Screening and affinity optimization of single domain antibody targeting the SARS-CoV-2 nucleocapsid protein
- Published
- Accepted
- Received
- Academic Editor
- Xin Zhang
- Subject Areas
- Biochemistry, Bioinformatics, Molecular Biology, Virology, COVID-19
- Keywords
- Phage display, Single domain antibody, SARS-CoV-2, Fusion protein
- Copyright
- © 2024 Yang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits using, remixing, and building upon the work non-commercially, as long as it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2024. Screening and affinity optimization of single domain antibody targeting the SARS-CoV-2 nucleocapsid protein. PeerJ 12:e17846 https://doi.org/10.7717/peerj.17846
Abstract
The coronavirus disease 2019 (COVID-19) pandemic, which caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), lead to a crisis with devastating disasters to global public economy and health. Several studies suggest that the SARS-CoV-2 nucleocapsid protein (N protein) is one of uppermost structural constituents of SARS-CoV-2 and is relatively conserved which could become a specific diagnostic marker. In this study, eight single domain antibodies recognized the N protein specifically which were named pN01–pN08 were screened using human phage display library. According to multiple sequence alignment and molecular docking analyses, the interaction mechanism between antibody and N protein was predicted. ELISA results indicated pN01–pN08 with high affinity to protein N. To improve their efficacy, two fusion proteins were prepared and their affinity was tested. These finding showed that fusion proteins had higher affinity than single domain antibodies and will be used as diagnosis for the pandemic of SARS-CoV-2.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, is a continuing global health crisis with over 6.9 million deaths worldwide as of July 2023 (World Health Organization, 2024). Severe acute respiratory syndrome corona-virus 2 (SARS-CoV-2) is the pathogen of COVID-19. Compared with other corona-virus categories, COVID-19 has the highest mortality because of its high transmission risk and variability (Han et al., 2023). The fourteenth meeting of the International Health Regulations (2005) Emergency Committee regarding the coronavirus disease (COVID-19) pandemic agreed that COVID-19 was still considered an extremely dangerous infectious disease that can cause significant damage to health systems on 27 January 2023 (WHO, 2023). Although some effective vaccines and drugs which reduce symptoms and decrease mortality of COVID-19 have been developed, concern about the possibility of a surge in infections in the future remain. Therefore, it is crucial to develop more effective COVID-19 diagnosis and treatment plans.
SARS-CoV-2, one of the genus Cytomegalovirus members within the Coronaviruses family, is a righteous single stranded RNA virus with a genome size between 27 and 32 KB (Nafian et al., 2023). SARS-CoV-2 genome encode four encoded structural proteins: spike (S) protein, envelope protein, nucleocapsid (N) protein, and membrane protein. The N protein is the most abundant and conserved protein in coronaviruses among them, and it is also the core component of virus particles (Cascarina & Ross, 2020). N protein plays a significant role in viral replication and transcription process, such as the RNA-binding and chaperone activities in N protein which can promote gRNA replication. It has been shown that the N protein has highly antigenic and induces antibodies to this antigen in patients, so the N protein is a suitable target for potential therapeutic drugs (Woo et al., 2020; Zinzula et al., 2021).
Up to now, World Health Organization have defined five SARS-CoV-2 variants, namely Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and Omicron (B.1.1.529), inducing waves of prevalence (Carter et al., 2020). Antibodies developed based on the spike of SARS-CoV-2 have good therapeutic prospects, but emerging variants show the possibility of virus escape (Hernandez et al., 2023). More conserved epitopes may become preferred targets because a low degree of variation indicates limited mutational ability. Recent studies have shown that the generation of anti-N protein antibodies could be used to reveal viral N protein functions in viral pathogenesis, and more importantly, the anti-N protein antibodies might be useful when applied in diagnostic assays for the detection of SARS-CoV-2 infection, such as serologic and antigen tests (Lu et al., 2021; Terry, Anderson & Scherman, 2021; Dangi et al., 2022). Currently, main detection methods for SARS-CoV-2 include reverse transcription-polymerase chain reaction (RT-PCR) test and rapid colorimetric antigen detection test. The above methods are time-consuming or less sensitive, hence novel, rapid and accurate analytical methods are required for SARS-CoV-2 detection. The development of antibodies targeting SARS-CoV-2 N protein may provide new ideas for SARS-CoV-2 detection methods.
Single domain antibody, also known as nanobody, is a new type of genetically engineered antibody that is obtained though cloning the variable region of heavy chain antibody which is naturally short of light chain in camelid animals (Hamers-Casterman et al., 1993). Compared with single domain antibody, traditional antibodies cannot always bind certain antigen surfaces because of the large size (Mustafa & Mohammed, 2023). Single domain antibody has the advantages such as small molecular weight, high stability, good solubility, strong penetrating ability and low immunogenicity (Zhang et al., 2022; Debie et al., 2020; Khilji et al., 2022). Taken together, single domain antibody possesses multiple favorable properties for COVID-19 prophylaxis, therapeutic and diagnostic purposes.
In our study, the M13 phage display system was employed to screen the single domain antibody targeting the N protein and find eight candidates against the N protein. Then, we designed two fusion proteins which contains multiple fusion fragments on the base of the single domain antibody against the N protein. Our findings suggest that fusion protein showed higher affinity than single domain antibody and it can effectively combine the N protein of the COVID-19. Fusion proteins may be able to reach the target more accurately, reducing the damage of the drug to the body and exerting a more powerful therapeutic effect (Chen et al., 2022; Dacon et al., 2022).
Materials and Methods
Chemicals and reagents
The HuSdL-2TM Phage Display Library were provided by Creative Biolabs (Shirley, NY, USA). The HRP-conjugated anti-M13 phage antibody was obtained from Abcam (Cambridge, UK). Bovine serum albumin (BSA), iso-propylthio-β-D-galactoside (IPTG) and triethylamine were purchased from Yeasen (Shanghai, China). The recombinant human SARS-CoV-2-N were purchased from Genscript. 3,3′,5,5′-Tetramethylbenzidine (TMB) was purchased from Solarbio (Beijing, China). All of the other inorganic chemicals and organic solvents were analytical reagent.
Biopanning
Biopanning of anti-SARS-CoV-2-N antibody was performed based on the instruction manual of phage library kit with a few modifications. In the first round of biopanning, 100 µL of SARS-CoV-2-N protein (13.6 µg/mL) dissolved in 0.01 M (pH 7.2) phosphate-buffered saline (PBS) was added into the MaxiSorp® ELISA plate (Nunc, Rochester, NY, USA) and then incubated at 4 °C for all night. Then, the plate was blocked with PBS containing 5% skim milk. After washing, the phage library (1010~1011 plaque forming units (PFU)) was incubated at 37 °C for 60 min, then washed. The bound phages were eluted using 100 µL triethylamine (100 mM) for 30 min at 37 °C, and the eluent was neutralized with 200 µL 1M Tris-buffer (pH 7.4). The eluent was amplified in 10 mL of exponentially in logarithmic phase for the next round of selection.
The inputted phages remained unchanged (1.0 × 1011 phage) in the next two rounds of biopanning, but SARS-CoV-2-N protein concentration was reduced to half compared to last round. The input and recovery of each selective step was assessed to evaluate efficiency of each step. The monoclonal phage DNA was extracted, purified and sequenced by Fuzhou Sunya Biotechnology Co., LTD (Fujian, China). Sequence translation software was employed for analysis of amino acid sequence.
Phage ELISA
A total of 96-well plate was coated with the SARS-CoV-2-N (100 μL, 5 μg/ mL) in coating buffer for all night at 4 °C, then blocked with 200 μL 5% BSA for 2 h at 37 °C. 100 μL of phage (cycle 1–4) was added to the blocked well and incubated at 37 °C for 2 h. The plates were washed three times with 200 μL washing buffer. After that, 5,000-fold diluted solution of anti-M13-HRP was added to the wells. The plate was incubated for at 37 °C 1 h followed by washing again. Then, 100 μL of TMB was added to each well. Finally, the plate was quenched with 1.0 M phosphoric acid, and absorbance was tested at 450 nm.
DNA sequence analysis
The obtained positive monoclonal phage single domain antibody genes were identified by PCR and DNA sequencing. The positive clones selected by PCR using the following primers: Forward Primer L1:5′-TGGAATTGTGAGCGGATAACAATT-3′, Reverse Primer S6: 5′-GTAAATGAATTTTCTGTATGAGG-3′. The sequences of the antibody against SARS-CoV-2-N were analyzed by DNA sequencing. Then, the sequences of positive clones were analyzed by Fuzhou Sunya Biotechnology Co., LTD (Fujian, China). Sequence translation software was used for determination of amino acid sequence.
Bioinformatics
Nucleotide sequence alignment
The pN01–pN08 gene sequences were compared using the comparison function of MEGA-X, and the aligned sequences were derived and analyzed. Positions of the complementarity determining regions of the variable domains were determined by IMGT/ V-QUEST tool (https://imgt.org/IMGT_vquest/user_guide) (Giudicelli, Brochet & Lefranc, 2011).
Structural construction of ligand and receptor
The pN01–pN08 Gene sequence selected by phage display library was translated into amino acids by Gene Runner software (Haghighi et al., 2019), and the amino acid sequence was analyzed by Expasy (https://www.expasy.org/) (Gasteiger et al., 2005), an online tool. The molecular weight, isoelectric point, instability coefficient and other biological information of different proteins were obtained, which was used as part of the reference basis for subsequent experiments. The amino acid sequences for 2019-nCoV nucleocapsid N-terminal domain protein (SARS-CoV-2-N-NTD; PDB code: 7CDZ) were downloaded from PDB. Then, all of the above the structures were predicted for Swiss Model. Among the many structures predicted by Swiss Model, the structural energy is the lowest the PDB file is saved for later use.
Docking studies
The selected ligand and receptor structures were uploaded in the virtual screening software interface HawkDock (http://cadd.zju.edu.cn/hawkdock/) (Weng et al., 2019). Structural prediction of protein-protein complex by using the global docking algorithm implemented in ATTRACT and the HawkRank scoring function. The best conformer was selected based on the docking score and better non-covalent bond interaction. The photographs of the docking pose and interactions were collected and saved. Then, the docking output files were analyzed for the interactions between ligand with receptor using Pymol software (version 3.0+) (Janson & Paiardini, 2021) and Ligplus (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/) (Laskowski & Swindells, 2011).
Expression of single domain antibody in E. coli and purification
The expression vector was constructed by fusing with the genes fragment of anti-SARS-CoV-2-N and pET-28a(+)/NdeI/XhoI. Then, E. coli strains BL21(DE3) was used as the expression host and were transformed with expression plasmid. Subsequently, the transformed bacteria were cultured grew overnight at 37 °C in LB medium whose Kanamycin concentration was 20 μg/mL. Then, the cultures were diluted at a ratio of 1:100 in fresh liquid LB medium containing 20 μg/mL kanamycin and cultured at 37 °C with shaking. When the absorbance at 600 nm of the cultures reached 0.6, the cultures were induced by 1 mM IPTG and harvested after 5 h by centrifugation at 10,000 g for 10 min.
Subsequently, the collected E. coli cell was resuspended in 100 mL TGE buffer (pH 7.9) which contained 50 mM Tris, 0.5 mM EDTA, 5% and glycine 50 mM NaCl. And then E. coli cell was disrupted by sonication on ice for 30 min, following with centrifugation at 10,000 g at for 10 min 4 °C. After preliminary inclusion body purification, the precipitate was resuspended and purified with HisPur™ Ni-NTA Magnetic Beads. Then the purified products were pooled and dialyzed in urea with different gradients at 4 °C.
Antibody binding affinity determination using ELISA
A total of 96-well Maxisorp plates (Nunc) were incubated with 100 μL of 0.7 μg/mL SARS-CoV-2-N at 4 °C overnight. Subsequently, the plates were washed two times with PBS containing 0.1% Tween-20 and then blocked for 2 h with 300 μL of 5% BSA. After washing, the plate was incubated with 6.8 μg/mL anti- SARS-CoV-2-N antibodies at 37 °C for 1 h. The absorbance of 450 nm was measured using an ELISA plate reader (Bio Tek, Winooski, VT, USA).
Fusion protein
We fused different single domain antibodies with (G4S)2 as the linker peptide to construct fusion proteins named as pN3-3 and pN3-1-3, respectively. The method of expression, purification and determination referred to 2.6 and 2.7 with slight modification. After washing, each group was coated with 3.4 μg/mL fusion protein in ELISA experiment.
Statistical analysis
All data were analyzed using GraphPad Prism version 8.0 (GraphPad Software, San Diego, CA, USA) and presented as the mean ± SD. were evaluated by two-tailed test. Multiple group comparisons and two groups were tested using one-way analysis of variance and two-tailed test, respectively. P < 0.05 was determined to be significant.
Results
Screening of specific single domain antibody against SARS-CoV-2-N
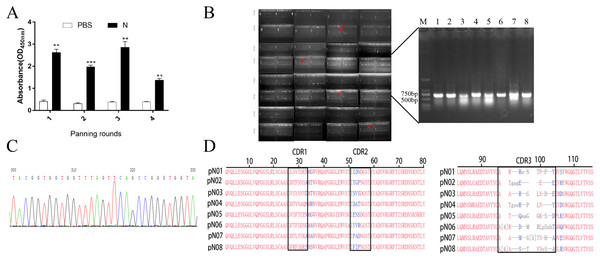
HuSdL-2™ phage library was screened for three rounds to obtain anti-SARS-CoV-2 N protein single domain antibody. The uncoated antigen wells were set as a negative control during each round of screening, and the titer of the eluted phages was tested after each round of screening. The recovery rate is an effective indicator for estimating the degree of screening enrichment. It was shown that with the number of screening rounds increasement, the recovery rates increased, which indicating the effectiveness of the enrichment screening. The affinity increased as the biopanning rounds went on in Fig. 1A, suggesting that the specificity of phages was effectively enriched (Table 1).
Figure 1: Screening and identification of single domain antibody targeting SARS-CoV-2-N.
(A) Detection of the phage enrichment degree and its ability to bind to SARS-CoV-2-N fragments were detected by phage ELISA. (B) Positive clones were screened by PCR, the size of the target DNA is about 700 bp. (C) Partial sequencing map. (D) Alignment of amino acid sequences of eight screened single domain antibodies. **P < 0.01; ***P < 0.001 vs PBS group.| Round of panning | N (μg/ml) | Input phage (PFU/well) |
Output phage (PEU/well) |
Output/Input ratio |
|---|---|---|---|---|
| 1st round | 14 | 1.14 × 1013 | 2.02 × 108 | 1.77 × 10−5 |
| 2nd round | 7 | 1.94 × 109 | 5.8 × 105 | 2.99 × 10−4 |
| 3rd round | 3.5 | 1.36 × 1012 | 6.2 × 106 | 4.56 × 10−6 |
Note:
The N protein, Nucleocapsid protein of the COVID-19; sdAb: Single-domain antibody; N: SARS-CoV-2-N; PFU, plaque forming unit.
DNA was amplified by PCR with primer L1 and primer S6 and the clones containing the complete sequence (approximately 700 bp) were verified by 1% nucleic acid gel (Fig. 1B). Positive clones were selected for DNA sequencing to identify their sequences (Fig. 1C). Finally, eight anti-SARS-CoV-2-N single domain antibody gene sequences were obtained named pN01-pN08, respectively. As is shown in Fig. 1D, the conserved residues at the positions of Val37, Gly44, Leu45 and Trp47 were all conservative disability amino acids in the eight different sequences.
Bioinformatics analysis of antibody against SARS-CoV-2-N
The gene sequence was converted into amino acid sequence by Gene Runner and online software Expasy (https://web.expasy.org/protparam/) was employed to calculate the relevant biochemical information of the sequence. The molecular mass of antibodies is about 13 kDa, the half-life in the mammalian cell is 0.8 h and the instability coefficients of all antibodies are less than 40 that means all single domain antibodies are stable protein. pN01, pN02, pN03 and pN04 are acidic proteins, pN07 is neutral protein, and pN05, pN06 and pN08 are basic proteins (Table 2). pN1 and pN3 were selected to construct fusion proteins named pN3-3 and pN3-1-3, respectively.
| Number of amino acids | M (kDa) | PI | Aliphatic index | Instability index | GRAVY | |
|---|---|---|---|---|---|---|
| pN01 | 120 | 13.04 | 4.96 | 75.50 | 34.38 | −0.388 |
| pN02 | 119 | 12.67 | 5.22 | 68.82 | 28.75 | −0.297 |
| pN03 | 119 | 12.82 | 4.73 | 76.13 | 31.56 | −0.318 |
| pN04 | 121 | 13.06 | 5.28 | 70.08 | 27.52 | −0.419 |
| pN05 | 121 | 12.94 | 7.87 | 69.26 | 34.97 | −0.367 |
| pN06 | 126 | 13.74 | 9.26 | 68.10 | 30.63 | −0.293 |
| pN07 | 124 | 13.34 | 6.77 | 74.76 | 26.05 | −0.256 |
| pN08 | 124 | 13.25 | 8.70 | 77.02 | 36.34 | −0.115 |
Binding pattern of the single domain antibody against SARS-CoV-2-N
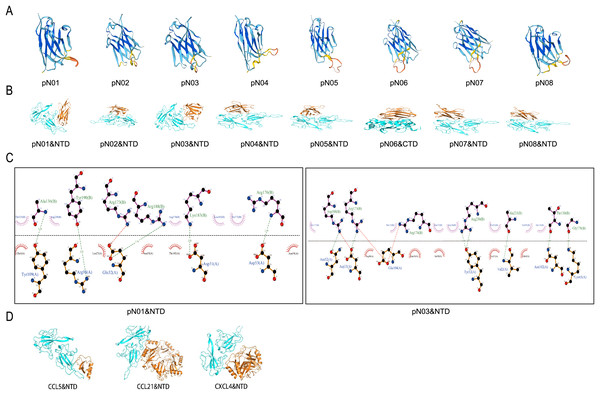
Swiss-model software was employed for constructing the 3-D structure of single domain antibody (Fig. 2A). The single-domain antibody 3D model presented a single antigen-binding variable domain (VHH) and classical immunoglobulin fold similar to that of the alpaca antibody, with two layers of antiparallel beta chain folded composition, forming a classic sandwich-like structure. Each nanobody had three highly variable rings called CDRS, which were major regions that undergo genetic changes to present antigen-specific binding surfaces. The single-domain antibody 3D model had the characteristic of “VHH structural antibody”, which does not require complex folding and can be expressed in the prokaryotic system. HawkDock (http://cadd.zju.edu.cn/hawkdock/) was employed to study the molecular docking between single domain antibody against the N-terminal domain protein of 2019-nCoV nucleocapsid (SARS-CoV-2-N-NTD; PDB code: 7CDZ) and the C-terminal domain protein of 2019-nCoV nucleocapsid (SARS-CoV-2-N-CTD; PDB code: 7CE0). The amino acid sequences for SARS-CoV-2-N-NTD and SARS-CoV-2-N-CTD were obtained from the Protein Data Bank (https://www.rcsb.org/). It was showed that pN06 mainly interacted with SARS-CoV-2-N-CTD and the other single domain antibodies mainly interacted with SARS-CoV-2-N-NTD (Fig. 2B).
Figure 2: Analysis of binding pattern between SARS-CoV-2-N and the single domain antibody against SARS-CoV-2-N.
(A) 3-D structures of single domain antibody against SARS-CoV-2-N. (B) Molecular docking of pN01-pN08 and SARS-CoV-2-N. Blue cartoons represent SARS-CoV-2-N-NTD (NTD) or SARS-CoV-2-N-CTD (CTD) and orange cartoons represent pN01-pN08. (C) The binding mode between pN01, pN03 and SARS-CoV-2-N-CTD or SARS-CoV-2-N-NTD. (D) The binding pattern of CCL5, CCL21 and CXCL4 to SARS-CoV-2-N-NTD.The protein-ligand complex was further analyzed and visualized through Pymol software and Ligplus. As shown in Fig. 2C and Table 3, five hydrogen bond interactions were observed between SARS-CoV-2-N-NTD and pN01, with Asp31 (2.00 Å), Glu32 (2.34 Å), Asp53 (2.90 Å), Arg98 (2.83 Å) and Tyr109 (3.29 Å), respectively. Moreover, pN01 formed hydrophobic interactions with SARS-CoV-2-N-NTD. Focusing on pN07 and pN08, the amino acids and hydrogen bonds involved in the binding interface of the two proteins are the same. According to previous studies, SARS-CoV-2-N had high binding affinity to 11 kinds of human chemokines (Alturaiki et al., 2023). Increase in chemokines can attract immune cells to the infected site, leading to inflammation and tissue damage (Catalán, Gómez-Ambrosi & Rodríguez, 2013). On the basis of the molecular docking map, the combination situations of CCL5, CCL21 and CXCL4 with SARS-CoV-2-N were similar to that of pN01-pN08. As shown in Fig. 2D, CCL5, CCL21 and CXCL4 mainly interacted with SARS-CoV-2-N-NTD. The binding affinity between protein complexes were calculated using HawkDock tool. The binding affinity of CCL5, CCL21 and CXCL4 were more than or equal to −9.0. The binding affinity of all the single domain antibodies were less than −9.0 except the pN05 (Table 4). It was suggested that there was a competitive relationship between pN01-pN08 and chemokines to bind SARS CoV-2-N, and pN01-pN08 might inhibit the combination between SARS-CoV-2-N and chemokines to reduce inflammatory response.
| Complex | Ligan | Receptor | Bond length (Å) | Number of non-covalent bonds |
|---|---|---|---|---|
| pN01& SARS-CoV-2-NTD | Asp31 | Lys183 | 2.00 | Five hydrogen bonds and multiple hydrophobic interactions |
| Glu32 | Arg188 | 2.34 | ||
| Asp53 | Arg176 | 2.90 | ||
| Arg98 | Tyr190 | 2.83 | ||
| Tyr109 | Ala136 | 3.29 | ||
| pN02& SARS-CoV-2-NTD | Asp62 | Thr45 | 2.63 | Five hydrogen bonds and multiple hydrophobic interactions |
| Asp62 | Thr45 | 2.48 | ||
| Glu103 | Val239 | 3.02 | ||
| Glu104 | Asn158 | 3.25 | ||
| Gly110 | Gln244 | 1.91 | ||
| pN03& SARS-CoV-2-NTD | Val2 | Ala233 | 3.12 | Seven hydrogen bonds and multiple hydrophobic interactions |
| Tyr32 | Arg230 | 2.43 | ||
| Tyr32 | Arg230 | 2.49 | ||
| Asp33 | Arg173 | 2.93 | ||
| Lys65 | Gly178 | 1.81 | ||
| Asn102 | Thr130 | 2.19 | ||
| Asn52 | Arg188 | 3.31 | ||
| pN04& SARS-CoV-2-NTD | Arg19 | Asp98 | 3.02 | Four hydrogen bonds and multiple hydrophobic interactions |
| Thr58 | Gly205 | 2.85 | ||
| Phe68 | Asn107 | 2.26 | ||
| Arg72 | Arg72 | 3.04 | ||
| pN05& SARS-CoV-2-NTD | Gly15 | Asn108 | 2.70 | Seven hydrogen bonds and multiple hydrophobic interactions |
| Arg19 | Asp98 | 2.70 | ||
| Gly56 | Thr102 | 2.93 | ||
| Gly56 | Thr102 | 3.24 | ||
| Asp55 | Arg149 | 2.84 | ||
| Asp55 | Arg149 | 2.58 | ||
| Asp55 | Arg149 | 2.42 | ||
| pN06& SARS-CoV-2-NTD | Gly8 | Ala167 | 3.06 | Two hydrogen bonds and multiple hydrophobic interactions |
| Thr119 | Lys117 | 3.29 | ||
| pN07& SARS-CoV-2-NTD | Leu11 | Asn104 | 2.37 | Six hydrogen bonds and multiple hydrophobic interactions |
| Glu13 | Asn2 | 2.95 | ||
| Gly19 | Asn31 | 2.96 | ||
| Gly19 | Asn31 | 1.64 | ||
| Lys76 | Thr30 | 1.51 | ||
| Asn84 | Asp98 | 2.51 | ||
| pN08& SARS-CoV-2-NTD | Leu11 | Asn104 | 2.37 | Six hydrogen bonds and multiple hydrophobic interactions |
| Glu13 | Asn2 | 2.95 | ||
| Gly19 | Asn31 | 2.96 | ||
| Gly19 | Asn31 | 1.64 | ||
| Lys76 | Thr30 | 1.51 | ||
| Asn84 | Asp98 | 2.51 |
| Complex | Binding affinity (kal/mol) |
|---|---|
| pN01&NTD | −12.2 |
| pN02&NTD | −13.9 |
| pN03&NTD | −11.0 |
| pN04&NTD | −9.0 |
| pN05&NTD | −7.8 |
| pN06&CTD | −15.5 |
| pN07&NTD | −10.3 |
| pN08&NTD | −10.3 |
| CCL5&NTD | −9.0 |
| CCL21&NTD | −7.9 |
| CXCL4&NTD | −8.5 |
Construction, expression, and purification of the single domain antibody against SARS-CoV-2-N
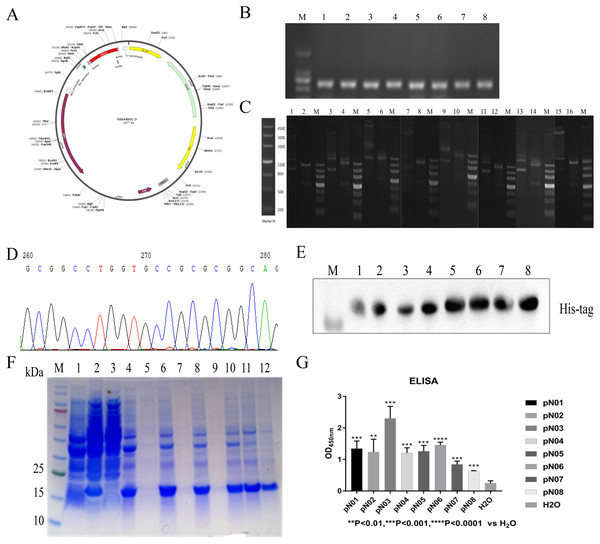
The vector was conducted with a C-terminal 6× His tag for purification (Fig. 3A). The recombinant plasmid was verified by double enzymatic digestion and digested to two bands, one of which was approximately 700 bp and the other band was approximately 4,500 bp (Fig. 3B). This indicated that the insertion of the gene was correct. DNA sequencing analysis result also suggested that plasmid was successfully constructed (Fig. 3C). Then, the ligated product was transformed into E. coli BL21 (DE3) cells. Colony PCR result showed a band of approximately 700 bp (Fig. 3D). The subsequent expression of the single domain antibody was induced with IPTG and the molecular weight of single domain antibody approximately was 13 kDa. The purification of single domain antibody was performed with HisPur™ Ni-NTA Magnetic Beads and the purity of target protein was more than 95% (Fig. 3E). The purified product was identified by western blot and the result showed that the purified product was target protein (Fig. 3F).
Figure 3: Construction, expression, purification and binding affinity of the anti-SARS-CoV-2-N single domain antibody.
(A) Anti-SARS-CoV-2-N single domain antibody prokaryotic expression plasmid schematic. (B) PCR of selected positive clones. (C) Verification of recombinant plasmid using double digested. (D) Partial DNA sequencing. (E) SDS-PAGE analysis of anti-SARA-CoV-2-N single domain antibody. Lane M: marker, Lane 1: before induction, Lane 2: after induction, Lane 3: supernatant after ultrasonication, Lane 4: precipitation after ultrasonication, Lane 5: supernatant treated with 1% DOC, Lane 6: precipitation treated with 1% DOC, Lane 7: supernatant treated with 2 M urea, Lane 8: precipitation treated with 2 M urea, Lane 9: supernatant treated with 3 M urea, Lane 10: precipitation treated with 3 M urea, Lane 11: dissolved supernatant, Lane 12: purified product. (F) Western blot assay to identify single domain antibodies against SARS-CoV-2-N. (G) ELISA results show the affinity of single domain antibodies to SARS-CoV-2-N compared to H2O.Binding activity evaluation of the single domain antibody against SARS-CoV-2-N
The binding activity of the antibodies to SARS-CoV-2-N were determined by ELISA. As shown in Fig. 3G, all antibodies could specifically bind to SARS-CoV-2-N. Furthermore, pN03 had best affinity with SARS-CoV-2-N in all single domain antibodies (Fig. 3G).
Binding pattern of SARS-CoV-2-N to fusion proteins against SARS-CoV-2-N
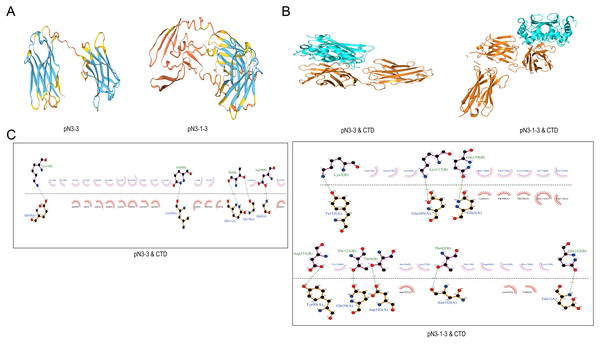
Swiss-model software was used to construct the 3D structures of fusion proteins (Fig. 4A). The molecular docking between fusion proteins and SARS-CoV-2-N-CTD or SARS-CoV-2-N-NTD were analyzed by HawkDock software and Ligplus. Figs. 4B and 4C showed the interaction between fusion proteins and SARS-CoV-2-N-CTD. As shown in Table 5, pN3-3 formed five hydrogen bond interactions and multiple hydrophobic interactions with SARS-CoV-2-N-CTD. The hydrogen bond interactions were observed at Thr9 (3.23 and 2.89 Å), Gln18 (2.47 Å), Lys113 (3.12 Å) and Asp199 (2.38 Å) of pN3-3. pN3-1-3 showed two hydrogen bond interactions with SARS-CoV-2-N-CTD at same amino acid of pN3-3 at Thr9 and Lys113. The binding affinity of pN3-3 and pN3-1-3 was −10.8 and −12.7, respectively (Table 6).
Figure 4: The binding pattern between SARS-CoV-2-N and fusion proteins.
(A) 3D structures of pN3-3 and pN3-1-3. (B) Molecular docking of fusion proteins obtained by HawkDock. The blue cartoon represents SARS-CoV-2-N-CTD (CTD), and the orange cartoon represents fusion proteins. (C) The binding pattern of fusion proteins to SARS-CoV-2-N-CTD.| Complex | Ligan | Receptor | Bond length (Å) | Number of non-covalent bonds |
|---|---|---|---|---|
| pN3-3& SARS-CoV-2-NTD | Thr9 | Gly110 | 3.23 | Five hydrogen bonds and multiple hydrophobic interactions |
| Thr9 | Gln111 | 2.89 | ||
| Gln18 | Leu229 | 2.47 | ||
| Lys113 | Glu105 | 3.12 | ||
| Asp199 | Gly42 | 2.38 | ||
| pN3-1-3& SARS-CoV-2-CTD | Lys3 | Tyr32 | 2.87 | Eight hydrogen bonds and multiple hydrophobic interactions |
| Thr9 | Asp103 | 1.86 | ||
| Thr42 | Asn102 | 2.58 | ||
| Lys117 | Glu105 | 1.91 | ||
| Thr121 | Gln39 | 2.20 | ||
| Asp153 | Tyr95 | 2.97 | ||
| Gln159 | Gln3 | 3.11 | ||
| Gln162 | Glu1 | 3.02 |
| Number of amino acids | M (kDa) | PI | Aliphatic index | Instability index | GRAVY | Binding affinity (kal/mol) | |
|---|---|---|---|---|---|---|---|
| pN3-3 | 247 | 26.20 | 4.61 | 34.70 | 73.36 | −0.324 | −10.8 |
| pN3-1-3 | 376 | 39.81 | 4.66 | 36.39 | 72.29 | −0.348 | −12.7 |
Note:
M, molecular weight; PI, isoelectric point; GRAVY, grand average of hydropathicity.
Construction, expression, and purification of the fusion proteins against SARS-CoV-2-N
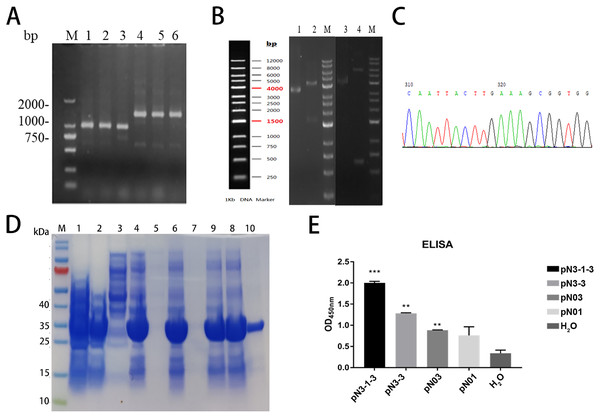
The construction process of the fusion proteins was similar to that of single domain antibody. In brief, the constructed product was transformed into competent cells. Colony PCR verified the inserted gene was correct. The PCR products of pN3-3 was approximately 800 bp and the PCR products of pN3-1-3 was approximately 1,500 bp (Fig. 5A). To confirm the cloning, plasmid extracted from transformed cells was digested by XhoI and MluI. Results showed pN3-1-3 was about 6,000 bp, and pN3-3 was about 4,500 bp (Fig. 5C). DNA sequencing analysis was consistent with the above results (Fig. 5B). SDS-PAGE results confirmed that the protein was greatly induced after the addition of IPTG. The molecular weight of pN3-3 was approximately 27 kDa and the molecular weight pN3-1-3 was approximately 40 kDa (Table 6). The recombinant fusion protein was purified by HisPur™ Ni-NTA Magnetic Beads, the purity of target protein was more than 95% (Fig. 5D). ELISA was performed to compare the binding activity of the two fusion proteins. As is shown in Fig. 5E, all fusion proteins could specifically bind to SARS-CoV-2-N and the affinity of fusion proteins is stronger than that of single domain antibody.
Figure 5: Construction, expression, and purification of the fusion proteins against SARS-CoV-2-N.
(A) Positive clones were screened by PCR. M: Marker; Lane1-3: pN3-3; Lane4-6: pN3-1-3. (B) Partial sequencing map of fusion protein. (C) The electrophoresis results of recombinant plasmid. Lane 1: pN3-3 plasmid, Lane 2: double digested pN3-3 plasmid, Lane 3: pN3-1-3 plasmid, Lane 4: double digested pN3-1-3 plasmid. (D) SDS-PAGE analysis of anti-SARS-CoV-2-N fusion proteins. Lane M: marker, Lane 1: before induction, Lane 2: after induction, Lane 3: supernatant after ultrasonication, Lane 4: precipitation after ultrasonication, Lane 5: supernatant treated with 1% DOC, Lane 6: precipitation treated with 1% DOC, Lane 7: supernatant treated with 2 M urea, Lane 8: precipitation treated with 2 M urea, Lane 9: supernatant treated with 3 M urea, Lane 10: precipitation treated with 3 M urea, Lane 11: dissolved supernatant, Lane 12: purified product. (E) The binding affinity of fusion proteins to SARS-CoV-2-N. **P < 0.01; ***P < 0.001 vs control group.Discussion
The global outbreak of COVID-19 caused by novel coronavirus impose a heavy burden on global health systems and is a serious threat to social stability (Qu et al., 2023). With SARS-CoV-2 spreading globally amidst a dearth of effective diagnostics, treatments, and reagents to tackle pandemic, it is more important than ever that tools for detection and research are developed and validated. In this study, eight effective anti-SARS-CoV-2-N single domain antibody sequences were screened from HuSdL-2TM human single-domain antibody library constructed based on camel-based human VH3 using phage display technology. These sequences were cloned into pET vector to construct eight single domain antibody prokaryotic expression strains a named pN01-pN08 and two fusion proteins named pN3-3, pN3-1-3. These single-domain antibodies and fusion proteins can be used in the future for rapid, accurate, and inexpensive diagnostic assays for bed-side diagnosis of COVID patients. Future studies can utilize these antibodies for studies determining N protein structure, intracellular interactions, diagnostic development, and potential therapeutics.
Recent studies have shown that SARS-CoV-19 has genetic homology with traditional herpes virus, syncytial virus and atypical pneumonia virus, with only a few nucleic acid changes. Hence the amino acid sequence of pN01–pN08 was compared with antibodies of viruses that cause respiratory disease. The data showed that pN01 was similar with the heavy chain variable region of anti-H7N9 IgG originated from human. H7N9 is a subtype of avian influenza virus which is not generally circulating in the human population, but it has significant mortality when it outbreaks in the human (Liu et al., 2022).
pN02, pN04, pN05, pN07 and pN08 were compared amino acid sequence with the anti-gliadin. Gliadin is related to occurrence of inflammation (Zainumi et al., 2022). Hyperactive macrophages and plenty of proinflammatory cytokines such as tumor necrosis factor-α, interleukin-6 and interleukin-1β were found in the lungs of patients with COVID-19 (Cai et al., 2022; Guarnieri et al., 2022). pN02 shows the active from anti-SARS-CoV-2-N candidate drugs, which helps in understanding important details of the process of COVID-19 development.
pN03 was compared amino acid sequence with neutralizing antibodies against SARS-CoV-2-S and RBD of the SARS-CoV-2-S1 subunit immunoglobulin neutralizing antibodies (Rogers et al., 2020). Neutralizing antibodies induced by viruses or vaccines play a crucial role in fighting virus infections, which be able to broadly neutralize the virus and its variants (Chen et al., 2023). The amino acid sequence of pN03 has similarity with neutralizing antibodies of SARS-CoV-2-S, which may show an incredibly broad neutralization spectrum against SARS-CoV-2.
pN06 was compared amino acid sequence with the anti-pneumococcal antibody (Chang, Zhong & Lees, 2002). Pneumococci cause pneumonitis in the lungs. The symptoms of pneumonitis are fever, chills, cough, bloody sputum and chest pain. The symptoms of patients with pneumonia may cover up SARS-CoV-2 infections. If the candidate drug can improve both pneumonia and COVID-19 in the meanwhile, it may reduce the SARS-CoV-2 patients, especially by reducing preventable pneumonia hospitalization rates and the accompanying risk of SARS-CoV-2 transmission in hospitals (McIntosh, Feemster & Rello, 2022). These data suggested that pN01 and pN03 are two separate binding proteins.
Bivalent or multivalent nanobodies may offer unique advantages compared with single domain antibody, such as stronger antigen binding ability, lower their toxicity and less immune escape of the virus (Chen et al., 2022). pN03 showed the highest binding ability against SARS-CoV-2-N among these single domain antibodies in ELISA assay. On the base of the single domain antibodies screening against SARS-CoV-2-N, two fusion proteins expression strains pN3-3 and pN3-1-3 were constructed, successfully expressed and purified using one-step. ELISA results indicated that pN3-3 and pN3-1-3 exhibited stronger binding ability than pN03 and pN01. Two fusion protein was constructed using single domain antibody with strong binding ability as a unit. Hence, there were more sites on the fusion protein that could bind to SARS-CoV-2-N than a single single-domain antibody. It was also proved that fusion protein had strong binding affinity than single-domain antibody in ELISA experiments using uncorrelated antibodies. It was similar to our results that Thébault et al. (2022) reported that F9-C2 fusion protein bind the spike receptor binding domain with stronger affinity than single αRep.
In conclusion, our study date showed the novel anti-SARS-CoV-2 fusion proteins which targeted multiple N protein epitopes, possessed highly affinity to the N protein in vitro and had the potential to develop antigen detection kits for SARS-CoV-2. It lays the foundation of further clinical studies with anti-SARS-CoV-2 fusion proteins. Moreover, the platform we conducted can also be used for screening single domain antibodies against novel viruses which laid a foundation for diagnosis of novel viruses.
Supplemental Information
The EC50 of single domain antibodies against SARS-CoV-2-N affinity analysis.
The binding mode between pN01- pN08 and SARS-CoV-2-N-CTD or SARS-CoV-2-N-NTD.
Raw data.
Pzfx files must be opened with GraphPadPrism software.