Chigno/CG11180 and SUMO are Chinmo-interacting proteins with a role in Drosophila testes somatic support cells
- Published
- Accepted
- Received
- Academic Editor
- Judith Yanowitz
- Subject Areas
- Cell Biology, Developmental Biology, Genetics, Molecular Biology
- Keywords
- Sex maintenance, Testis, Spermatogenesis, Germline, Germ cell tumors, Cancer, Infertility, Chinmo, CG11180/Chigno, SUMO/Smt3
- Copyright
- © 2024 Rinehart et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits using, remixing, and building upon the work non-commercially, as long as it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2024. Chigno/CG11180 and SUMO are Chinmo-interacting proteins with a role in Drosophila testes somatic support cells. PeerJ 12:e16971 https://doi.org/10.7717/peerj.16971
Abstract
Stem cells are critical for replenishment of cells lost to death, damage or differentiation. Drosophila testes are a key model system for elucidating mechanisms regulating stem cell maintenance and differentiation. An intriguing gene identified through such studies is the transcription factor, chronologically inappropriate morphogenesis (Chinmo). Chinmo is a downstream effector of the Jak-STAT signaling pathway that acts in testis somatic stem cells to ensure maintenance of male stem cell fate and sexual identity. Defects in these processes can lead to infertility and the formation of germ cell tumors. While Chinmo’s effect on testis stem cell behavior has been investigated in detail, there is still much to be learned about its structure, function, and interactions with other proteins. Using a two-hybrid screen, we find that Chinmo interacts with itself, the small ubiquitin-like modifier SUMO, the novel protein CG11180, and four other proteins (CG4318, Ova (ovaries absent), Taf3 (TBP-associated factor 3), and CG18269). Since both Chinmo and CG11180 contain sumoylation sites and SUMO-interacting motifs (SIMs), we analyzed their interaction in more detail. Using site-directed mutagenesis of a unique SIM in CG11180, we demonstrate that Chinmo’s interaction with CG11180 is SUMO-dependent. Furthermore, to assess the functional relevance of both SUMO and CG11180, we performed RNAi-mediated knockdown of both proteins in somatic cells of the Drosophila testis. Using this approach, we find that CG11180 and SUMO are required in somatic cells of adult testes, and that reduction of either protein causes formation of germ cell tumors. Overall, our work suggests that SUMO may be involved in the interaction of Chinmo and CG11180 and that these genes are required in somatic cells of the adult Drosophila testis. Consistent with the CG11180 knockdown phenotype in male testes, and to underscore its connection to Chinmo, we propose the name Chigno (Childless Gambino) for CG11180.
Introduction
Stem cells are critical for organ function and tissue homeostasis. Their capacity to self-renew while producing daughter cells that differentiate into functional cell types is critical for the continuous replacement of cells lost to death or damage over time. While our understanding of these important cells has increased dramatically, there is still much to be learned. Due to their relatively simple structure and defined stem cell niche, Drosophila testes are a key model for uncovering mechanisms controlling stem cell behavior in other systems (de Cuevas & Matunis, 2011; Fuchs, Tumbar & Guasch, 2004; Herrera & Bach, 2019; Leatherman, 2013; Singh, 2015). In addition to elucidating mechanisms controlling tissue homeostasis, defects in Drosophila reproductive stem cell behavior provide insight into infertility, cancer, and aging (Han et al., 2017; Hou & Singh, 2017; Salz, Dawson & Heaney, 2017; Spradling et al., 2011; Voog & Jones, 2010).
The Drosophila testis forms a coiled tube with the germline stem cell (GSC) niche at its apex, and differentiated spermatids at the distal end that is connected to the seminal vesicle (Fuller, 1993; Hardy et al., 1979). The GSC niche is made up of a tight cluster of quiescent somatic cells, collectively called the hub, which acts as a signaling center to control adjacent stem cells (de Cuevas & Matunis, 2011; Demarco et al., 2014; Greenspan, De Cuevas & Matunis, 2015 PMID; Herrera & Bach, 2019). Two populations of stem cells, the GSCs and somatic cyst stem cells (CySCs), are docked at the hub (Hardy et al., 1979). Asymmetric division of GSCs oriented away from the hub results in production of a GSC daughter that remains associated with the niche, and a gonialblast daughter that is displaced from the hub (Inaba et al., 2010; Yamashita, Jones & Fuller, 2003). The gonialblast continues to divide with incomplete cytokinesis; making a 16-cell spermatogonia that is pushed further down the testis coil and initiates meiosis to produce 64 individualized sperm (Fabian & Brill, 2012; Fuller, 1993). Similar to GSCs, CySCs undergo self-renewing divisions to produce a CySC that remains associated with the hub, and a daughter cyst cell that functions to nurture spermatogenic differentiation (Zoller & Schulz, 2012). During spermatogenesis, two non-mitotic somatic cyst cells wrap around each gonialblast and then grow in size to envelope the differentiating spermatogonia throughout spermatogenesis (Fuller, 1993; Zoller & Schulz, 2012).
Signaling between hub cells, CySCs, and GSCs is critical for niche homeostasis and continued fertility (Greenspan, De Cuevas & Matunis, 2015; Herrera & Bach, 2019; Leatherman, 2013). Key pathways involved in GSC and CySC maintenance include the Janus kinase-signal transducer and activator of transcription (Jak-STAT), Hedgehog (Hh), and bone morphogenetic protein (BMP) pathways. Jak-STAT and Hh signaling in CySCs are induced via secretion of activating ligands from the hub to promote CySC maintenance (Amoyel et al., 2013; Kiger et al., 2001; Leatherman & DiNardo, 2010; Michel et al., 2012; Tulina & Matunis, 2001; Zhang et al., 2013), while secretion of BMP activating ligands from hub cells and CySCs promote GSC self-renewal (Kawase et al., 2004; Leatherman & DiNardo, 2008; Leatherman & DiNardo, 2010; Michel et al., 2011; Shivdasani & Ingham, 2003). Thus, defects in hub and CySC function can lead to either loss of GSCs or the expansion of undifferentiated GSCs as germline tumors. Because cyst cells produced by CySCs are required for spermatogenic differentiation, defects in cyst cell differentiation can also cause germ cell tumors and infertility (Hudson et al., 2013; Kiger, White-Cooper & Fuller, 2000; Lim & Fuller, 2012; Matunis et al., 1997; Schulz, SI & Fuller, 2002; Tran, Brenner & Di Nardo, 2000).
Jak-STAT signaling was among the first pathways shown to control testis stem cell maintenance (Kiger et al., 2001; Tulina & Matunis, 2001). In the germline, Jak-STAT activation promotes adhesion of GSCs to the hub (Leatherman & DiNardo, 2010), while it acts in CySCs to repress cyst differentiation by altering gene transcription (Herrera & Bach, 2019). Among the genes induced by the Jak-STAT pathway in CySCs is the transcription factor, chronologically inappropriate morphogenesis (Chinmo). Chinmo is evolutionarily conserved as ZFP509/ZNF509 in mammals and functions to regulate hematopoiesis, neural and eye development, as well as GSC and CySC behavior in Drosophila (Flaherty et al., 2010; Grmai et al., 2018; Grmai et al., 2021; Ma, Wawersik & Matunis, 2014; Tseng et al., 2022). In CySCs, chinmo has been shown to promote CySC maintenance, with over-expression of Chinmo in somatic support cells causing CySC expansion that leads to GSC over-proliferation (Flaherty et al., 2010). Chinmo is also known to regulate maintenance of male CySC sexual identity. Indeed, loss of chinmo function in the adult testis has been shown to cause conversion of male CySCs to ovarian follicle stem cells (FSCs) (Grmai et al., 2018; Ma, Wawersik & Matunis, 2014). Under these conditions, CySCs fail to splice mRNA encoded by the evolutionarily conserved double-sex (dsx) gene into its male form (Grmai et al., 2018), causing formation of germ cell tumors and infertility. Interestingly, loss of the mammalian dsx homolog, double-sex/mab-3 related transcription factor-1 (Dmrt1), in post-natal mouse testes, causes feminization of somatic Sertoli cells that support spermatogenesis in a manner similar to Drosophila cyst cells (Matson et al., 2011). Thus, not only does Chinmo regulate CySC self-renewal and sex-maintenance, but it acts to promote sex-maintenance through mechanisms that are conserved between flies and mammals.
To better understand mechanisms by which Chinmo impacts CySC behavior, other aspects of Drosophila development, and to understand the roles of ZFP509 and other putative Chinmo orthologs in mammals, structure function analyses are critical. Inspection of the Chinmo cDNA and ORF reveals several notable features. Genomic analyses indicate the existence of six distinct chinmo transcripts encoding a 604 amino acid and an 840 amino acid protein, respectively (https://flybase.org/). Chinmo is a member of the BTB-ZF (broad-complex, tramtrack and bric-à-brac - zinc finger) protein family, which frequently plays a role in transcriptional regulation (Siggs & Beutler, 2012). In mammalian BTB-ZF proteins, the ZF domain determines sequence specificity while the BTB domain promotes oligomerization and the recruitment of transcriptional repressors (Siggs & Beutler, 2012). The various mammalian BTB-ZF domain proteins are involved in lymphocyte development, fertility regulation, skeletal morphogenesis, and neural development. In Drosophila, BTB-ZF genes other than chinmo, such as fruitless, abrupt, and lola, are involved in neurological development and control a variety of processes, such as cell differentiation, organ formation, and sex-specific behavior (Ito et al., 1996; Sato et al., 2019; Zhu et al., 2006). We find that Chinmo also contains several putative motifs for covalent or noncovalent interaction with the post-translational modifier SUMO (Small Ubiquitin-like Modifier) (Kerscher, 2007). SUMO can be covalently linked to specific target proteins to modify their activity, localization, interactions, and half-life (Kerscher, 2007; Kerscher, Felberbaum & Hochstrasser, 2006). Functionally, proteins that undergo modification by SUMO perform essential roles including those in transcription, mRNA processing, DNA replication, intracellular transport, DNA-damage response, and stress tolerance (Flotho & Melchior, 2013; Ryu, Ahn & Hochstrasser, 2020) . One example of a sumoylated protein that functions in testes CySCs is Cubitus interruptus (Ci), which acts in the Hh signaling pathway and is critical for CySC maintenance (Lv et al., 2016). The sumoylation sites found in Chinmo thus raise the possibility that SUMO targets Chinmo for modification and regulates its interactions and essential functions.
Here, we show that Chinmo interacts with itself, SUMO, and five other proteins, 3 of which are novel in function (CG4318, Ova (Ovaries absent), Taf3 (TBP-associated factor 3), CG18269, and CG11180). CG11180 encodes an uncharacterized Drosophila protein orthologous to human PINX1 which functions to regulate ribosome biogenesis and telomerase activity (annotations on https://flybase.org/; Larkin et al., 2021). CG11180 was identified six times in our Chinmo interactor screen and was thus chosen for further analysis. We show that the interaction between Chinmo and CG11180 requires CG11180′s SIM, indicating SUMO-dependency. Furthermore, Chinmo’s interaction with both CG11180 and SUMO are functionally relevant in vivo as knockdown of either of these two proteins in somatic cells of the adult Drosophila testis yields germ cell tumor phenotypes similar to those observed after somatic chinmo knockdown. Due to its interactions with Chinmo and its tumor associated germ cell differentiation defects, we propose to name CG11180, Chigno (Childless Gambino). Together, our data suggest that SUMO may functionally link the activity of Chinmo and CG11180/Chigno in somatic cells of the adult Drosophila testis, thereby playing a key role in male fertility and spermatogenic differentiation.
Materials and Methods
Portions of this text were previously published as part of a preprint: https://doi.org/10.1101/2021.11.03.467147.
Yeast two-hybrid assays
Two-hybrid screening and assays were performed as described in the Matchmaker™ Pretransformed Libraries User Manual (Takara PT3183-1) using a Drosophila Mate & Plate™ Library—Universal Drosophila (Normalized A+ RNA isolated from embryo (∼20 hr), larval, and adult stage Drosophila melanogaster) Cat. No. 630485. Briefly, a fusion of Chinmo(native) with the Gal4 DNA Binding domain (BD) was expressed in yeast (AH109) and then mated to yeast two-hybrid library of normalized Drosophila melanogaster cDNA clones transformed in a MATa GAL4 reporter strain (Y187). Chinmo interactors were identified as colonies that robustly activated the ADE2, HIS3 reporter genes and formed blue colonies on SD/-Ade/-His/-Leu/-Trp/X-alpha-Gal media. Individual clones were isolated, sequenced, and retested (see Fig. 1C). Yeast strains and plasmids are listed in Table S2.
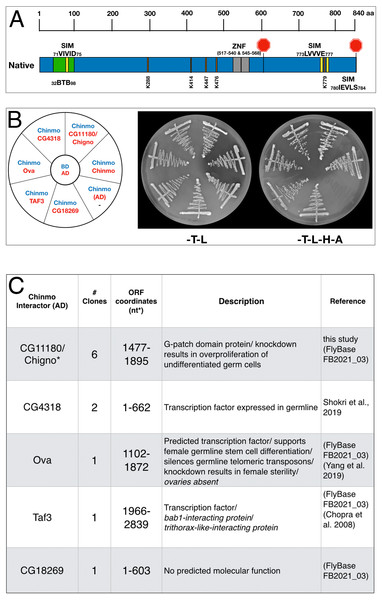
Figure 1: Chinmo protein domains and interactors.
(A) Chinmo contains a BTB protein interaction domain spanning residues 32 to 98 and two zinc finger domains (ZNF) spanning from 517 to 540 and 545 to 568. Two stop codons in the cDNA terminate translation at residue 604, or, if read-through occurs, at residue 840. Furthermore, SUMO interaction motifs and sumoylation sites are shown in yellow and orange respectively. Note that a putative SIM domain in Chinmo is contained within the BTB protein-interaction domain. (B) Two-hybrid analysis shows that Chinmo interacts with five novel proteins (encoded by CG4318, Ova (Ovaries absent), Taf3 (TBP-associated factor 3), CG11180, and CG18269) and other Chinmo proteins. Left: graphic representation of BD-Chinmo (blue) and its interactors (red). BD-Chinmo/AD-empty vector is a negative control. Middle: The presence of BD-Chinmo (in pOBD2/TRP1) and the indicated AD constructs (in pOAD/LEU2) was confirmed by growth on growth media lacking tryptophan and leucine (-T-L). Right: The interaction between Chinmo constructs and Chinmo interactors is confirmed by growth on media lacking lacking tryptophan, leucine, histidine, and adenine (T-L-H-A). Plates were imaged after three days of growth. (C) Shown are the gene/protein name, approximate coordinates of the cDNA fragment identified, description, and reference.Protein domain prediction
Prediction of SIMs and sumoylation consensus sites was done using GPS SUMO (https://sumo.biocuckoo.cn/). Only high confidence SIMs and sumoylation sites are reported (Zhao et al., 2014). Other domains in Chinmo and CG1180/Chigno were mapped using https://prosite.expasy.org (Sigrist et al., 2013). Sequences and transcript analyses were retrieved from flybase.org (Larkin et al., 2021).
Cloning and site-directed mutagenesis
BD-Chinmo clones were derived using a commercially available cDNA clone (SD04616) as template and PCR primers specific for the long and short Chinmo open-reading frame. The resulting Chinmo amplicons were recombined into two-hybrid plasmid pOBD2 (www.addgene.org; pOBD2 (Plasmid #196826) http://depts.washington.edu/sfields/protocols/pOBD2.html). Mutagenic primers and the Q5 site-directed mutagenesis kit (NEB E0554S) were used to generate the BD-Chinmonon-stop and the CG11180/Chigno-sim* mutant. All clones were confirmed using sequence analysis and, in the case of Chinmo, western blotting.
Fly stocks
c587-Gal4 (Kai & Spradling, 2003) and eyaA3-Gal4 (Leatherman & Dinardo, 2008) were used to drive UAS-transgene expression in the somatic CySCs and early cyst cells (the CySC lineage). UAS- lines include: UAS-CG11180-RNAi (Bloomington stock #28629; TRiP.JF03044;), UAS-Smt3-RNAi (Bloomington stock #28034; TRiP.JF02869), UAS-CG5694-RNAi (Bloomington stock #62485;TRiP.HMJ23969), UAS-CG4318-RNAi (Bloomington stock #51720;TRiP.HMC03257), UAS-Bip2/Taf3-RNAi (Bloomington stock #43174;TRiP.GLI01516). y,w1118 flies were used as wild type, with controls generated by mating c587-Gal4 or EyaA3-Gal4 to y,w1118 flies unless otherwise noted. Fly stocks were obtained from Bloomington Stock Center (http://flystocks.bio.indiana.edu/) unless otherwise specified.
Testis collection & immunostaining
Flies were raised at 25 °C unless otherwise indicated. To prevent Gal4-UAS activity during development, flies were reared at 18 °C and allowed to eclose as adults for 48–72 h, after which flies were shifted to 25 °C. Adult testes were dissected, fixed and immunostained at designated ages as described (Matunis et al., 1997). At least two replicas of each immunostaining experiment were performed with data for quantitative analyses. Antibodies used for immunostaining were Rabbit anti-Vasa at 1:500 (R. Lehmann) or Rat anti-Vasa at 1:60 (A.C. Spradling/D. Williams; DSHB), mouse anti-Hts/1B1 (H. Lipshitz; DSHB) at 1:4, and goat anti-rabbit Alexa 568 (Invitrogen; A-11011), goat anti-rat RRX (Jackson Labs; 112-295-068), or goat anti-mouse Alexa 488 (Jackson Labs; 205-545-108) at 1:500. Nuclei were stained using DAPI at 1 µg/mL (Roche) for 3 min.
Microscopy & phenotype classification
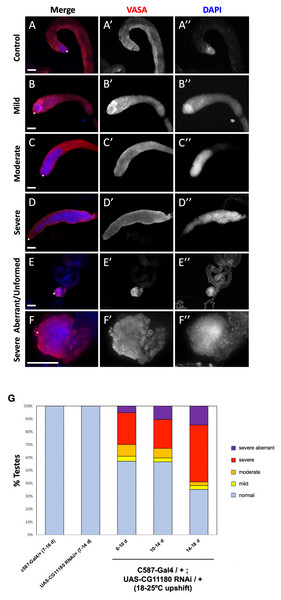
Samples were mounted in 70% glycerol containing 2.5% DABCO (Sigma) and p- phenylenediamine anti-fade agent (Sigma) at a final concentration of 0.2 mg/mL. Slides were viewed with an Olympus BX51 microscope equipped with a DSU spinning disc confocal system and Q-imaging RETIGA-SRV CCD camera. Images were captured and analyzed with Slidebook software by 3I. For scoring of phenotype severity, wild type testes with high-level DAPI staining restricted to the testis apex where less differentiated germ cells reside (Tulina & Matunis, 2001) were scored as normal. An increase in DAPI-bright region extending up to 10% the length of the testes coil was scored as mild, 10–30% scored as moderate, and greater than 30% of the testes coil scored as severe. Testes with expansion of less differentiated DAPI-bright germ cells but lacking an obvious testes coil were scored as severe aberrant/underdeveloped. Data shown in graphs represent pooled numbers from at least two replicates of each genotype/treatment.
Results
Identification of Chinmo interactors
Chinmo contains a BTB protein interaction domain (amino acids (aa) 32–98), two Zinc Finger Domains (aa 517–545 and 545–573), three putative SUMO-interacting motifs (SIM) (aa 71–75, 773–777, 780–784), and at least 5 SUMO consensus sites (K288, K414, K447, K476, K779) (Fig. 1A). These domains and motifs raise the possibility that Chinmo interacts with multiple different proteins, (e.g., other Chinmo proteins or transcriptional repressors), SUMO, and sumoylated proteins. However, additional analyses are required to identify Chinmo interacting proteins that could provide functional clues. Therefore, we used a two-hybrid screen to identify these proteins and investigate Chinmo’s interactions in detail. To perform this analysis, we obtained a Chinmo 4259 nucleotide (nt) cDNA (SD04616) to clone the Chinmo ORF in frame with the Gal4 DNA Binding domain (BD) into the pOBD2 two-hybrid vector. Inspection of the Chinmo coding sequence in this cDNA clone revealed an 1,815 nt ORF, terminating in a single STOP codon. This ORF translates into a 604 aa Chinmo protein containing all indicated protein interaction domains and motifs (Fig. 1A). Curiously, we noticed that omission of the STOP codon (potentially by read-through) yields a 2,529 nt ORF encoding a previously reported 840 aa long Chinmo protein isoform (http://flybase.org/reports/FBgn0034528). To assess potential differences between the short and long Chinmo ORFs, we generated the following BD-Chinmo clones: BD-Chinmonative (with the Stop at nt 1,813), and BD-Chinmonon-stop (lacking the Stop codon at position 1813), and BD-ChinmoSTOP (omitting sequences after the Stop codon at position 1,813) (Fig. S1A). Western blots showed that all versions of BD-Chinmo expressed fusion proteins were of the expected size, ∼90 kDa for BD-Chinmonative and BD-ChinmoSTOP compared to ∼135 kDa for the BD-Chinmonon-stop (Rinehart, 2017).
To identify Chinmo interacting proteins, we used the BD-Chinmonative clone, which expresses a fusion of the Gal4-BD to 604 aa of Chinmo corresponding to the shorter transcript variant, to screen a two-hybrid library of D. melanogaster cDNAs fused to the Gal4 Activation Domain (AD-cDNA) (See materials and methods). In this screen, we identified 5 different AD-cDNA fusions (CG11180, CG4318, Ova, Taf3, and CG18269). These constructs resulted in reporter gene activation (ADE2, HIS3) when co-expressed with BD-Chinmo and robust growth on media lacking adenine and histidine (referred to as -H-A) (Fig. 1B). As a negative control, BD-Chinmo alone (Chinmo/AD) did not result in reporter gene activation as expected (see Figs. 1B and 1C). Two Chinmo-interactors identified in our screen (CG4318 and Ova) are transcription factors functionally linked to germline differentiation (FlyBase, 2021; Yang et al., 2019), and Taf3 is involved in transcriptional activation and regulation (Chopra et al., 2008). CG11180 and CG18269 encode proteins of unknown function (Fig. 1C). While an interaction between CG4318 and Chinmo was previously identified in a high throughput interactome study (Shokri et al., 2019), interactions between Chinmo and the other four proteins are novel.
As part of our screen for Chinmo interactors, we also included an AD-Chinmo clone and found that it interacts with the BD-Chinmonative clone used for library screening. In support of choosing BD-Chinmonative for our 2-hybrid screen, we did not discern a difference in the interaction between the different BD-Chinmo clones (Native, Non-Stop, and Stop) and AD-Chinmo (Fig. S1B). In summary, our 2-hybrid screen identified 5 novel Chinmo-interacting proteins and also revealed that Chinmo dimerizes with itself.
Chinmo interaction with SUMO
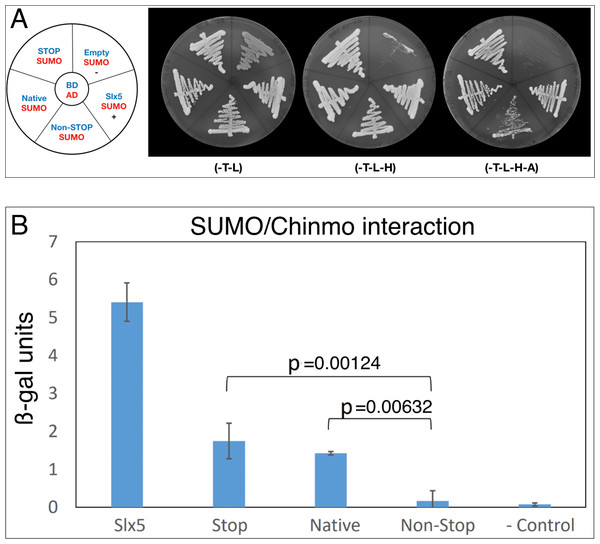
Because Chinmo has several SIMs (Fig. 1A) and we did not identify SUMO in our screen, we also directly assayed Chinmo’s predicted interaction with SUMO. SUMO/Smt3 is a structurally highly conserved protein with the yeast and Drosophila orthologs bearing 42% sequence identity and 57% similarity. Thus, we tested the interaction of ScSmt3 from yeast (AD-SUMO) with all three BD-Chinmo variants. In our assay, AD-SUMO interacted with all three BD-Chinmo variants (BD-Chinmonative, BD-Chinmonon-stop, and BD-ChinmoSTOP) on media lacking histidine. BD-Slx5, a SUMO interacting protein with multiple SIMs, served as a positive control and as expected, AD-SUMO alone (empty/SUMO) did not result in reporter gene activation. Using a more stringent selection (SD-H-A) recapitulated our finding that SUMO interacts robustly with BD-Chinmonative and BD-ChinmoSTOP. However, the interaction of Chinmonon-stop with SUMO was weaker, as apparent from reduced growth on SD-H-A media (Fig. 2A). An ONPG ß-galactosidase assay to quantitate the interaction of SUMO and Chinmo also showed that the STOP and Native Chinmo clones showed equivalent reporter gene activation, while the interaction of AD-SUMO and the Chinmonon-stop was reduced to background levels (Fig. 2B). In summary, our data show that both BD-Chinmonative and BD-ChinmoSTOP interact with SUMO, and that this interaction may depend on the SIM (aa 71–75) in Chinmo.
Figure 2: Chinmo interacts with SUMO.
(A) Two-hybrid analysis shows that BD-Chinmo Native and BD-Chinmo Stop (and to a lesser degree BD-Chinmo Non-Stop) interact with AD-SUMO. Left: Graphic representation of the three BD-Chinmo clones (Native, Stop, and Non-Stop) (blue) and AD-SUMO (red). Negative (BD-empty vector with AD-SUMO) and positive controls (BD-Slx5 with AD-SUMO) are as indicated. Middle: The presence of the indicated BD-Chinmo constructs (in pOBD2/TRP1) and AD-SUMO (in pOAD/LEU2) was confirmed by growth on growth media lacking tryptophan and leucine (-T-L). Right: Interactions are confirmed by growth on media lacking tryptophan, leucine, histidine, and adenine (T-L-H and -T-L-H-A). Plates were imaged after three days of growth. (B) Quantification of the interaction of SUMO with the indicated Chinmo proteins using a quantitative ONPG assays and strains shown in A. Tukey Kramer multiple comparison procedure was used to determine significant differences in SUMO interaction.The interaction of Chinmo with CG11180/Chigno depends on SUMO
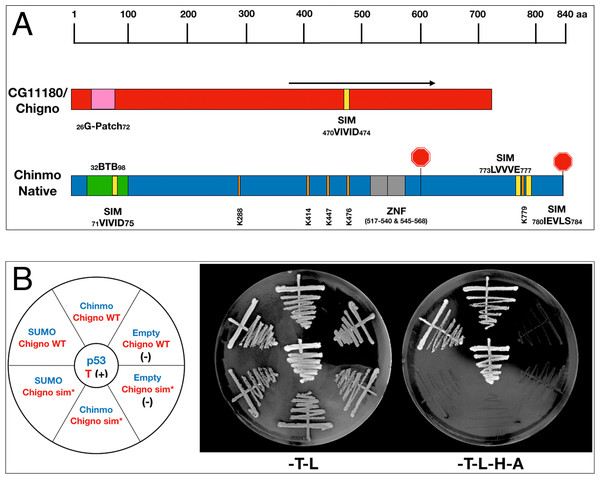
Both Chinmo and the partial cDNA of CG11180/Chigno (aa 373–621) isolated in our screen contain SIM domains (Fig. 3A). Therefore, we also assessed if Chinmo’s interaction with CG11180/Chigno is SUMO-dependent. For this analysis, we focused on the functional role of a SIM mutant (sim*) in AD-CG11180/Chigno for two reasons: (i) One of Chinmo’s SIMs resides within the BTB domain and a sim* mutant may affect its dimerization, and (ii) Chinmo has multiple sumoylation sites that potentially interact with the SIM in CG11180/Chigno (unlike the Chigno clone). For our analyses, we created an AD-CG11180/Chignosim∗ mutant by site-directed mutagenesis, replacing the SIM in CG11180/Chigno (VIVID) with alanine’s (AAAAA). We then tested both the AD-CG11180/Chignosim∗ mutant and the AD-CG11180/Chigno clone for the ability to interact with BD-SUMO, BD-Chinmo, and an empty BD control plasmid. We found that AD-CG11180/Chigno interacts with both BD-SUMO and BD-Chinmo. In contrast, the AD-CG11180/Chignosim∗ mutant failed to interact with both BD-SUMO and BD-Chinmo (Fig. 3B). As a negative control, the empty BD vector failed to elicit reporter gene activation with both the AD-CG11180/Chignosim∗ mutant and the AD-CG11180/Chigno clone. Additionally, the positive control (p53/T) showed reporter gene activation, as expected. Together, these results suggest that the VIVID sequence in CG11180/Chigno constitutes a bona-fide SIM that could be required for the interaction with Chinmo.
Figure 3: Chinmo’s interaction with CG11180/Chigno depends on a SIM in Chigno.
(A) Comparison of protein domains and motifs in CG11180/Chigno (red) and Chinmo (blue). Coordinates of SIMs and sumoylation consensus sites as shown. Note that the SIM domain in Chinmo is contained within the BTB protein-interaction domain. The black arrow demarcates the CG11180/Chigno protein fragment encoded by the cDNA isolated in our screen (373–621). (B) Left: Graphic representation of BD-Chinmo constructs (blue) and AD-CG11180/Chigno, either containing or lacking the SIM at residues 71 to 75 (red). Negative controls (empty pOBD2/TRP1) and a positive control (p53/T) are as indicated. Middle: The presence of the indicated BD constructs (blue: pOBD2/TRP1) and either AD-CG11180/Chigno WT or AD-CG11180/Chigno sim* (pOAD/LEU2) was confirmed by growth on growth media lacking tryptophan and leucine (-T-L). Right: Interactions are confirmed by growth on media lacking tryptophan, leucine, histidine, and adenine (-T-L-H-A). Plates were imaged after three days of growth.CG11180/Chigno and SUMO are required in somatic cells of the adult Drosophila testis
We next assessed the functional significance of identified Chinmo-interacting proteins in vivo. Because Chinmo functions in somatic CySCs of adult Drosophila testes (Flaherty et al., 2010; Grmai et al., 2018; Ma, Wawersik & Matunis, 2014), we performed RNAi knockdown of Chinmo-interacting proteins in somatic support cells using the Gal4-UAS system (Brand & Perrimon, 1993). Specifically, UAS-RNAi constructs against Chinmo-interactors identified through our screen, as well as SUMO/Smt3, were expressed using the c587-Gal4 driver (Kai & Spradling, 2003) which is frequently used to induce high-level gene expression in CySCs and early cyst cells (Demarco et al., 2014; Lv et al., 2016; Ma, Wawersik & Matunis, 2014). Adult testes were then isolated and processed for fluorescence immunostaining. Because functional CySCs and cyst cells are critical for normal germ cell differentiation, and inhibition of Chinmo results in germ cell differentiation defects associated with over-proliferation of under-differentiated germ cells, testes were assayed for germ cell differentiation defects in our initial analysis presented here.
We find that somatic knockdown of CG4318, ova, or Taf3 show no germ cell differentiation defects in 0–4 day old adult testes (Fig. S2). Similar results were obtained for 7–12 day old testes, though ova knockdown caused lethality in aged flies and these data will be reported elsewhere. However, RNAi knockdown of CG11180/Chigno and SUMO/Smt3 using the c587-Gal4 driver caused developmental lethality in flies reared at 25 °C. For technical reasons, we were unable to test a UAS-RNAi construct for the impact of CG18269.
To assess if CG11180/Chigno functions in adult testes, experiments were adapted to prevent developmental lethality. Specifically, flies carrying the somatic c587-Gal4 driver and a UAS-CG11180-RNAi construct were reared at lower temperature (18 °C) to reduce Gal4 function. Upon emergence from their pupal cases (eclosion), adult male flies were then upshifted to 25 °C to permit full Gal4 activation. Under these conditions, we found that knockdown of CG11180/Chigno in somatic cells of the adult testis results in formation of germ cells tumors marked by over-proliferation of undifferentiated germ cells that are not observed in controls carrying only the c587-Gal4 driver or UAS-Chigno-RNAi construct (Fig. 4; Table S1). Severity of phenotype varied (Fig. 4), with ∼38% of 6–10 day old testes showing mild to severe germ cell over-proliferation phenotypes (Figs. 4B–4D, 4G; Table S1), and ∼4% displaying a severely aberrant/unformed phenotype where testes lack their normal coiled structure and show an absence of spermatogonial differentiation (Figs. 4E–4G; Table S1). Defects in spermatogonial differentiation and the over-proliferation of undifferentiated germ cells was confirmed by immunostaining for fusomes, a germline specific organelle that shows distinct morphology in GSCs and gonialblasts compared to differentiating spermatogonia (Fig. S3). Consistent with adult induction of RNAi knockdown after temperature upshift, phenotype severity increases with age, with ∼44% of testes showing a phenotype 6–10 days after somatic knockdown while ∼65% are phenotypic at 14–18 days (Fig. 4G; Table S1).
Figure 4: Somatic knockdown of the Chinmo interactor, CG11180/Chingo, in adult testes impacts germ cell proliferation and differentiation.
(A–F) Adult testes 7–10 days after eclosion immunostained with anti-Vasa (red) and DAPI (blue) to reveal germ cells or nuclei respectively. Vasa and DAPI shown alone in black and white (A’–F’ and A”–F”, respectively). Testes apex containing the GSC niche indicated with an asterisk. Scale bars = 70 µm. (A) c587-Gal4/+ control testis showing presence of undifferentiated germ cells with spherical morphology and bright DAPI staining near the GSC niche and differentiating spermatogonia with dim DAPI staining extending away from the niche. (B–F) c587-Gal4/+; UAS-CG11180-RNAi/+ flies reared at 18 °C then upshifted to 25 °C showing defects in germ cell differentiation marked by expansion of undifferentiated germ cells that display spherical morphology and bright DAPI staining. Representative images of testes with mild (B), moderate (C), and severe (D) phenotype are shown. A severe aberrant/unformed testis at low (E) and high magnification (F) lacking an obvious testes coil while showing expansion of undifferentiated germ cells is also shown. (G) Graph showing percentage of control and c587-Gal4/+; UAS-CG11180-RNAi/+ testis displaying phenotype at specific ages, with phenotype severity increasing over time after somatic CG11180 knockdown. Sample sizes are: c587-Gal4/+ (7-16d), n = 56; UAS-CG11180 RNAi/+ (7-14d), n = 22; c587-Gal4/+; UAS-CG11180 RNAi/+ (6-10d), n = 77; c587-Gal4/+; UAS-CG11180 RNAi/+ (10-14d), n = 67; c587-Gal4/+; UAS-CG11180 RNAi/+ (14-18d), n = 34.To determine if SUMO/Smt3 functions in somatic cells of adult testes, we used another common CySC/early cyst-Gal4 driver (EyaA3-Gal4; Demarco et al., 2014; Leatherman & Dinardo, 2008) to induce expression of the UAS-SUMO/Smt3-RNAi transgene. Under these conditions, flies were viable whether reared at 25 °C, or at 18 °C then upshifted to 25 °C. Furthermore, under both conditions, adult males displayed a similar phenotype as CG11180/Chigno knockdown testes, with germ cell tumors marked by over-proliferation of undifferentiated germ cells (Figs. 5A–5B). Like CG11180/Chigno knockdown testes, this phenotype was observed at varying degrees (Fig. 5; Table S1), with ∼96% of testes showing a phenotype in samples reared at 25 °C (Figs. 5A, 5D; Table S1). Phenotype induction was also observed after temperature upshift, with 1–3 day old testes lacking a phenotype, but ∼93% of testes developing a phenotype 7 days after upshift (Fig. 5B, 5D; Table S1). Confirmation of germline differentiation defects and over-proliferation of undifferentiated germ cells was confirmed via fusome staining (Fig. S3). Taken together, these data suggest that CG11180/Chigno and SUMO/Smt3 play an important role in the regulation of somatic cell behavior in adult testes, with indirect impacts on germ cell proliferation and differentiation. However, it should be noted that the phenotypes observed after somatic SUMO knockdown should not necessarily be interpreted as solely related to Chinmo since SUMO depletion is likely to affect a variety of other processes, including Hh signaling in the testis CySCs (Lv et al., 2016). Specific mechanisms by which these genes function in the testis soma, and their connection to each other as well as Chinmo, are topics for future studies.
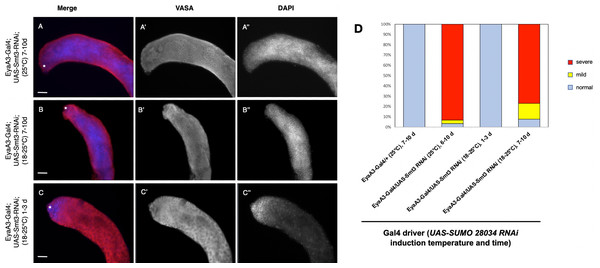
Figure 5: Somatic knockdown of SUMO/Smt3 in the adult testis causes proliferation of undifferentiated germ cells.
(A–C) EyaA3-Gal4; UAS-Smt3-RNAi adult testes immunostained with anti-Vasa (red) and DAPI (blue) to detect germ cells and nuclei respectively. Vasa (A’–C’) and DAPI (A”–C”) are displayed individually in black and white. The testes apex containing the GSC niche is indicated with an asterisk. Scale bar = 70 µm. (A) 7-10d EyaA3-Gal4; UAS-Smt3-RNAi testis reared at 25 °C displaying severe overproliferation of undifferentiated germ cells as indicated by expansion of spherical germ cells with strong DAPI fluorescence (compare to control in Fig. 4A). (B) 7-10d EyaA3-Gal4; UAS-Smt3-RNAi testis upshifted from 18 to 25 °C for 7 days displaying a similar phenotype marked by overproliferation of undifferentiated germ cells. (C) 1-3d Eya A3-Gal4; UAS-Smt3-RNAi testis raised at 18 °C then upshifted for 1 day is similar to controls. (D) Graph showing variance in phenotype severity in EyaA3-Gal4; UAS-Smt3-RNAi testes with different treatments as compared to controls. Samples sizes are: EyaA3-Gal4/+ (25 °C) 7-10d, n = 36; EyaA3-Gal4/UAS-Smt3 RNAi (25 °C) 6-10d, n = 28; EyaA3-Gal4/UAS-Smt RNAi (18°–25 °C) 1-3d, n = 16; EyaA3-Gal4/UAS-Smt3 RNAi (18–25 °C) 7-10d, n = 12.DISCUSSION
In this study we set out to identify Chinmo-interacting proteins using a two-hybrid screen and targeted two-hybrid assays. Our results suggest that Chinmo, a putative BTB-zinc finger transcription factor, interacts directly with three uncharacterized proteins, several transcriptional regulators, SUMO/Smt3, and itself. Chinmo dimerization is not surprising as BTB-zinc finger transcription factors are known to homo and hetero-dimerize via their BTB domains. Because most BTB-domain proteins are transcription factors, their homo-dimerization is predicted to increase affinity for their targets and modulate transcriptional repression properties. In contrast, some BTB domain proteins also act as substrate adapters for ubiquitin E3 ligases, targeting heterologous substrate proteins they bind for ubiquitination (Perez-Torrado, Yamada & Defossez, 2006).
Among the Chinmo-interacting proteins, CG4318, Ova, Taf3, and SUMO stand out for their previously identified roles in Drosophila reproductive tissues. Specifically, CG4318 encodes a putative transcription factor expressed in the germline that was previously identified as a Chinmo-interacting protein through a two-hybrid screen designed to map the Drosophila transcription factor interactome (Shokri et al., 2019). Identification of CG4318 in this study, therefore, validates our experimental approach. Additionally, Ova is required for fertility in female flies and functions to support ovarian germline stem cell differentiation (Yang et al., 2019), while Taf3 is required for the expression of germline genes in primordial germ cells (Yatsu et al., 2008) and is known to interact with proteins, like Chinmo, that contain BTB domains (Pointud et al., 2001). In the present study, we did not observe an obvious phenotype for the knockdown of CG4318, Ova, and Taf3 in somatic cells of the testis. It is, therefore, possible that these proteins interact with Chinmo in the germline, or in other tissues with Chinmo expression such as the nervous system where Chinmo regulates neuronal temporal identity during post-embryonic brain development (Yu & Lee, 2007).
Since Chinmo has multiple SUMO-interacting motifs and SUMO consensus sites, we also predicted to identify SUMO or sumoylated proteins in our screen. While we failed to isolate a Drosophila SUMO clone in our initial screen, we confirmed the interaction of Chinmo with SUMO in a two-hybrid assay with yeast ScSMT3. It is, therefore, possible that the Drosophila cDNA prey library we used did not contain a functional dmSmt3 clone or that dmSmt3 interactions and conjugation fail to take place in budding yeast cells. Supporting the latter, an AD-dmSmt3 construct we cloned and expressed in the yeast smt3-331ts strain fails to complement this temperature-sensitive mutant at 37 °C (data not shown). Accordingly, human HsSUMO3 (which is related to fly DmSmt3) also fails to complement a deletion of ScSmt3 in yeast, reportedly because hsSUMO3 is not compatible with precursor processing and E1 activation (Newman et al., 2017; Ureña et al., 2016). Therefore, a notable caveat to our data is that interactions between Chinmo, Chigno and Drosophila Smt3 are subject to confirmation through ongoing studies. Consistent with a role for Drosophila SUMO functioning in concert with Chinmo, however, SUMO and sumoylation genes are expressed in the Drosophila germline during spermatogenesis (Hashiyama, Shigenobu & Kobayashi, 2009). As previously noted SUMO has also been shown to play a role in CySC regulation in Drosophila testes. Specifically, SUMOylation promotes CySC self-renewal via upregulation of the Hedgehog (Hh) signaling pathway (Lv et al., 2016). Indeed, the Hh pathway transcription factor, Cubitus interruptus (Ci), interacts directly with the SUMO E2-conjugating enzyme, Lesswright (Lwr). Additionally, inhibition of Lwr in somatic support cells causes a reduction in CySC number and pre-mature cyst cell differentiation (Lv et al., 2016). Interestingly, a reduction of CySCs might be expected to cause defects in GSC maintenance, rather than the over-proliferation of undifferentiated germ cells we observe after somatic knockdown of SUMO/Smt3. Therefore, while Lv et al. clearly show a role for sumoylation in modulating Hh-mediated CySC self-renewal, our observations indicate that SUMO must play additional roles in somatic support cells. Our data suggest that one of these roles may be non-covalent binding of SUMO to Chinmo to facilitate interactions with other proteins that are sumoylated or contain SIMS, though future analyses are required to verify these interactions in vivo.
Less is known about function of the novel proteins, CG18269 and CG11180/Chigno. While we did not yet assess the impact of CG18269 knockdown, there is some evidence suggesting the gene product of CG18269 may function in the testis. Specifically, CG18269 is downregulated in testis where germ cells are subjected to oxidative stress (Tan, Suda & Baeg, 2018). Furthermore, while CG18269 expression levels are low in adult Drosophila tissues, RNA-Seq indicates an enrichment in the adult testis (Chintapalli, Wang & Dow, 2007). Thus, it is possible that CG18269 functions alongside Chinmo in the adult germline and/or somatic cells of the testis niche.
The main focus of analysis of Chinmo interactors fell on CG11180/Chigno because somatic knockdown of CG11180/Chigno caused formation of germ cell tumors. This provides evidence that CG11180/Chigno functions in somatic support cells where Chinmo regulates somatic sex maintenance and stem cell renewal. This notion is further supported by a whole-genome RNAi screen where CG11180/Chigno knockdown in the CySC lineage leads to male infertility marked by lack of germ cell differentiation (Fairchild, Islam & Tanentzapf, 2017). Further discussion of potential roles for CG11180/Chigno in adult testes and the significance of its interactions with Chinmo are presented below. A model by which SUMO may coordinate function of Chinmo-Chinmo as well Chinmo-Chigno interactions is also presented (Fig. 6).
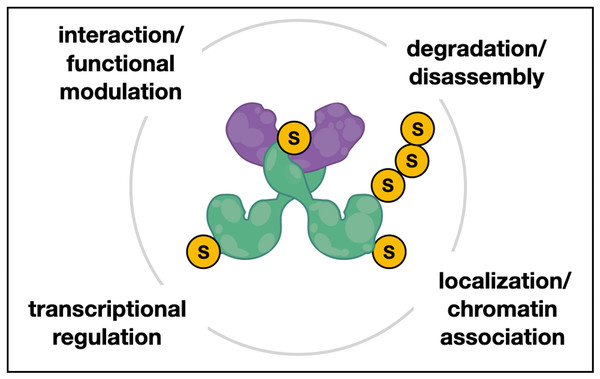
Figure 6: Hypothetical model of Chinmo/SUMO/Chigno interaction in testes.
Hypothetical model of interactions between the conserved BTB-ZF protein Chinmo (green dimer), the PINX1-related protein Chigno/CG11180 (purple), and SUMO (S). Both Chinmo and Chigno/CG11180 have SUMO interacting motifs (SIMs) for non-covalent SUMO binding that may aid in their interaction or the interaction with other SUMO-associated and/or sumoylated proteins. Additionally, Chinmo has several putative sumoylation sites that may facilitate SUMO-related processes such as regulation of Chinmo’s transcriptional activity, it’s localization or association with chromatin, and possibly disassembly and/or turnover of this complex.CG11180/Chigno function and the significance of Chinmo-Chigno interactions
As previously indicated, formation of germ cell tumors marked by germline over-proliferation and germ cell differentiation defects after somatic knockdown of CG11180/Chigno provides physiological evidence for CG11180/Chigno function in somatic cells of the testes. The direct interaction between CG11180/Chigno and Chinmo also suggests that these two proteins can function together in this tissue. Indeed, a CG11180/Chigno clone encompassing amino-acids 373–621 was identified six times in our 2-hybrid screen. Given Chinmo’s function as a transcription factor, it is tempting to propose that Chigno/Chinmo interactions serve to modulate Chinmo’s function in gene regulation. In support of CG11180/Chigno as a transcripitonal regulator, an interactome screen for modulators of insulin signaling identified a potential interaction between CG11180/Chigno and the transcription factor, FOXO (Vinayagam et al., 2016). Additionally, the conserved G-patch domain present in CG11180/Chigno has been linked to transcriptional regulation (Bohnsack et al., 2021). If CG11180/Chigno does function in somatic support cells to modulate Chinmo function, it will be interesting to determine if CG11180/Chigno serves to positively or negatively regulate Chinmo function. Indeed, while loss of Chinmo function in CySCs results in germ cell tumors due the conversion of CySCs to ovarian FSCs (Ma, Wawersik & Matunis, 2014), germ cell tumors with undifferentiated germ cells are also observed after Chinmo over-expression due to CySC over-proliferation (Flaherty et al., 2010). Further research must, therefore, be performed to directly examine the role of CG11180/Chigno in regulating CySC self-renewal, differentiation, and sex-maintenance, while also exploring genetic interactions between Chinmo and CG11180/Chigno that might affect these behaviors.
Novel functions of a Chigno/Chinmo complex may also be predicted based on presence of CG11180/Chigno’s conserved G-patch domain and function of its closest mammalian ortholog, PINX1. As previously noted, PINX1 is a tumor suppressor and telomerase inhibitor that has also been implicated in regulation of mRNA splicing and ribosome biogenesis (Chen et al., 2014; Guglielmi & Werner, 2002; Mouffok et al., 2021; Zhou & Lu, 2001). The conserved G-patch domain has also been linked to other functions including regulation of DEAH-box RNA helicases which control critical processes such as RNA metabolism, mRNA splicing, and ribosome biogenesis that are also linked to PINX1 function (Bohnsack et al., 2021 PMID 33857358). Thus, it is possible that a Chigno/Chinmo complex could act to regulate RNA processing to alter cell behavior, though it is also possible that CG11180/Chigno may function alone or in complex with other proteins to mediate these functions. Studying how dysregulation of these processes impact CySC and cyst behavior, and the role of CG11180/Chigno and Chinmo in RNA processing is an interesting line of inquiry for the future.
Role of SUMO in regulation of CG11180/Chigno and Chinmo
Sumoylation plays a pivotal role in transcriptional repression and the formation of protein complexes via protein sumoylation and SIMs (Kerscher, 2007; Wilkinson & Henley, 2010). As we have identified SUMO as a potential Chinmo interacting protein, it is, therefore, likely that sumoylation of Chinmo on one or several consensus site lysines regulates its activity and interactions as a transcription factor. Indeed, SUMO (ScSmt3) interacts strongly with the native and STOP variants of Chinmo that both produce ∼90 kDa proteins (Fig. 2A and Rinehart, 2017). However, deletion of a Chinmo STOP codon at position 604 (non-stop) results in expression of an artificially-lengthened Chinmo protein of ∼135 kDa that fails to interact strongly with SUMO. Whether this longer Chinmo protein exists in vivo and whether this observation is physiologically relevant or merely indicates the misfolding of an artificially-lengthened protein in yeast is currently not clear.
In our analysis we also noted that a bona-fide SIM is situated within the BTB dimerization domain of Chinmo. This suggests that SUMO-binding of this SIM could alter Chinmo’s conformation or interfere with its ability to dimerize and/or hetero-dimerize. Indeed, CG11180/Chigno also contains a SIM motif that is located at amino-acids 470–474 that facilitates binding to Chinmo; suggesting that SUMO binding might alter Chinmo-Chigno interactions. Consistent with this, we found that (1) CG11180/Chigno also interacts with SUMO and (2) mutation of CG11180/Chigno’s SIM completely abolished the interaction of CG11180/Chigno with both SUMO and Chinmo (Fig. 3). This may suggest that sumoylation of Chinmo could be required for its interaction with CG11180/Chigno. SUMO could, therefore, be utilized to recruit the Chinmo monomer to chromatin-associated CG11180/Chigno, or vice versa. A hypothesized model depicting the capacity of SUMO binding to modulate Chinmo-Chigno as well as Chinmo-Chinmo interactions is shown in Fig. 6. The functional significance of SUMO’s interactions with Chinmo and Chigno and whether SUMO binding mediates formation of Chinmo homodimers and/or Chinmo-Chigno interactions in vivo remains to be determined.
Conclusions
We hypothesized that a structure/function analysis of Chinmo and its interacting proteins will further our understanding of mechanisms controlling stem cell renewal and sex-maintenance in Drosophila. In summary, our study provides evidence for a complex relationship of Chinmo with other transcription factors and several proteins, such as SUMO, involved in somatic and germline regulation. Specifically, our structure-function analyses, combined with RNAi studies suggest that a Chinmo/Chigno/SUMO complex may function in somatic support cells to ensure fertility and prevent germ cell tumors formation. However, it is also likely that some of the Chinmo interactions we have identified are not specific to the testis soma, but rather play important regulatory roles in germ cells or other tissues (e.g., neurons). Future studies will investigate details of the subcellular localization of Chigno, provide a more detailed structure-function analysis of the predicted Chinmo/Chigno/SUMO complex, and focus on its precise role in the Drosophila testes and other tissues.
Supplemental Information
Analysis of the interaction of Chinmo variants
(A) Native, Stop, and Non-Stop based on cDNA clone SD04616. Chinmo Native and Stop constructs are predicted to encode a 604 aa proteins. Chinmo Non-Stop is predicted to encode a 842 aa Chinmo protein. Domains in all contractors as indicated: BTB protein interaction domain (residue 32 to 98) and two zinc finger domains (ZNF) spanning (residues 517 to 540 and 545 to 568), as indicated. SUMO interacting motifs (SIMs) and SUMO consensus sites are shown in yellow and orange respectively. Note that the SIM domain in Chinmo is contained within the BTB protein-interaction domain. (B) Left : Graphic representation of AD and BD pairings of the three different Chinmo clones (Native, Stop, and Non-Stop). Negative (BD-empty vector/AD-Chinmo Stop) and positive controls (p53/T) are as indicated. Middle : The presence of the indicated BD constructs (in pOBD2/TRP1) and AD constructs (in pOAD/LEU2) was confirmed by growth on growth media lacking tryptophan and leucine (-T-L). Right : The interaction of the Chinmo Native, Stop, and Non-Stop with other Chinmo proteins is confirmed by growth on media lacking tryptophan, leucine, histidine, and adenine (-T-L-H-A). Plates were imaged after three days of growth.
Somatic knockdown of the Chinmo interactors, CG4318, Taf3, and Ova, does not impact germ cell behavior
Adult testes reared at 25 °C immunostained with anti-Vasa (red) and DAPI (blue) to display germ cells or nuclei respectively. Vasa and DAPI shown alone in black and white (A’–D’ and A”–D”, respectively). The GSC niche at the testis apex is marked with an asterisk. Scale bar = 70 µm. (A) 7-10 day c587-Gal4/+ control testes have undifferentiated germ cells with spherical morphology and strong DAPI staining near the niche, and weaker DAPI stain further down the testis coil. 0–3 day (B) c587-Gal4; UAS-CG4318-RNAi, (C) c587-Gal4; UAS-Taf3-RNAi, and (D) c587-Gal4; UAS-Ova-RNAi testes are similar to control. Samples size <20 for all genotypes.
Inhibition of CG11180 and Smt3 in somatic support cells cause over-proliferation of undifferentiated germ cells
Adult testes aged 7–10 days after eclosion immunostained with anti-Vasa (magenta) and anti-1B1 (green) to reveal germ cells and fusomes respectively. Vasa and 1B1 shown alone in black and white as indicated. Testes apex indicated with an asterisk. Scale bars = 40 µm. (A) Control c587-Gal4/+ testes show undifferentiated germ cells with dot and bar/elongated fusomes at the testes apex and branched fusomes in differentiating spermatogonia away from the hub (see inset). Testes with either (B) CG11180 or (C) Smt3 knocked down in somatic support cells show undifferentiated germ cells with dot and bar/elongated germ cells along the length of the testes and away from the niche (see insets). n <20 for all genotypes.
Percentage and degree of phenotype observed in adult testes of specific genotypes under different rearing conditions
Calculations and analysis of data for Fig. 2B
Quantification of the interaction of SUMO with the indicated Chinmo proteins using a quantitative ONPG assays and strains shown in A. Tukey Kramer multiple comparison procedure was used to determine significant differences in SUMO interaction.
Data used for graphing Fig. 4
Graph showing percentage of control and c587-Gal4/+; UAS-CG11180-RNAi/+ testis displaying phenotype at specific ages, with phenotype severity increasing over time after somatic CG11180 knockdown (c587-Gal4/+ (7-16d), n = 56; UAS-CG11180 RNAi/+ (11-14d), n = 11; c587-Gal4/+; UAS-CG11180 RNAi/+ (6-10d), n = 77; c587-Gal4/+; UAS-CG11180 RNAi/+ (10-14d), n = 64; c587-Gal4/+; UAS-CG11180 RNAi/+ (14-18d), n = 34).
Calculations and analysis of data in Fig. 5
Graph of data showing variance in phenotype severity in EyaA3-Gal4; UAS-Smt3-RNAi testes with different treatments as compared to controls (EyaA3-Gal4/+ (25 °C) 7-10d, n = 23; EyaA3-Gal4/UAS-Smt3 RNAi (25 °C) 6-10d, n = 13; EyaA3-Gal4/UAS-Smt RNAi (18 °–25 °C) 1-3d, n = 8; EyaA3-Gal4/UAS-Smt3 RNAi (18–25 °C) 7-10d, n = 4).