). Therefore, researchers have developed several computational models based on artificial neural network (ANN) to predict the N-linked sites (Le, Sandag & Ou, 2018; Butt et al., 2016; Alkuhlani et al., 2021). Although, few reviews exist on N-linked prediction model, but they mainly focus on algorithm used to train the model and less focused on the feature set construction and performance metric, as shown in Table 1. These studies only analyzed the models developed up to 2019.
| Article Ref. No. | Focus | Year | Survey approach | Quality assessment | N-linked model (Tool) | Feature construction | Training algorithm | Organism type | Performance metric (ACC, SN, SP) | Target repository |
|---|---|---|---|---|---|---|---|---|---|---|
| Alkuhlani et al. (2021) | Glycosylation sites prediction tool using AI. | 2021 | Informal | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Shek, Kotidis & Betenbaugh (2021) | Experimental and computation method for PTM site prediction | 2021 | Informal | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ |
| Kuo-Chen (2019) | PTM sites prediction model develop using Chou’s 5 step model. | 2019 | Informal | ✗ | ✗ (other PTM) | ✓ | ✓ | ✗ | ✗ | ✗ |
| He, Wei & Zou (2019) | Research progress in PTM site prediction. | 2019 | Informal | ✗ | ✗ (glyco type not specified) | ✓ | ✓ | ✗ | ✗ | ✗ |
| Audagnotto & Dal Peraro (2017) | Tools used for PTM. | 2017 | Informal | ✗ | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ |
| This survey | N-linked site prediction tool including training algorithm, and feature approach which helps to construct an efficient model for other PTM. | 2021 | Systematic Review | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 5 |
The glycosylated region of N-linked sites appears at the specific location within the protein sequence, as protein sequence consists of the chain of amino acid and each amino acid out of known 20 is represented by specific alphabetic character (Qiu et al., 2018; Yang et al., 2019; Kumari, Kumar & Kumar, 2018). In computational approach, it is required to extract some useful information from these sequences to construct the feature vector (Butt, Rasool & Khan, 2017; Chien et al., 2020; Hamby & Hirst, 2008; Naseer et al., 2021b). The feature vectors of glycosylated and non-glycosylated N-linked sites have certain pattern of protein sequences and these patterns have identified through the various technique (algorithm) of machine learning method (Taherzadeh et al., 2019; Tran, Pham & Ou, 2021; Hayat & Khan, 2011; Park et al., 2019; Xiang, Zou & Zhao, 2021; Dimeglio et al., 2020). The evidence of organism type also helps in the successful identification of such sites (Huang & Li, 2018).
The existing reviews are compared on various perspectives such as quality assessment scores, availability of N-linked model, feature set construction method, training model algorithm, specie type, performance metric and target repositories as shown in Table 1. The proposed study only focused on the review articles accepted in recognized journals because of reliability (Barukab et al., 2019). This comparison helps the need to build the survey.
The rational of our work is to provide the comprehensive systematic literature review on the identification of N-linked sites to bring out the detail of exiting computational models. The researchers have performed numerous efforts to identify such sites computationally in the recent past. The work presented by these researchers has been reviewed by few authors to ensure the effectiveness of the proposed prediction model to identify the N-linked sites (Shek, Kotidis & Betenbaugh, 2021; Alkuhlani et al., 2021; Audagnotto & Dal Peraro, 2017). The authors primarily focused on the feature set construction algorithm and training algorithm, and less or no focus on quality assessment criteria, performance metric evaluation and the type of species of the reviewed articles used to predict the N-linked sites. The proposed systematic review provides novel features such as targeting channel, quality assessment score, new classification criteria, and performance evaluation based on accuracy, sensitivity, and specificity metric after evaluating studies empirically.
This SLR will help the medical scientists in the targeted identification of cancer, type I diabetic cell for treating the patients, and help the pharmacists in effective drug development by opting the accurate predictor of N-Linked sites. Furthermore, it will facilitate the researchers to develop more accurate and efficient predictive model by analyzing the techniques used by existing researchers.
The proposed article is presented in the following sequence: the methodology adopted to conduct survey along with objectives and research questions is presented in “Survey methodology”. The analysis of the research question is described in “Assessment and discussion”. The “Discussion and future direction” presents synthesis of reviewed literature. Finally, the article has been concluded in “Conclusion”.
Survey methodology
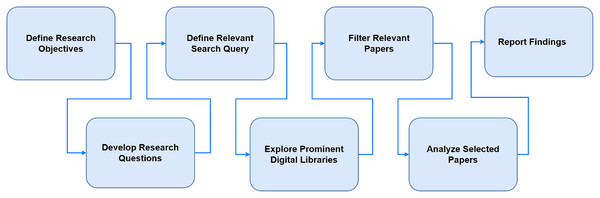
The survey methodology consists of three phases: plan, conduct of review and conclusion as shown in Fig. 1.
Figure 1: Research strategy.
Review plan
The process involved to conduct the review is shown in Fig. 2.
Figure 2: Research strategy.
Review conduct
The steps involved to conduct the review were: (a) Search of relevant primary study from different search venues. (b) Selection of relevant research articles from searched articles obtained in previous step through predefined inclusion/exclusion criteria. (c) The selected articles were then assigned score based on their defined quality parameters. (d) Backward snowballing to include the important articles.
Automated search in digital library
The relevant research articles have been extracted through system search. Therefore, automatic, and manual search has been performed. The google scholar is used as digital venue to get the relevant research articles.
Google Scholar (http://scholar.google.com/)
IEEE Xplore (https://ieeexplore.ieee.org/search/advanced)
Springer Link (https://link.springer.com/)
Bioinformatics (https://academic.oup.com/bioinformatics)
PLOS ONE (https://journals.plos.org/plosone/)
To get appropriate and relevant search result, keyword based search has been applied on the digital venue. Based on the RQs mentioned in Table 2, keyword are selected for primary and secondary term. The Boolean operator ‘AND’ and ‘OR’ are used to build query string. The search query based on keyword is shown in Fig. 3. The search query is grouped into three groups where each group contain the similar keyword to ensure maximum relevant studies as mentioned in Table 1. Using the Boolean operators (OR, AND) final search query is designed in which AND operator is applied in different groups and OR operator is with in different keywords of a group.
| RQ | Research question | Research objective/motivation |
|---|---|---|
| RQ1 | Which are the relevant publishing channel for N-Linked glycosylation research? Which channel type and geographical area target this research? | To identify • High quality publishing venue. • Research published during 2017–till October-2021. • Scentometric analysis based on meta information including research type, approaches and validation methods. |
| RQ2 | Which are the exiting prediction model (tool) used for the identification of N-linked Glycosylation sites and for which kind of species these sites are identified? | To help the researchers to identify diseases i.e., cancer detection, type 1 diabetic and also drug discoveries through cost effective and time saving approach. |
| RQ3 | Which algorithm or method are used to construct N-Linked feature vector? | To understand the in-depth structure of protein sequences to extract useful information to train model. |
| RQ4 | Which algorithm or method are used to train N-Linked model? | To develop efficient tool to predict the N-linked sites through computational approach. |
| RQ5 | How effective are the existing model to predict the N-Linked sites? | By evaluating the 1. Availability of data set. 2. Availability of tool. 3. Determining the Accuracy measure including Accuracy, Sensitivity and Specificity metrics. 4. Result comparison with existing studies. |
Figure 3: Keyword used to develop query string.
Listening 1 [“n linked” OR “Post translation modification”] AND [“Glycosylation sites” OR “Glycan”] AND [“prediction model” OR “Artificial Intelligence” OR “Neural Network” OR “Deep Learning”]
Primary keywords were selected as a key identifier for N-linked prediction models. Primary keywords along with the secondary and additional keywords were chosen. Combination of keywords and Boolean operators have developed as mentioned in Table 3.
| Digital library | Search query | Applied filter |
|---|---|---|
| IEEE Xplore | (“n linked” OR “Post translation modification”) AND (“prediction model” OR “Artificial Intelligence” OR “Neural Network” OR “Deep Learning”) | 2017–2021 |
| Springer link | (“n linked” OR ”Post translation modification”) AND (“Glycosylation sites” OR “Glycan”) AND (“prediction model” OR “Artificial Intelligence” OR “Neural Network” OR “Deep Learning”) | 2017–2021 |
| Bioinformatics | (n linked OR Post translation modification) AND (Glycosylation sites OR Glycan) AND (prediction model OR Artificial Intelligence OR Neural Network OR Deep Learning) | 2017–2021 |
| PLOS ONE | (“n linked”) AND (“Glycosylation”) AND (“Neural Network” OR “Deep Learning”) | 2017–2021 |
| Google scholar | (“n linked” OR “Post translation modification”) AND (“Glycosylation sites” OR “Glycan”) AND (“prediction model” OR “Artificial Intelligence” OR “Neural Network” OR “Deep Learning”) | 2017–2021 |
Inclusion and exclusion criteria for selection
Inclusion Criteria
The article included in review must contain prediction of N-linked glycosylation sites or Glycosylation sites.
-
It must target any of the research question mentioned in Table 2.
It is published in journal or in preprint repository since 2017.
It should contain computation or semi computational approach for prediction.
2. Exclusion Criteria
Eliminate articles that do not address the N-linked glycosylation or glycosylation.
Eliminate articles that purely identify N-linked sites through biological experimentation.
Eliminate the books appeared in the result of search query.
Quality assessment as selection criteria
The quality assessment (QA) is the major step to conducting any systematic review. In this study, questionnaire has been designed to measure the quality of selected articles. The score is computed on the following criteria:
The study has awarded score (1) if N-linked predictive tool has developed, otherwise scored (0).
The study has awarded score (2) if the method developed to extract feature from data based on computational approach, score (1) for hybrid approach and score (0) in-case of experimental approach.
The study has awarded score (1) if the computation method for training has provided, otherwise scored (0).
The score (1) has been awarded if the data set used is available otherwise scored (0).
The score (1) has been awarded if the organism type is available otherwise scored (0).
The studies were rated by taking conference and journal rating list into account. The possible score for publication is shown in Table 4.
| Publication source | +4 | +3 | +2 | +1 | 0 |
|---|---|---|---|---|---|
| Journals | Q1 | Q2 | Q3 | Q4 | No JCR ranking |
| Conference | CORE A* | CORE A | CORE B | CORE C | Not in CORE ranking |
The resultant score has been calculated for each study by aggregating the points of all question. Article achieving minimum score (5) has been included in the review.
Selection based on snowballing
After performing the quality assessment, back-word snowballing to extract the relevant articles from the references of the selected articles. The articles by Kumar et al. (2020) and Ilyas et al. (2019) have been shortlisted after performing the inclusion exclusion criteria and quality assessment.
Review report
The glycosylation sites especially N-Linked identification is very important domain, therefore in this review, systematic and empirical method is adopted to extract the relevant article from the digital libraries mentioned in Table 3, using query string as shown in Listening 1. Almost 800 articles are left after removing the articles before 2017.
The shortlisted articles are then filtered based on title, abstract, introduction and examined the full article if required for each search result. The article contains less than four pages and irrelevant articles were eliminated. The results of primary search, filtering and inspection phase, covering five digital libraries, are presented in Table 5.
| Phase | Selection | Selection criteria | PLOS ONE | Bioinformatics | Springer link | IEEE Xplore | Google scholar | Total articles |
|---|---|---|---|---|---|---|---|---|
| 1 | Search | Keyword (Fig. 2) | 21 | 3 | 47 | 4 | 770 | 845 |
| 2 | Filtering | Title | 15 | 3 | 18 | 3 | 212 | 251 |
| 3 | Filtering | Abstract | 10 | 3 | 13 | 3 | 160 | 189 |
| 4 | Filtering | Introduction and conclusion | 6 | 2 | 7 | 3 | 125 | 143 |
| 5 | Inspection | Full article | 1 | 1 | 2 | 2 | 62 | 68 |
After the preprocessing of articles, inclusion/exclusion test has been performed and after that quality assessment score has been computed. The article having at least five score have included in this study and it is total of 70 in count as given in Table 6.
| Sr. No. | Ref. No. | P.Year | P.Channel | Research type | Empirical type | Species | PTM type | Feature set method | Model training algorithm | Model | (a) | (b) | (c) | (d) | (e) | (f) | SCORE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Akmal, Rasool & Khan (2017) | 2017 | Journal | Solution | Computational | Human | N-linked | Position relative and Statistical Moments | ANN/Back propagation | - | 0 | 2 | 1 | 1 | 1 | 4 | 9 |
| 2 | Chien et al. (2020) | 2020 | Journal | Solution | Computational | Human and Mouse | N-linked | Sequence, Structure and Function feature | XGBOOST | N-GlycoGo | 1 | 2 | 1 | 0 | 1 | 4 | 9 |
| 3 | Taherzadeh et al. (2019) | 2019 | Journal | Solution | Computational | Human and Mouse | N-linked and O-linked | Sequence and Structure | Deep ANN and SVM | Sprint-Gly | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 4 | Tran, Pham & Ou (2021) | 2021 | Journal | Solution | Computational | Human and Mouse | N-linked | Word embedding Vector Technique | RM, KNN, SVM and XGBoost. | - | 0 | 2 | 1 | 1 | 0 | 4 | 8 |
| 5 | Liu et al. (2019) | 2019 | Journal | Solution | Computational | Human | N-linked | Sequence | ANN | NetGlyco (Exiting) | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 6 | Li et al. (2019) | 2019 | Journal | Solution | Computational | Human | N-linked (and C/O-linked) | Sequence and Structure Feature | PA2DE using AlphaMax | GlycoMine_PU | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 7 | Bojar et al. (2021b) | 2021 | Journal | Solution | Hybrid | Eukaryote | Glycosylation | Sequence feature | Recurrent NN (LSTM) | SweetOrigin | 0 | 2 | 1 | 1 | 1 | 4 | 9 |
| 8 | Thomès, Burkholz & Bojar (2021) | 2021 | Journal | Solution | Computational | Animal | N-linked and O-linked | - | - | GlycoWork | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 9 | Carpenter et al. (2022) | 2021 | bioRxiv | Solution | Computational | Not Mention | Glycosylation | Fingerprint Encoding | MNN (ADAM) | GlyNet | 1 | 2 | 1 | 0 | 1 | 0 | 5 |
| 10 | Pitti et al. (2019) | 2019 | Journal | Solution | Computational | Human | N-linked | Similarity voiting and Gap Peptide | SVM | NGlyDE | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 11 | Lundstrøm et al. (2022) | 2021 | bioRxiv | Solution | Computational | Human | Glycosylation | Protein-Glycan Sequence Feature | Grpah CNN | LectinOracle | 0 | 2 | 1 | 1 | 1 | 0 | 5 |
| 12 | Burkholz, Quackenbush & Bojar (2021) | 2021 | Journal | Solution | Computational | Human | Glycosylation | Graph and Statistical feature | Graph NN | SweetNet | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 13 | Kotidis & Kontoravdi (2020) | 2020 | Journal | Solution | Hybrid | Human | N-linked | - | ANN/Kinetic Model | - | 0 | 0 | 1 | 1 | 1 | 4 | 7 |
| 14 | Lee et al. (2021) | 2021 | Journal | Solution | Experimental | Mammalian | Glycosylation | - | MS | - | 0 | 0 | 0 | 0 | 1 | 4 | 5 |
| 15 | Alkuhlani et al. (2021) | 2021 | Journal | Review | Computational | Human | Glycosylation | Computational | AI | - | 1 | 2 | 1 | 1 | 4 | 9 | |
| 16 | Adolf-Bryfogle et al. (2021) | 2021 | Journal | Solution | Computational | Not Mention | N-linked | KDE | Glycan Tree Modler | Rosetta Carbohydrate Framework | 1 | 2 | 1 | 1 | 0 | 0 | 5 |
| 17 | Sha et al. (2019) | 2019 | Journal | Solution | Experimental | Human | N-linked | Flux Balance Analysis | Kinetic | - | 0 | 1 | 0 | 1 | 1 | 2 | 5 |
| 18 | Zhang et al. (2021a) | 2021 | Journal | Solution | Experimental | Human | N-linked | MS | - | - | 0 | 1 | 0 | 0 | 1 | 4 | 6 |
| 19 | Park et al. (2019) | 2019 | Journal | Solution | Computational | Human | N-linked and O-linked | Sequence and Structure | Clustring | Glycan Reader and Modeler | 1 | 2 | 1 | 0 | 0 | 4 | 8 |
| 20 | Zhang et al. (2021c) | 2021 | Journal | Solution | Experimental | Human | N-linked | - | - | - | 0 | 0 | 0 | 0 | 1 | 4 | 5 |
| 21 | Xiang, Zou & Zhao (2021) | 2021 | Journal | Solution | Computational | Human | Glycosylation (O-linked) | feature set selected using SVM them mRmR | SVM,RF and NB | VPTMdb | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 22 | Antonakoudis et al. (2021) | 2021 | Journal | Solution | Hybrid | Human | N-linked | Stochiometric | ANN | - | 0 | 1 | 1 | 1 | 1 | 4 | 8 |
| 23 | Huang et al. (2017) | 2017 | Journal | Solution | Experimental | Mammalian | N-linked | - | - | - | 0 | 1 | 0 | 0 | 1 | 4 | 6 |
| 24 | Naseer et al. (2021b) | 2021 | Journal | Solution | Computational | Not Mention | PTM (Amidation) | PseAAC | CNN | IAmideV-deep | 1 | 2 | 1 | 1 | 0 | 2 | 7 |
| 25 | Hwang et al. (2020) | 2020 | Journal | Solution | Hybrid | Human | N-linked | IQ-GPA human plazma protein | DNN | - | 0 | 1 | 2 | 0 | 4 | 7 | |
| 26 | Coff et al. (2020) | 2020 | Journal | Solution | Computational | Human and Avian | Glycosylation | Frequent Subtree mining and mRMR | Regression Classifier | CCARL | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 27 | Le, Sandag & Ou (2018) | 2018 | Journal | Solution | Computational | Human | PTM (including N linked) | Statistical Moment and F score | RBF Network | PTM Transporter | 1 | 2 | 1 | 1 | 1 | 2 | 8 |
| 28 | He, Wei & Zou (2019) | 2019 | Journal | Review | Computational | Not Mention | N-linked | Provided | Provided | Provided | 1 | 2 | 1 | 0 | 0 | 4 | 8 |
| 29 | Audagnotto & Dal Peraro (2017) | 2017 | Journal | Review | Computational | Not Mention | N-linked | - | Provided | Provided | 1 | 0 | 1 | 1 | 0 | 4 | 7 |
| 30 | Krasnova & Wong (2019) | 2019 | Journal | Review | Experimental | Human | N-linked and O-linked | - | - | - | 0 | 0 | 1 | 4 | 5 | ||
| 31 | Kellman & Lewis (2021) | 2020 | Journal | Solution | Experimental | Human | Glycan (including N) | - | - | - | 1 | 1 | 4 | 6 | |||
| 32 | Huang et al. (2021) | 2021 | Journal | solution | Computational | Not Mention | Glycosylation (O-linked) | Sequence feature | RF | OGP-Based | 1 | 2 | 1 | 1 | 0 | 4 | 9 |
| 33 | Shek, Kotidis & Betenbaugh (2021) | 2021 | Journal | Review | Computational | Not Mention | Glycosylation | - | Provided | Provided | 1 | 2 | 1 | 0 | 0 | 4 | 8 |
| 34 | Mondragon-Shem et al. (2020) | 2020 | Journal | solution | Hybrid | Human | N-linked | MS | - | Existing Tool | 1 | 1 | 0 | 1 | 1 | 4 | 8 |
| 35 | Wilson et al. (2021) | 2021 | Journal | solution | Experimental | Human | Glycosylation (including N) | - | - | - | 0 | 1 | 0 | 1 | 1 | 2 | 5 |
| 36 | Zhang et al. (2021b) | 2021 | Journal | solution | Computational | Mammalian | N-linked | Unknown Parameter and Structure | Baysen Network | - | 0 | 1 | 1 | 0 | 1 | 4 | 7 |
| 37 | Hua11 (2019) | 2019 | Conference | Solution | Computational | Human | Protein Prediction | Frequency Feature of AA and EH Method | SVM and NN | PPSNN | 1 | 2 | 1 | 0 | 1 | 0 | 5 |
| 38 | Zhao et al. (2020) | 2020 | Journal | Solution | Experimental | Human | N-linked | - | MS | - | 0 | 0 | 1 | 0 | 1 | 4 | 6 |
| 39 | Wang et al. (2017) | 2017 | Journal | solution | Hybrid | human | N-linked | CfsSubSetEval | SVM | - | 0 | 1 | 2 | 0 | 1 | 4 | 8 |
| 40 | Badgett et al. (2018) | 2018 | Journal | Solution | Experimental | Human | N-linked | - | MS | - | 0 | 0 | 1 | 1 | 1 | 2 | 5 |
| 41 | Suga, Nagae & Yamaguchi (2018) | 2018 | Journal | Solution | Hybrid | Human | N-linked | Structural Feature | Maturation | - | 0 | 1 | 1 | 1 | 1 | 4 | 8 |
| 42 | Bao et al. (2019) | 2019 | Journal | Solution | Computational | Human | Glycosylation and Phosphorylation | Membrane Buried, Confrontational and average Flexible Indices | NN+ELM+SVM | CMSENN | 1 | 2 | 1 | 0 | 1 | 3 | 8 |
| 43 | de Souza et al. (2019) | 2019 | Journal | Solution | Experimental | human | N-linked | - | MS | - | 0 | 0 | 0 | 1 | 1 | 4 | 6 |
| 44 | Jiang et al. (2018) | 2018 | Journal | Solution | Computational | Not Mention | Glycosylation (O-linked) | KPCA and FUS | Rotation Forest | OGLYCPred | 1 | 2 | 1 | 1 | 4 | 9 | |
| 45 | Kuo-Chen (2019) | 2019 | Journal | Review | Computational | Human | Non-Glycosylation | KC Chou’s 5 step | - | Povided | 1 | 1 | 0 | 1 | 1 | 1 | 5 |
| 46 | Dimeglio et al. (2020) | 2020 | Journal | Solution | Hybrid | Human | N-linked | Statistical Moment | ANN | THETA Model | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 47 | Dobson, Zeke & Tusnády (2021) | 2021 | Journal | Solution | Hybrid | Human | Protein Traffic membrane (N and O) | Topology and Putative SLiMs | CNN with Adam | PolarportPred | 1 | 1 | 1 | 0 | 1 | 4 | 8 |
| 48 | Kumar et al. (2020) | 2020 | Conference | Solution | Hybrid | Human | PTM | Psycho-Chemical, structural and PTM | ML | - | 0 | 2 | 1 | 1 | 1 | 0 | 5 |
| 49 | Ilyas et al. (2019) | 2019 | Journal | solution | Computational | Human | PTM | Chou’s 5-steps | ANN | - | 0 | 2 | 1 | 1 | 1 | 2 | 7 |
| 50 | Yang & Han (2017) | 2017 | Journal | Solution | Computational | Mammalian | Glycosylation (O-linked) | Protein factor base Features | KNN | - | 0 | 2 | 1 | 0 | 1 | 2 | 6 |
| 51 | Magaret et al. (2019) | 2019 | Journal | Solution | Hybrid | Not Mention | N-linked | Sequences | RM, Super Learner and Glmnet | - | 0 | 1 | 1 | 1 | 0 | 4 | 7 |
| 52 | Sugár et al. (2021) | 2021 | Journal | Solution | Experimental | Human | N-linked | - | RanoLC-MS | - | 1 | 1 | 0 | 1 | 1 | 4 | 8 |
| 53 | Campbell (2017) | 2017 | Journal | review | Hybrid | Human | Glycosylation | Partial Mentioned | Partial Mentioned | - | 1 | 1 | 1 | 0 | 1 | 1 | 5 |
| 54 | Jia, Zuo & Zou (2018) | 2018 | Journal | Solution | Computational | Not Mention | Glycosylation (O-linked) | FUS and KPCA | KNN,RM,SVM and NB, SVM outperform | rgb 0.141, 0.125, 0.129O-GlcNAcPRED-II | 1 | 2 | 1 | 1 | 0 | 4 | 9 |
| 55 | Ferreira et al. (2021) | 2021 | Journal | Solution | Experimental | Human | N-linked | - | MS | - | 0 | 1 | 0 | 0 | 1 | 4 | 6 |
| 56 | Ye & Vakhrushev (2021) | 2021 | Journal | Solution | Experimental | Human | Glycosylation | - | MS | - | 0 | 0 | 0 | 0 | 1 | 4 | 5 |
| 57 | Bojar et al. (2021a) | 2021 | bioRxiv | Solution | Hybrid | Human | N-linked | Sequence | ML | - | 0 | 2 | 1 | 1 | 1 | 0 | 5 |
| 58 | Desaire, Patabandige & Hua (2021) | 2021 | Journal | Solution | Hybrid | Not Mention | Glycosylation | MS | SVM | - | 0 | 1 | 1 | 1 | 0 | 4 | 7 |
| 59 | Chen et al. (2021) | 2021 | Journal | Solution | Computational | Human | PTM | Binary Encoding,AAC,EAAC and Dipeptide | Deep Learning | CNNrgb | 1 | 2 | 1 | 1 | 1 | 4 | 10 |
| 60 | Zou et al. (2017) | 2017 | Conference | Solution | Computational | Human | Glycosylation (O-linked) | Vector Word | SVM | GLycoCell | 1 | 2 | 1 | 1 | 1 | 0 | 6 |
| 61 | Perpetuo et al. (2021) | 2021 | Journal | Solution | Computational | Human | PTM | Sequences | AI | - | 0 | 2 | 1 | 0 | 1 | 3 | 7 |
| 62 | Li et al. (2020) | 2020 | Journal | Solution | Computational | Not Mention | Protein | AAC, PseAAC,NC, PseKNC | adaboost and random forest | PPAI | 1 | 2 | 1 | 0 | 0 | 4 | 8 |
| 63 | Lei, Tang & Du (2017) | 2017 | Journal | Solution | Hybrid | Not Mention | PTM (S-sulfenylated) | Psysciochemical and Clustring Method | Ensemble Classifier | - | 0 | 1 | 2 | 1 | 0 | 1 | 5 |
| 64 | Murad et al. (2021) | 2021 | bioRxiv | Solution | Computational | Not Mention | PTM (Ubiquitination) | Statistical Moment | Random Forest | UBISites-SRF | 1 | 2 | 1 | 0 | 1 | 0 | 5 |
| 65 | Qiu et al. (2018) | 2018 | Journal | Solution | Computational | Not Mention | PTM (Lipoylation) | Biprofile Bayes Encoding | SVM | LipoPred | 1 | 1 | 1 | 0 | 0 | 3 | 6 |
| 66 | Yang et al. (2019) | 2019 | Journal | Solution | Hybrid | Human | PTM | SNP | - | Awesome | 1 | 1 | 1 | 1 | 1 | 4 | 9 |
| 67 | Liu et al. (2021) | 2021 | Journal | Solution | Computational | Human | PTM | UbiSite-XGBoost | Extreme gradient boosting classifier | UbiSite=XGBoost | 1 | 2 | 1 | 1 | 1 | 3 | 9 |
| 68 | Ruiz-Blanco et al. (2017) | 2017 | Journal | Solution | Computational | Human | N-linked | ProDCal | Jrip Classifier | Sequon | 0 | 2 | 1 | 1 | 1 | 4 | 9 |
| 69 | Kumari, Kumar & Kumar (2018) | 2018 | Journal | Solution | Computational | Human | Palmitoylation | PSSM | SVM | RAREPalm | 1 | 2 | 1 | 0 | 1 | 3 | 8 |
| 70 | Huang & Li (2018) | 2018 | Journal | Solution | Computational | Human | PTM | Sequence, Structure | KNN | - | 0 | 2 | 1 | 1 | 1 | 4 | 9 |
Assessment and discussion
In this section, the research questions have been analyzed based on 70 primary studies.
Assessment of q1:
Which are the relevant publishing channel for N-linked glycosylation research? Which channel type and geographical area target this research?
To find the relevant publishing channel, channel type and geographical aspects for the N-linked glycosylation sites requires the meta information. To achieve this purpose, channel type, publishing year and demographical distribution is presented for the analysis of selected studies.
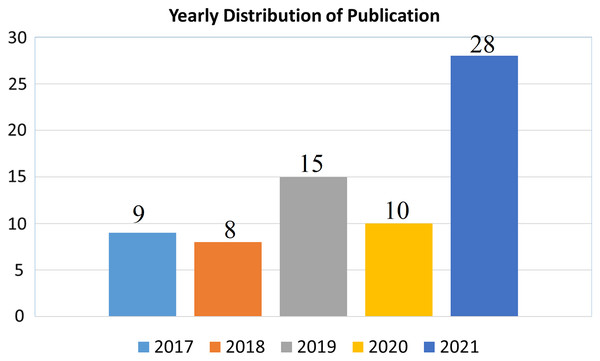
The importance of selected topic can be evaluated from the yearly publication on the relevant domain. The 28 out of 70 articles has been published in 2021 which also of 40% of selected article as shown in Fig. 4.
Figure 4: Year wise distribution of publication.
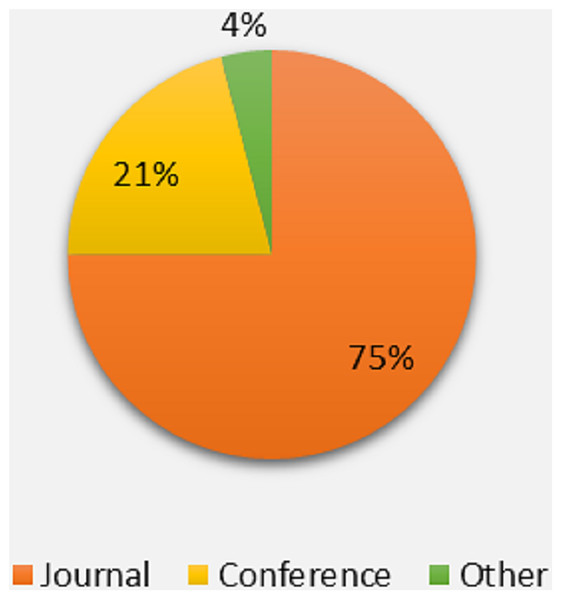
It is clear from Fig. 5 that the maximum portion of studies belong to the recognized journal followed by international conferences.
Figure 5: Percentage of publication channel.
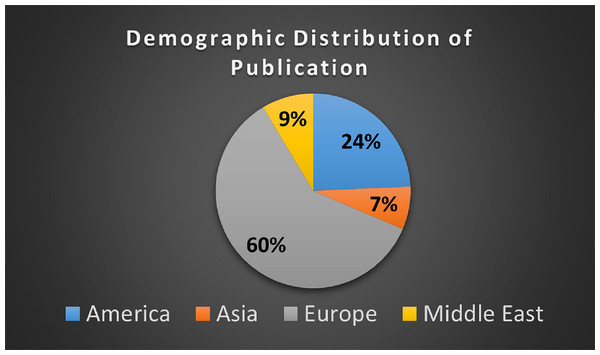
It is observed, 42 out of 70 studies have been published in the different regions of the Europe as shown in Fig. 6.
Figure 6: Demographical distribution of publication.
Quality assessment score for each finalized study awarded according to defined criteria in quality assessment score section, shown in Table 7. It is clearly observed that only studies qualifying minimum threshold are listed. The article published in Q1 quality journal achieve highest score, it will help researchers to find the relevant publishing venues for the N-linked and other glycosylation site prediction studies. Almost 50% of the studies achieve eight score or above which shows the relevancy of the selected studies through developed query string.
The overall classification result and QA studies have presented in of Table 6. The finalized articles have classified based on seven parameters: research type (solution proposed or review article), empirical type (computational approach, experimental approach based on biological studies or hybrid approach based on computational and biological study), glycosylation type, specie type, method (used for feature extraction), Algorithm (used to train predictive model) and tool (developed for prediction).
Furthermore, the sources of finalized studies, and total number/percentage of studies per publication source mentioned in Table 8.
| Publication source | Reference | Count | %age |
|---|---|---|---|
| Amino Acids | Ruiz-Blanco et al. (2017) | 1 | 1 |
| Analytical and Bioanalytical Chemistry | Desaire, Patabandige & Hua (2021) | 1 | 1 |
| Bioinformatics | Taherzadeh et al. (2019), Jiang et al. (2018), Dimeglio et al. (2020), Dobson, Zeke & Tusnády (2021), Jia, Zuo & Zou (2018) | 5 | 7 |
| bioRxiv | Carpenter et al. (2022), Lundstrøm et al. (2022), Adolf-Bryfogle et al. (2021), Bojar et al. (2021a), Murad et al. (2021) | 5 | 7 |
| Biotechnology and Bioengineering | Zhang et al. (2021b) | 1 | 1 |
| BMC Bioinformatics | Li et al. (2019), Coff et al. (2020), Li et al. (2020) | 3 | 4 |
| Briefings in Bioinformatics | Xiang, Zou & Zhao (2021) | 2 | 3 |
| Briefings in Functional Genomics | He, Wei & Zou (2019) | 1 | 1 |
| Cell Host Microbe | Bojar et al. (2021b) | 1 | 1 |
| Cell Reports | Burkholz, Quackenbush & Bojar (2021) | 1 | 1 |
| Chemometrics and Intelligent Laboratory Systems | Bao et al. (2019), Qiu et al. (2018) | 2 | 3 |
| Computational and Structural Biotechnology Journal | Audagnotto & Dal Peraro (2017) | 1 | 1 |
| Computational Biology and Chemistry | Le, Sandag & Ou (2018), Yang & Han (2017) | 2 | 3 |
| Computers Chemical Engineering | Antonakoudis et al. (2021) | 1 | 1 |
| Computers in Biology and Medicine | Tran, Pham & Ou (2021) | 1 | 1 |
| Current Bioinformatics | Alkuhlani et al. (2021), Huang & Li (2018) | 2 | 3 |
| Current Genomics | Ilyas et al. (2019) | 1 | 1 |
| Current Opinion in Chemical Engineering | Shek, Kotidis & Betenbaugh (2021) | 1 | 1 |
| Environmental Microbiology | Zhang et al. (2021c) | 1 | 1 |
| Expert Review of Proteomics | Perpetuo et al. (2021) | 1 | 1 |
| Frontiers in Endocrinology | Zhang et al. (2021c) | 1 | 1 |
| Fuzzy Systems and Data Mining | Hua11 (2019) | 1 | 1 |
| Genomics, Proteomics Bioinformatics | Huang et al. (2021), Ferreira et al. (2021) | 2 | 3 |
| Glycobiology | Thomès, Burkholz & Bojar (2021), Park et al. (2019), Suga, Nagae & Yamaguchi (2018) | 3 | 4 |
| IEEE Access | Chien et al. (2020) | 1 | 1 |
| IEEE International Conference on Machine Learning and Applied Network Technologies (ICMLANT) | Kumar et al. (2020) | 1 | 1 |
| International Conference of Pioneering Computer Scientists, Engineers and Educators. Springer, Singapore, | Zou et al. (2017) | 1 | 1 |
| Journal of Bimolecular Techniques | Badgett et al. (2018) | 1 | 1 |
| Journal of Computational Biology | Kumari, Kumar & Kumar (2018) | 1 | 1 |
| Journal of Molecular Graphics and Modelling | Liu et al. (2021) | 1 | 1 |
| Journal of Proteomics | Zhao et al. (2020), de Souza et al. (2019) | 2 | 3 |
| Journal of the American Chemical Society | Huang et al. (2017), Krasnova & Wong (2019) | 2 | 3 |
| Letters in Organic Chemistry | Lei, Tang & Du (2017) | 1 | 1 |
| Mathematical Bioscience | Liu et al. (2019) | 1 | 1 |
| Metabolic Engineering Communications | Kotidis & Kontoravdi (2020) | 1 | 1 |
| Molecular Cellular Proteomic | Ye & Vakhrushev (2021) | 1 | 1 |
| Nature Communications | Wang et al. (2017) | 1 | 1 |
| Nucleic Acids Research | Yang et al. (2019) | 1 | 1 |
| PLoS Computational Biology | Magaret et al. (2019) | 1 | 1 |
| PLOS ONE | Akmal, Rasool & Khan (2017) | 1 | 1 |
| Processes | Sha et al. (2019) | 1 | 1 |
| Scientific Reports | Pitti et al. (2019), Hwang et al. (2020), Mondragon-Shem et al. (2020), Sugár et al. (2021) | 4 | 6 |
| Symmetry | Naseer et al. (2021b) | 1 | 1 |
| The American Journal of Human Genetics | Wilson et al. (2021) | 1 | 1 |
| Trends Artifi. Intell | Kuo-Chen (2019) | 1 | 1 |
| Trends in Biochemical Science | Kellman & Lewis (2021) | 1 | 1 |
| Trends in Glycoscience and Glycotechnolog | Campbell (2017) | 1 | 1 |
| Trends in Microbiology | Lee et al. (2021) | 1 | 1 |
Assessment of q2:
Which are the exiting prediction model (tool) for the identification of N-linked Glycosylation sites and for which kind of species these sites are identified?
The available tool to identify the N-Linked glycosylation sites and for which kind species it can identify the relevant site is the parameter of this study. There is hierarchy of N-Linked Glycan to PTM. Where PTM is classified into various type and Glycosylation in one of them and glycosylation is further classified into five group and N-linked is one of them.
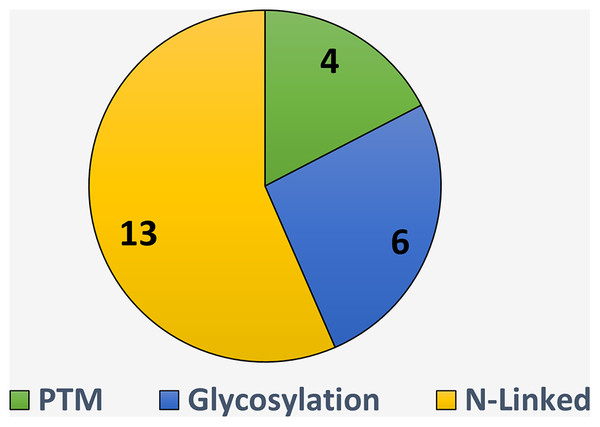
The summarized detail of eight is represented in Fig. 7. It is observed, there are 13 studies including (Chien et al., 2020; Taherzadeh et al., 2019; Liu et al., 2019; Li et al., 2019; Thomès, Burkholz & Bojar, 2021; Pitti et al., 2019; Adolf-Bryfogle et al., 2021; Park et al., 2019; He, Wei & Zou, 2019; Audagnotto & Dal Peraro, 2017; Mondragon-Shem et al., 2020; Dimeglio et al., 2020; Ruiz-Blanco et al., 2017) which have developed the tool specific to the N-Linked site identifications, few studied developed tool for glycosylation sites identification irrespective of the specific type including (Bojar et al., 2021b; Carpenter et al., 2022; Lundstrøm et al., 2022; Burkholz, Quackenbush & Bojar, 2021; Coff et al., 2020; Shek, Kotidis & Betenbaugh, 2021) and some authors (Le, Sandag & Ou, 2018; Liu et al., 2021; Yang et al., 2019; Campbell, 2017) develop tool without mentioning the type of PTM. These all tools have list down in the Table 9.
Figure 7: Tool available for N-linked sites identification.
| Ref. | P.Year | Species | Tool | Finding |
|---|---|---|---|---|
| Bojar et al. (2021b) | 2021 | Eukaryote | SweetOrigin | The model develop to identify Glycosylation sites using Hybrid approach on Eukaryotes. |
| Thomès, Burkholz & Bojar (2021) | 2021 | Animal | GlycoWork | The computational model used to identify both N and O-linked in Animal. |
| Carpenter et al. (2022) | 2021 | Not mention | GlyNet | The computational model used to identify glycosylation protein sequences. |
| Lundstrøm et al. (2022) | 2021 | Human | LectinOracle | The computational model used to identify glycosylation protein sequences for human. |
| Burkholz, Quackenbush & Bojar (2021) | 2021 | Human | SweetNet | The computational model used to identify glycosylation protein sequences for human. |
| Adolf-Bryfogle et al. (2021) | 2021 | Not mention | Rosetta Carbohydrate Framework | The computational model used to identify N-linked sites and species are not mentioned. |
| Shek, Kotidis & Betenbaugh (2021) | 2021 | Not mention | Provided | The computational model used to identify glycosylation sites and species are not mentioned. |
| Chen et al. (2021) | 2021 | Human | CNNrgb | The computational model used to identify PTM sites for human protein. |
| Liu et al. (2021) | 2021 | Human | UbiSite = XGBoost | The computational model used to identify PTM sites for human protein. |
| Chien et al. (2020) | 2020 | Human and Mouse | N-GlycoGo | The computational model used to identify N-Linked sites for human and mouse protein sequences. |
| Coff et al. (2020) | 2020 | Human and avian | CCARL | The computational model used to identify glycosylation sites for human and avian protein sequences. |
| Mondragon-Shem et al. (2020) | 2020 | Human | Existing Tool | The hybrid model consists of both experimental and computational approach to develop N-linked site identification on human protein |
| Dimeglio et al. (2020) | 2020 | Human | THETA Model | The hybrid model consists of both experimental and computational approach to develop N-linked site identification on human protein |
| Taherzadeh et al. (2019) | 2019 | Human and Mouse | Sprint-Gly | The computational model used to identify both N and O-linked in human and Mouse. |
| Liu et al. (2019) | 2019 | Human | NetGlyco (Exiting) | The computational model used to identify N-linked sites in human. |
| Li et al. (2019) | 2019 | Human | GlycoMine_PU | The computational model used to identify N, O and C-linked in human. |
| Pitti et al. (2019) | 2019 | Human | NGlyDE | The computational model used to identify N-linked in human. |
| Park et al. (2019) | 2019 | Human | Glycan Reader and Modeler | The computational model used to identify both N and O-linked in human. |
| He, Wei & Zou (2019) | 2019 | Not mention | Provided | The computational model used to identify N-linked sites while specie is not mentioned. |
| Yang et al. (2019) | 2019 | Human | Awesome | The hybrid approach develop to identify PTM sites for human. |
| Le, Sandag & Ou (2018) | 2018 | Human | PTM Transporter | The computational approach developed PTM sites including N-Linked sites for human. |
| Audagnotto & Dal Peraro (2017) | 2017 | Not mention | Provided | The computational model used to identify N-linked sites while specie type is missing. |
| Ruiz-Blanco et al. (2017) | 2017 | Human | Sequon | Computational method to identify N-Linked sites for human. |
It is important to specify for which kind of species these tools will be operating, therefore to achieve this purpose the information is also extracted from the selected studies. Some authors (He, Wei & Zou, 2019; Audagnotto & Dal Peraro, 2017; Shek, Kotidis & Betenbaugh, 2021; Carpenter et al., 2022) did not mention the organism type while other mentioned it and it is observed most of them use human data for site identification as mention in Table 9.
Assessment of q3:
Which algorithm or method are used to construct N-Linked feature vector?
The data is the major component to develop any machine learning model (Mahmood et al., 2020; Naseer et al., 2020a, 2020b; Khan et al., 2020b). In bioinformatics, there are two major sources of data on which model can be developed, one is existing repositories such as UniProt (protein repository), GenBank (nucleotide sequence) etc. and other is experimental data which obtain from specific biological experiments. The dataset obtained from any source needs preprocessing to construct the feature vector. The more accurate feature helps to develop efficient model (Barukab et al., 2019; Butt & Khan, 2019; Hussain, Rasool & Khan, 2020; Shah & Khan, 2020). For this purpose, feature method used to predict the N-Linked sites in the selected articles have taken as a parameter of this study.
Most of the authors used the computational feature extraction approach while few used the experimental data obtained from mass spectrometry, human plasma and psycho-chemical method as mentioned in Table 10. It is observed, mostly researcher (Akmal, Rasool & Khan, 2017; Chien et al., 2020; Taherzadeh et al., 2019; Liu et al., 2019; Li et al., 2019; Bojar et al., 2021b; Lundstrøm et al., 2022; Park et al., 2019; Le, Sandag & Ou, 2018; Suga, Nagae & Yamaguchi, 2018; Dimeglio et al., 2020; Magaret et al., 2019; Kumar & Gilula, 1986; Perpetuo et al., 2021; Huang & Li, 2018) used the statistical moment method based on combination of protein sequence, structure and functions along with some other parameters like position relevance of sequences using the protein dataset to construct the feature matrix. The other computational method used to construct features selected article are word embedding vector technique, UbiSite-XGBoost, Similarity voting, CfsSubSetEval, Kernel Density Estimate, correlation subset and graph method as mentioned in Table 10.
| Ref. | Glyotype | Method for feature | Finding |
|---|---|---|---|
| Tran, Pham & Ou (2021) | N-Linked | Word embedding Vector Technique | Word embedding technique to efficiently predict N-linked glycosylation sites in ion channels. |
| Adolf-Bryfogle et al. (2021) | N-Linked | KDE | Kernel Density Estimation based feature extracted. |
| Antonakoudis et al. (2021) | N-Linked | Stoichiometric | Hybrid method that used the experimental data using stoichiometric. |
| Zhang et al. (2021b) | N-Linked | Unknown Parameter and Structure | Protein structure feature and some undefined features used to construct feature vector. |
| Bojar et al. (2021a) | N-Linked | Sequence | Sequence based features computed. |
| Chien et al. (2020) | N-Linked | Sequence, Structure and Function feature | sequence, structure and function base feature set of human and mouse used to predict site on imbalance dataset. |
| Hwang et al. (2020) | N-Linked | IQ-GPA human plasma protein | IQ-GPA procedure was used to obtain data from human plasma. |
| Mondragon-Shem et al. (2020) | N-Linked | MS | Hybrid method based on Mass Spectrometry used data used for training. |
| Liu et al. (2019) | N-Linked | Sequence | Sequence based protein sequences have computed. |
| Pitti et al. (2019) | N-Linked | Similarity voting and Gap Peptide | Similarity Voting method and gap peptide method used to construct features. |
| Magaret et al. (2019) | N-Linked | Sequences | Sequence based protein sequences have computed. |
| Suga, Nagae & Yamaguchi (2018) | N-Linked | Structural Feature | Structure based protein sequences have computed. |
| Akmal, Rasool & Khan (2017) | N-Linked | Position relative and Statistical Moments | Position relative features and statistical moment based features have computed. |
| Wang et al. (2017) | N-Linked | CfsSubSetEval | Patients with different drug responses |
| Ruiz-Blanco et al. (2017) | N-Linked | ProDCal | ProtDCal method used to get protein features. |
| Dimeglio et al. (2020) | N-Linked | Statistical Moment | Statistical Moments computed to construct feature vector. |
| Li et al. (2019) | N-Linked (and C/O-Linked) | Sequence and Structure Feature | Sequence and structure based protein sequences have computed. |
| Taherzadeh et al. (2019) | N-Linked and O-Linked | Sequence and Structure | Sequence and structure based protein sequences have computed. |
| Park et al. (2019) | N-Linked and O-Linked | Sequence and Structure | Sequence and structure based protein sequences have computed. |
| Bojar et al. (2021b) | Glycosylation | Sequence feature | Develop models for glycans that are trained on a curated dataset of 19,299 unique glycans and used sequence based features. |
| Carpenter et al. (2022) | Glycosylation | Fingerprint Encoding | Feature vector based on Fingerprint encoding method for Predicting Protein-Glycan Interaction |
| Lundstrøm et al. (2022) | Glycosylation | Protein-Glycan Sequence Feature | The sequence feature of combined protein and glycan are used to extract feature vector based on sequence features. |
| Burkholz, Quackenbush & Bojar (2021) | Glycosylation | Graph and Statistical feature | Graph algorithm and statistical moments are used to construct feature matrix for glycan. |
| Desaire, Patabandige & Hua (2021) | Glycosylation | MS | Hybrid method based on Mass Spectrometry used data used for training. |
| Coff et al. (2020) | Glycosylation | Frequent Subtree mining and mRMR | frequent subtree mining and mRMR used for feature vector construction. |
| Perpetuo et al. (2021) | PTM | Sequences | Sequence based features used for feature vector construction. |
| Liu et al. (2021) | PTM | UbiSite-XGBoost | Pseudo ACC, K-spaced Acid Pair, Adapted Normal Distribution bi-profile Bayes, AA Index, Encoding Based Group Weight, LASSO, SMOTE and eXtreme Gradient Boosting features methods are used. |
| Kumar et al. (2020) | PTM | Psycho-Chemical, structural and PTM | Psycho-Chemical, structure moment of protein and PTM sequence features were used. |
| Ilyas et al. (2019) | PTM | Chou’s 5-steps | Chou’s 5-steps based feature vector was used. |
| Yang et al. (2019) | PTM | SNP | Single Nucleotide Polymorphism approach used to compute features. |
| Huang & Li (2018) | PTM | Sequence, Structure | Sequence and Structure based protein sequences have computed. |
| Chen et al. (2021) | PTM | Binary Encoding, AAC, EAAC and Dipeptide | Various features have extracted including binary encoding, Amino Acid Composition, Enhanced AAC and Dipeptide. |
| Le, Sandag & Ou (2018) | PTM (including N Linked) | Statistical Moment and F score | Statistical moment used and then F-Score was computed |
Assessment of q4:
Which algorithm or method are used to train N-Linked computation model?
The choice of algorithm to train any predictive model is most important factor which impact the performance of any model (Butt & Khan, 2019; Hussain, Rasool & Khan, 2020; Malebary & Khan, 2021). Therefore, it is required to know which type of algorithm are being used to develop the N-linked prediction model. For this purpose, algorithm used for training models in the selected article has been noted as the parameter of this review article as mentioned in Table 11.
| Ref. | Model training algorithm | PTM type | Finding |
|---|---|---|---|
| Akmal, Rasool & Khan (2017) | ANN/Back propagation | N-Linked | Prediction of N-linked glycosylation sites using position relative features and statistical moments through multilayered ANN using back propagation approach. |
| Chien et al. (2020) | XGBOOST | N-Linked | Extreme Gradient Boost method was used to predict site on imbalance dataset. |
| Tran, Pham & Ou (2021) | RF, KNN, SVM and XGBoost | N-Linked | Various classifiers were used for prediction including Random Forest, K-Nearest Neighbor, Support Vector Machine and XGBoost but RM outperform. |
| Liu et al. (2019) | ANN | N-Linked | Artificial Neural Network algorithm used to identify N-linked site in Influenza virus using existing model on dataset. |
| Pitti et al. (2019) | SVM | N-Linked | N-GlyDE: a two-stage N-linked glycosylation site prediction incorporating gapped dipeptides and pattern-based encoding using SVM after collecting feature vector through two stages. |
| Kotidis & Kontoravdi (2020) | ANN/Kinetic Model | N-Linked | artificial neural networks and Kinetic model used for predicting protein glycosylation. |
| Adolf-Bryfogle et al. (2021) | Glycan Tree Modler | N-Linked | prediction based on Tree method. |
| Sha et al. (2019) | Kinetic | N-Linked | a two-component modeling framework integrating FBA and glycosylation kinetic model was used for prediction. |
| Antonakoudis et al. (2021) | ANN | N-Linked | predict N linked sites using features computed by stoichiometric and then train model using ANN with forward propagation. |
| Hwang et al. (2020) | DNN | N-Linked | N linked site using DNN which later used to classify fucosylation |
| Zhang et al. (2021b) | Baysen Network | N-Linked | Probabilistic model by Bayesian network for the prediction of antibody glycosylation in perfusion and fed-batch cell cultures |
| Wang et al. (2017) | SVM | N-Linked | Drug responses identified using SVM method. |
| Dimeglio et al. (2020) | ANN | N-Linked | New genotypic approach for predicting HIV-1 CRF02-AG using ANN |
| Magaret et al. (2019) | RM, Super Learner and Glmnet | N-Linked | Protein sequence and biological data used to identify N-linked sites using super learner algorithm. |
| Bojar et al. (2021a) | ML | N-Linked | Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities |
| Ruiz-Blanco et al. (2017) | Jrip Classifier | N-Linked | Novel “extended sequons” of human N-glycosylation sites improve the precision of qualitative predictions: an alignment-free study of pattern recognition using ProtDCal protein features. |
| Park et al. (2019) | Clustering | N-O Linked | CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. |
| Dobson, Zeke & Tusnády (2021) | CNN with Adam | N-O Linked | Novel mechanism to collect dataset using polarization and then train on CNN model. |
| Taherzadeh et al. (2019) | Deep ANN and SVM | N-O Linked | Predicting N-and O-linked glycosylation sites of human and mouse proteins by using sequence and predicted structural properties through DNN and SVM |
| Li et al. (2019) | PA2DE using AlphaMax | N-C-O Linked | Positive-unlabeled data set used to predict sites using AlphaMax algorithm |
| Bojar et al. (2021b) | Recurrent NN (LSTM) | Glycosylation | develop deep-learning using Recurrent NN models used for glycans that are trained on a curated dataset of 19,299 unique glycans and can be used to study and predict glycan functions. |
| Carpenter et al. (2022) | MNN (ADAM) | Glycosylation | A Multi-Task Neural Network using ADAM algorithm used for Predicting Protein-Glycan Interaction |
| Lundstrøm et al. (2022) | Graph CNN | Glycosylation | LectinOracle, a model combining transformer-based representations for proteins and graph convolutional neural networks for glycans to predict their interaction. |
| Burkholz, Quackenbush & Bojar (2021) | Graph NN | Glycosylation | sing graph convolutional neural networks to learn a representation for glycans. |
| Coff et al. (2020) | Regression Classifier | Glycosylation | frequent subtree mining and mRMR used for feature selection then train on regression classifier for glycan motifs. |
| Desaire, Patabandige & Hua (2021) | SVM | Glycosylation | The local-balanced model for improved machine learning outcomes on mass spectrometry data sets and other instrumental data |
| Le, Sandag & Ou (2018) | RBF Network | PTM | prediction of transport protein (including N linked) into three classes and six families using RBF Network. |
| Chen et al. (2021) | Deep Learning | PTM | nhKcr: a new bioinformatics tool for predicting crotonylation sites on human non histone proteins based on deep learning |
| Lei, Tang & Du (2017) | Ensemble Classifier | PTM | Predicting S-sulfenylation sites using physicochemical properties difference and ensemble classifier. |
| Murad et al. (2021) | Random Forest | PTM | Ubiquitination Sites Prediction Using Statistical Moment with Random Forest Approach. |
| Kumar et al. (2020) | ML | PTM | Machine Learning techniques to identify potential drug targets for Anti-epileptic drugs |
| Perpetuo et al. (2021) | AI | PTM | artificial intelligence be used for peptidomics |
| Qiu et al. (2018) | SVM | PTM | Predicting protein lysine methylation sites by incorporating single-residue structural features into Chou’s pseudo components. |
| Liu et al. (2021) | Extreme gradient boosting classifier | PTM | Prediction of protein ubiquitination sites via multi-view features based on eXtreme gradient boosting classifier. |
| Huang & Li (2018) | KNN | PTM | Feature extractions for computationally predicting protein post-translational modifications |
It is observed from the selected articles that most of the authors (Akmal, Rasool & Khan, 2017; Taherzadeh et al., 2019; Liu et al., 2019; Lundstrøm et al., 2022; Burkholz, Quackenbush & Bojar, 2021; Kotidis & Kontoravdi, 2020; Antonakoudis et al., 2021; Hwang et al., 2020; Dimeglio et al., 2020; Dobson, Zeke & Tusnády, 2021; Ilyas et al., 2019; Chen et al., 2021) used the Artificial Neural Network (ANN) or the variant of ANN such as Deep ANN, Graph NN, Convolution NN and Recurrent NN. The second most used algorithm is Support Vector Machine (SVM) used by authors (Tran, Pham & Ou, 2021; Pitti et al., 2019; Wang et al., 2017; Desaire, Patabandige & Hua, 2021; Qiu et al., 2018) and remaining authors used Random Forest, XGBOOST, Baysen Network, Regression Classifier, Radial Base Function and some used customized method as mention in Table 11.
Assessment of q5:
How effective are the existing model to predict the N-Linked sites?
The result comparisons are used to present the performance to various based on which conclusion can be drawn with respect specific dimension. In this systematic review, the performance comparison of N-linked model in the selected articles has performed. The parameter used for the performance consists of (a) availability of data set. (b) accuracy metric (c) sensitivity metric. (d) specificity (e) availability of developed tool (f) comparison on independent data and the type of glycosylation as mentioned in Table 12. It is observed, most of the authors (Kotidis & Kontoravdi, 2020; Sha et al., 2019; Park et al., 2019; Antonakoudis et al., 2021; Zhang et al., 2021b; Wang et al., 2017; Kumar et al., 2020; Ilyas et al., 2019; Sugár et al., 2021; Bojar et al., 2021a; Perpetuo et al., 2021; Huang & Li, 2018) did not provide the results or they did not follow provided performance metrics in their research. The authors (Akmal, Rasool & Khan, 2017; Chien et al., 2020; Taherzadeh et al., 2019; Tran, Pham & Ou, 2021; Liu et al., 2019; Li et al., 2019; Pitti et al., 2019; Hwang et al., 2020; Le, Sandag & Ou, 2018; Dimeglio et al., 2020; Magaret et al., 2019; Ruiz-Blanco et al., 2017) mentioned most of the performance metrics specific to N-Linked sites identification and out of these, authors Chien et al. (2020) and Hwang et al. (2020) has not provide the data set on which experiments have performed.
| Ref. | Glycosylation type | Result comparison on | Tool | Dataset | ACC (%) | SN (%) | SP (%) | Finding |
|---|---|---|---|---|---|---|---|---|
| Akmal, Rasool & Khan (2017) | N-Linked | Yes | No | Yes | 99.9 | 99.8 | 99.9 | Detail Comparison has perform and also present metrics but tool is not available |
| Chien et al. (2020) | N-Linked | Yes | Yes | No | 84.7 | 82.8 | 84.8 | Detail Comparison has performed and also present metrics. But data set is not available |
| Taherzadeh et al. (2019) | N-Linked and O-Linked | Yes | Yes | Yes | 97.5 | 98 | – | Detail Comparison has performed and also present metrics. |
| Tran, Pham & Ou (2021) | N-Linked | Yes | No | Yes | 93.4 | 98.6 | 92.8 | Detail comparison has perform and also present metrics but tool is not available |
| Liu et al. (2019) | N-Linked | No | Yes | Yes | 50 | – | – | Not compare the result properly. |
| Li et al. (2019) | N-Linked (and C/O-Linked) | Yes | Yes | Yes | 88.6 | – | – | Detailed comparison has performed but SN and SP not computed |
| Bojar et al. (2021b) | Glycosylation | No | No | Yes | 75 | – | – | Result not compare properly and also missing few metrics. |
| Carpenter et al. (2022) | Glycosylation | No | Yes | No | 75 | – | – | Tool is available but data set is missing and did not perform all performance metric |
| Pitti et al. (2019) | N-Linked | Yes | Yes | Yes | 74 | 49 | – | Detailed comparison has performed but SP. |
| Lundstrøm et al. (2022) | Glycosylation | No | No | Yes | 72 | – | – | Result are not performed properly as missing metrics and tool. |
| Burkholz, Quackenbush & Bojar (2021) | Glycosylation | No | Yes | Yes | 85 | – | – | Detailed comparison performed but missing few metrics |
| Kotidis & Kontoravdi (2020) | N-Linked | No | No | Yes | – | – | – | Did not specify results. |
| Sha et al. (2019) | N-Linked | No | No | Yes | – | – | – | Did not specify results. |
| Park et al. (2019) | N-Linked and O-Linked | No | Yes | No | – | – | – | Did not specify results. |
| Antonakoudis et al. (2021) | N-Linked | No | No | Yes | – | – | – | Did not specify results. |
| Hwang et al. (2020) | N-Linked | No | No | No | 99 | 100 | – | Achieved almost full accuracy but result comparison with independent data set, data set and tool is missing. |
| Coff et al. (2020) | Glycosylation | No | Yes | Yes | 89 | – | – | Achieve good result but comparison on independent data set is missing and glycosylation type is not specified. |
| Le, Sandag & Ou (2018) | PTM (including N Linked) | Yes | Yes | Yes | 92 | – | – | Detailed comparison has performed and achieved good results but not specify the PTM type. |
| Zhang et al. (2021b) | N-Linked | Yes | No | No | – | – | – | Did not specify results. |
| Wang et al. (2017) | N-Linked | Yes | No | No | – | – | – | Did not specify results. |
| Dimeglio et al. (2020) | N-Linked | Yes | Yes | Yes | 88 | 86 | 89 | Detailed comparison has performed and also achieved good results. |
| Kumar et al. (2020) | PTM | Yes | No | Yes | – | – | – | Did not specify results. |
| Ilyas et al. (2019) | PTM | Yes | No | Yes | – | – | – | Did not specify results. |
| Magaret et al. (2019) | N-Linked | Yes | No | Yes | 86 | 97 | 39 | Detailed comparison has performed and also achieved good results. |
| Sugár et al. (2021) | N-Linked | No | Yes | Yes | – | – | – | Did not specify results. |
| Bojar et al. (2021a) | N-Linked | No | No | Yes | – | – | – | Did not specify results. |
| Desaire, Patabandige & Hua (2021) | Glycosylation | No | No | Yes | 98 | – | – | Achieve good result but glycosylation type is not specified and missing few metrics |
| Chen et al. (2021) | PTM | Yes | Yes | Yes | 85 | 62 | 90 | Detailed comparison has performed and achieved good result, but it is generic for PTM as specific type was not mentioned |
| Perpetuo et al. (2021) | PTM | No | No | No | – | – | – | Did not specify results. |
| Liu et al. (2021) | PTM | No | Yes | Yes | 97 | – | – | Detailed comparison has performed and achieved good result, but SN and SP are missing |
| Ruiz-Blanco et al. (2017) | N-Linked | No | No | Yes | 99 | 82 | – | Detailed comparison has performed and achieved good result, but data set is missing. |
| Huang & Li (2018) | PTM | No | No | Yes | – | – | – | Did not specify results. |
Discussion and future direction
This section summarizes and discuss the detail of this systematic literature review regarding the identification of N-linked sites.
Taxonomy hierarchy
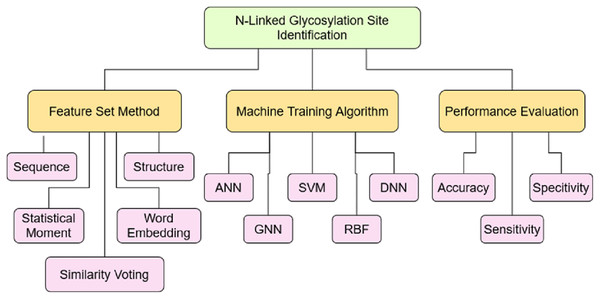
The objective of this study was to analyze the current progress to identify the N-linked glycosylation sites. To achieve this objective, a taxonomy has built based on the coding scheme as mentioned in Table 13 after critically analyzing 70 articles, selected through a systematic approach. The coding developed on the various aspects related to this study such as: Feature set construction method, machine model training algorithm and performance evaluation. These aspects are further divided into the sub-level showing the depth of each aspect and their role in the efficient identification of N-linked sites. The coding scheme helped to construct the taxonomy as shown Fig. 8 to further investigate domain and sub-domains identified through it.
Figure 8: Taxonomy of N-Linked site identification perspective.
General observation and future direction
Several possible observations can be made in the finding of this SLR based on the taxonomy as shown in Fig. 8. Various RQs were developed which plays a key factor in the identification of N-linked sites. The trends and finding can be observed while the identification of such sites. These include the following observation along with future direction.
(a) Feature set construction method The performance of computational model deeply depends on the quality of feature set extracted from the data set which later used for training the machine learning model (Saeed, Mahmood & Khan, 2018; Khan et al., 2019; Naseer et al., 2021a). The discriminating features helps the model to learn proficiently and then perform the right prediction. Therefore, it is significant to discover the techniques which extract the useful information from the dataset. The various methods have been used by authors to construct the feature set, the widely used are: protein sequence feature, protein structure feature, statistical moments, word embedding technique and similarity voting. The majority of the authors (Liu et al., 2019; Bojar et al., 2021b; Magaret et al., 2019; Bojar et al., 2021a) only used the sequence based information of protein to train the model. It has also observed, the authors (Akmal, Rasool & Khan, 2017; Taherzadeh et al., 2019; Li et al., 2019; Park et al., 2019; Murad et al., 2021) applied the combination of multiple features such as sequence, structural and statistical to construct feature vector. More than 50% of the research article selected in this study, which got 10 points based on quality assessment score used combination of various features as mentioned above. The new techniques adopted in recent research articles are word embedding vector, graph statistical feature along with similarity voting and Chou’s five step method. The researchers can use these feature extraction techniques to improve the performance of N-linked prediction model or any PTM site identification model.
(b) Machine training algorithm The most significant part of computational model after the feature extraction method is to develop the method to train the machine model (Hussain, Rasool & Khan, 2020; Barukab et al., 2022; Khan et al., 2020a). The performance of model impacted most by the technique used for training the machine. The appropriate learning algorithm along with fine feature extraction method, results highly adequate model that predicts the independent data with great accuracy. Therefore, the development of appropriate machine learning method is very much essential. The researchers proposed various methods to predict the N-linked sites accurately. The most widely used methods include: Artificial Neural Network (ANN), Support Vector Machine (SVM), Deep Neural Network (DNN), Graph Neural Network (GNN) and Radial Basis Function (RBF) Network. The research article published in Q1 journal according to the JCR, used the ANN (Akmal, Rasool & Khan, 2017; Liu et al., 2019; Dimeglio et al., 2020) widely along with SVM (Taherzadeh et al., 2019; Pitti et al., 2019) method. It has also been analysed the research article (Taherzadeh et al., 2019; Le, Sandag & Ou, 2018; Ruiz-Blanco et al., 2017) in which web server has provided and present the accuracy above 90% used the Jrip Classifier, DNN, SVM and RBF algorithm. The authors (Akmal, Rasool & Khan, 2017; Tran, Pham & Ou, 2021; Hwang et al., 2020; Magaret et al., 2019; Desaire, Patabandige & Hua, 2021) who proposed prediction model without providing the webserver and also have accuracy above 90% used ANN, SVM, DNN and RF algorithms. The researchers can use these algorithms to improve the performance of N-linked prediction model or any PTM site identification model.
(c) Performance evaluation Once the model has trained, it then validated on the independent data to evaluate the performance. There are various techniques to measure the validity of model, the most significant metrics to evaluate the performance are Accuracy metric, Sensitivity and Specificity metric. The sensitivity test measures the true positive accuracy of a model while specificity measures the true negative accuracy of the model. In this study, the performance has evaluated on aforementioned metrics. Around 40% of the authors have not validated their model on any of above mentioned performance metrics. Only 20% of the authors have performed each of the defined performance metrics. The predictive models in which PTM type is specialized to N-linked have better accuracy as compared to those in which PTM type is not specified or are the generalized ones. The highest accuracy of −99% was achieved by author Akmal, Rasool & Khan (2017) based on these evaluation criteria. It also presents the sensitivity and specificity measures of the model which were 99.8% and 99.9% respectively, but it did not provide the web server. The author Hwang et al. (2020) claims the accuracy of 99% along with the sensitivity of 100%, but did not provide the working tool, dataset, and result comparisons with other predictors. The most efficient predictive models with available web server are Sequon model Ruiz-Blanco et al. (2017) and Sprint-Gly model Taherzadeh et al. (2019) with the accuracy of 97.5% and 97% respectively. The Sequon model has trained on the human protein sequence only while Sprint-Gly is equally effective for both human and rat species. Therefore, Sprint-Gly considered to be a reliable model out of the currently available web servers.
Future direction
Bioinformatics is an emerging filed, there are lot of problems that needs the computational solution over the experimental. As it was mentioned earlier, the researchers have identified almost ∼ 200 types of PTM which plays key role in various biological functions. Apart from N-linked glycosylation, the other types of glycosylation such as O-linked and C-linked also play vital role in protein functioning and various drug discovery techniques. Therefore, it is the opportunity for the researchers, pharmaceutical and academia to develop the efficient computational model to solve the problem that needs better computational solution. Few of the existing problem that needs to be addressed are given below
(a) Identify the O-linked glycosylation sites for threonine and serine using ANN.
(b) How the performance of C-linked glycosylation can be enhanced through exiting neural network classifiers.
(c) Develop a comprehensive predictive model to classify the type of glycosylation.
(d) How effective are the exiting classifier to predict the other PTM sites?
Conclusion
The significance of N-linked glycosylation promotes the discovery of such sites using computational methods instead of experimental method due to its limitations. In this systematic study, existing information to identify such sites was studied which covered the possible challenges and their solutions through systematic method. The research articles, related to the keywords associated with N-linked glycosylation were evaluated through five major digital libraries. In the result of search query applied to digital libraries, more than 800 articles have found and after filtering process 70 article were remained for further analysis. The results show that approximately 75% of the articles were published in recognized journals and rest belong to top conferences. It was observed that more than 40% of articles were published in the American journal followed by the Middle East with 20%. Most of the selected studies focused on the feature construction method and training algorithm, but less focused on the performance evaluation criteria and development of tool or web server.
The major shortcomings of any SLR primarily are related to search strategy, poor classification, and inaccurate data extraction. In this SLR, these deficiencies were overcome by applying the search query on five major digital libraries to reduce biasness. The results of search queries were then filtered through well-defined inclusion/exclusion criteria.