morloc: a workflow language for multi-lingual programming under a common type system
- Published
- Accepted
- Received
- Academic Editor
- Alexander Bolshoy
- Subject Areas
- Data Science, Scientific Computing and Simulation, Programming Languages
- Keywords
- Workflow language, Functional programming, Polyglot, Strongly typed
- Copyright
- © 2025 Arendsee
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ Computer Science) and either DOI or URL of the article must be cited.
- Cite this article
- 2026. morloc: a workflow language for multi-lingual programming under a common type system. PeerJ Computer Science 12:e3435 https://doi.org/10.7717/peerj-cs.3435
Abstract
A conventional scientific workflow consists of many applications with untyped data flowing between them. Each application builds an internal data structure from input files, performs an operation, and writes output files. In practice, there are many data formats and many flavors of each. Compensating for this requires either highly flexible parsers on the application side or extra interface code on the workflow side. Further, the idiosyncrasies of wrapped applications limit the expressiveness of workflow languages; higher order functions, generics, compound data structures, and type checking are not easily supported. An alternative approach is to create a workflow within a single programming language using native functions rather than standalone applications. This offers programmatic flexibility, but mostly limits usage to one language. To address this problem, we introduce morloc: a language that supports efficient function composition between languages under a common type system. morloc gives the workflow designer the power of a modern functional language while allowing the nodes of the workflow to be written freely in any supported language as idiomatic functions of native data types.
Introduction
Computational researchers use a wide range of specialized programs on a daily basis. In the life sciences, these include tools for genome assembly, sequence similarity search, sequence alignment, phylogenetics, and protein modeling in addition to general data analysis, statistics and visualization. The heavy algorithms are typically implemented in high-performance languages like C and used as command line tools. Workflow-specific analytic steps are often written in higher languages like Python or R and applied as interpreted scripts within the pipeline or in notebooks that build on pipeline output. While applications may be strung together with shell scripts, dedicated workflow managers are often preferred for better scaling and reproducibility (Wratten, Wilm & Göke, 2021). These include graphical workflow managers such Galaxy (The Galaxy Community, 2024), build tools like Snakemake (Mölder et al., 2021), specification languages like the Common Workflow Language (CWL) (Crusoe et al., 2022), and domain specific languages like Nextflow (Di Tommaso et al., 2017), BioShake (Bedő, 2019) and Cuneiform (Brandt, Reisig & Leser, 2017).
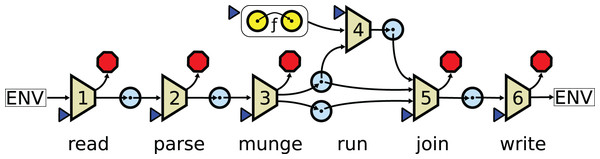
What these managers have in common is that nodes in the workflow are individually responsible for interpreting input and formatting output. Data between nodes is passed as files and each node must agree on the file format and be capable of reading and writing it. In this paradigm, scientific algorithms must be wrapped in applications that handle many layers of complexity beyond the pure algorithm itself (see Fig. 1). The complexity of these applications and the data they operate on lead to many problems, including those listed below.
Figure 1: In a conventional pipeline a function is wrapped in six layers of extra complexity.
The numbered trapezoids are auxiliary functions. The blue triangles are points where parameters may be passed in from the user. The red octagons are points where errors may be raised. The blue circles are data domains. is the core algorithm. The read layer maps the user arguments and files from the system to a set of raw inputs. The parse layer transforms raw inputs into internal structures. The munge layer extracts the data needed for the core functions. The run layer applies the core function to the extracted data structure. The join layer prepares the final results, possibly merging them with data that was removed in the munge layer. The write layer formats output data and writes it to the system.Format problem. A node in a workflow must parse input data into native structures, operate on these structures, and format the results into output files. A given data structure may be formatted in many ways. Tabular data may be formatted as TAB-delimited files, Excel spreadsheets, Parquet, or JavaScript Object Notation (JSON). In bioinformatics, there are many popular formats for storing biological data (sequences, alignments, trees, protein structures, etc). These formats are often creatively overloaded with custom information. For example, color palette information unique to one graphical program may be appended to a text field in a phylogenetics output file. Designing software with good format support is a hard task that must be repeated and specialized for every application.
Agreement problem. The formatted output of one node must be readable by downstream nodes. Since many possible formats and format variants may store a specific data structure, nodes must agree on a shared representation. However, the formatting logic is hard-coded into the applications and the applications are usually written by different groups. Communities, then, must converge on formatting conventions and faithfully follow them. To avoid conflicts, applications may need to support many formats, automatically guess formats, and handle misformatted files. Remaining conflicts must be resolved by adding extra glue code to the wrapper. New data with unexpected formatting conventions or uncommon patterns (like an apostrophe in a country name) can easily break pipelines.
Monolith problem. Writing a dedicated application, with robust file handling and a full user interface, for every function is impractical. For this reason, applications often cluster many independent functions behind one interface. The application may then play many different roles in a workflow. Command line arguments are required to select and specialize the different roles. Applications may have dozens or even hundreds of such options. These options may overlap across functions, over-ride one another, or be mutually exclusive. Testing and documenting all combinations is often infeasible and usage descriptions become bloated. Further, use of any function in the monolith imports the dependencies of all functions. Changes in any part of the monolith may trigger application-wide version increments. Bugs anywhere in the monolith may cause the entire system to fail.
Text problem. Bioinformatics pipelines often rely on direct operations on structured textual formats. Writing a dedicated application for these manipulations that parses the file into a well-defined data structure and applies safe operators to transform the data, would require extensive coding (see Fig. 1). So instead, it is common to rely on regular expressions over the literal text. For example, the second element in the header of a comma delimited table may be replaced with “foo” using the command line expression sed ‘1s/^\([^,]*\),\([^,]*\)/\1,foo/’. Such operations are hard to maintain and commonly lead to bugs.
Annotation problem. Data is typically passed between applications in formats that represent annotated collections of elements rather than the elements themselves. DNA sequence, for example, is usually passed as files that contain many sequences that each have an associated string annotation. If an element is changed in a pipeline, then the annotation may need to be updated. Alternatively, if new data is inferred for an element, it should be added to the element metadata. But when annotations are free text, with only conventions defining their form, there is no natural way to update or extend them. This hinders metadata propagation across the workflow.
Collection problem. When many records are passed within one file, the application must define a strategy for processing the elements. Should element input order be preserved? Should all elements be streamed or fully loaded into memory? Should intermediate results be cached (and how)? Should log entries be written for each? Should elements be processed in parallel? The best choice in each of these circumstances depends on the usage context. When elements are independent, the application could be simplified to operate on just one element, the base case, instead of the whole collection. Then collection handling logic could be managed externally and applied consistently across the pipeline. But support for standard, multi-entry formats and high application overhead prevent this approach.
Substitution problem. The design space for an application is huge: different conventions may be used for the interface; different formats may be supported; different choices may be made in caching, parallelism, and streaming. For this reason, even if two applications are fundamentally isomorphic (e.g., two sequence aligners), they cannot generally be substituted without refactoring pipeline code and risking the creation of new bugs. The high cost of substitution favors continual use of legacy algorithms even when better alternatives exist.
Abstraction problem. Most standard programming abstractions are not possible in workflow languages. Applications cannot generally be passed as arguments due to the substitution problem. Compound data structures cannot consistently be passed between applications since this would require format agreement. Polymorphism is usually limited to ad hoc additions to textual fields that are interpreted in special ways by certain tools or groups. Even basic function composition, the most fundamental prerequisite of all workflow programs, is not possible in general without context-specific wrappers around the application.
This paradigm leads to brittle, unreliable workflows that are hard to maintain and hard to extend. Each application in a workflow is responsible for many separate concerns (see Fig. 1). The interfaces between the applications are complicated by idiosyncratic parameters and lack of format agreement. Resolving conflicts often requires unsafe textual processing of structured files. Annotations must be propagated and extended without knowing the annotation format. Applications must consider parallelization, data propagation, and caching without knowing the usage context. Logically identical applications cannot be easily or safely substituted. Vague formats and inconsistent application behavior prevent expressive programming styles. Workflow managers may be effective for large-scale batch processing of pipelines with a small number of large nodes that consume clean consistent data, but they deal poorly with complexity and delegate most work to the application creator.
Complexity scaling can be improved by developing a workflow within a single language. Within the bioinformatics community, there are several language-specific projects designed to facilitate end-to-end analysis. These include Bioperl (Stajich et al., 2002), Biopython (Cock et al., 2009) and Bioconductor (R language) (Gentleman et al., 2004). The nodes of a single-language workflow are simply functions that communicate through native data structures in program memory. This resolves most of the problems discussed above. However, usage is limited to one language and no one language is best in all cases.
As a third approach, we present morloc. The purpose of morloc can be summarized by two core goals. First, the programmer should be free to focus on writing functions rather than applications. They should be free to write in their favored language and without responsibility for wrappers, formatting, user interfaces, or application programming interfaces (APIs). Second, the workflow designer should be free to seamlessly compose these functions using an expressive functional language.
morloc is a language that supports efficient function composition across languages under a common type system. The nodes in a morloc workflow are native functions imported from independent external libraries. All code needed for interoperability is automatically generated by the morloc compiler. The workflow is implemented in a simple functional programming language with full support for generics, parameterized types, type classes, and higher-order functions. morloc modules may be directly compiled into interactive command line tools or imported into other morloc programs.
In the following sections, we introduce the core features of the language, describe the compiler architecture, evaluate performance, demonstrate usage through a deep case study, compare morloc to conventional systems, and finally discuss the future of morloc.
Language design
Here we introduce the morloc language and show how it supports typed multi-lingual function composition. We will provide a practical description of the language and leave formal specification for future work.
Functions may be composed across languages
In morloc, functions are sourced from foreign languages and unified under a common type system. Every sourced function is given a type signature that specifies the general types of the function’s inputs and output. These general types describe language-independent structures. They may be mapped to many different language-specific native types. Non-function types additionally map to a common morloc binary form. The morloc compiler generates code in native languages that transforms native types to and from this shared binary form. Thus native types in different languages with the same general type can be automatically interconverted, allowing communication between languages.
The morloc programmer may import functions and develop programs through composition without knowing anything but the function’s general type. The source programmer may develop functions of native data structures without handling serialization or using any morloc-specific idioms, dependencies, or syntax. These native functions may be exported to the morloc ecosystem with no extra boilerplate beyond the general type signature. These signatures serve as the interface between the two programmers.
Functions may be sourced as shown below:
source Cpp from "foo.hpp" ("map", "sum", "snd")
source Py from "foo.py" ("map", "sum", "snd")Here C++ and Python implementations of the three functions map, sum and snd are loaded. The files “foo.hpp” and “foo.py” will be imported into the generated code and must provide definitions for the three functions. For the Python file, “foo.py”, only snd needs to be explicitly defined, since map and sum are already in scope as Python builtins with correct name and type. So the “foo.py” file needs exactly two lines:
def snd(pair):
return pair[1]For the C++ source file “foo.hpp”, all three functions may be implemented in a simple header file. These files contain no morloc-specific syntax or dependencies and operate only on native data types.
Next, the morloc programmer must specify the general types of the sourced functions by adding type signatures to the morloc script:
map a b :: (a -> b) -> [a] -> [b]
snd a b :: (a, b) -> b
sum :: [Real] -> RealThe type signatures loosely follow Haskell syntax conventions. The main difference is that generic terms ( and here) must be introduced explicitly on the left. Bracketed terms (e.g., [a]) represent lists and comma separated terms in parentheses represent tuples. Arrows represent functions. (a -> b) is a function from generic type a to generic type b. The map type ((a->b)->[a]->[b]) can be seen as a function that takes two input arguments—the function (a->b) and the list [a]—and returns the list [b].
Removing the list and tuple syntactic sugar, the signatures become:
map a b :: (a -> b) -> List a -> List b
snd a b :: Tuple2 a b -> b
sum :: List Real -> RealList, Tuple2 and Real are general types. Since these general types may each map to multiple native types in a given language, explicit mappings are needed to avoid ambiguity. These mappings may be provided as language-specific type functions that evaluate to representations of the native type. Here are examples for C++ and Python:
1 type Cpp => List a = "std::vector<$1>" a
2 type Cpp => Tuple2 a b = "std::tuple<$1,$2>" a b
3 type Cpp => Real = "double"
4 type Py => List a = "list" a
5 type Py => Tuple2 a b = "tuple" a b
6 type Py => Real = "float"Type names in source languages, such as “list”, are quoted, since they may be syntactically illegal in morloc. General types, like Real, map to native types in the source language, such as "float". Parameterized types, like (List a), map to parameterized native types where parameters may be substituted to make the final type. For example, morloc represents the C++ type vector<double> as ("vector<$1>” “double"). The morloc representation shows that double is the parameter that vector expects (this is needed for typechecking) and $1 shows where the parameter appears in the C++ source type (it is inserted between the angle brackets).
Sourced functions may be composed to create more complex functions. The following composition, sumSnd, sums the second values in a list of pairs:
sumSnd xs = sum (map snd xs)This composition extracts the second value from a list of pairs and then sums them. In this case, the morloc compiler can infer the type, so no explicit type signature is required. The compiler internally erases all morloc compositions, such as sumSnd, rewriting them in terms of sourced functions. sumSnd will be rewritten as the anonymous function
\ xs -> sum (map snd xs)To the morloc programmer, however, these functions are in all ways identical to sourced functions. The sumSnd function may be further simplified, as shown below, since morloc supports partial function application, eta-reduction, and the dot operator for function composition:
sumSnd = sum . map sndOne term may have many definitions
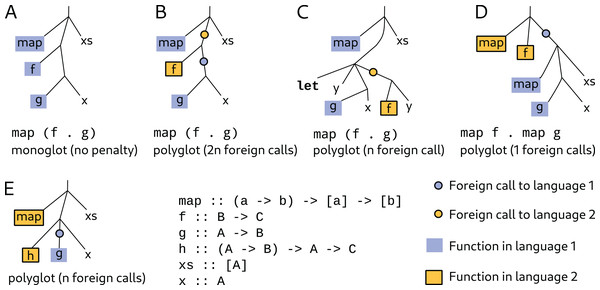
An unusual aspect of morloc is that one term may have multiple definitions. This is seen in morloc libraries where families of common functions are sourced from many supported languages. For example, the morloc module base exports functions such as operators over collections (e.g., map and fold), arithmetic and logical operators, and function combinators. These form a common functional vocabulary that may be composed to build complex programs that are polymorphic over language. When a specialized monoglot function is used, the polyglot context will adapt to optimize compatibility (see Fig. 2). A term may also be assigned to multiple morloc expressions. Thus any term in the internal morloc abstract syntax tree may contain many alternative subtrees.
Figure 2: The overhead cost of morloc is proportional to the number of required foreign calls.
The best-case performance of morloc is comparable to native code in the target language. This case occurs when all functions in the final tree are in the same language (A). In this case, the generated code uses within-language function calls. The worst-case performance is when every call is a foreign call (B). Foreign calls must message the foreign language server and may require data reformatting and copying. The compiler attempts to minimize foreign calls. In (C), the compiler reduces the number of foreign calls by moving evaluation of g x to the parent language context. Alternatively, compositions may be written so that fewer foreign calls are necessary. In (D), and are “unfused” into two mapping operations, each using a language-specific map function. When foreign functions that are not fully applied are passed as arguments, no optimizations are possible (E). With respect to data transfer, workflow languages have performance similar to morloc’s worst case, since all function calls require full serialization cycles.In the example below, the function mean is given three definitions:
1 import base (sum, div, size, fold, add)
2 import types
3 source Cpp from "mean.hpp" ("mean")
4 mean :: [Real] -> Real
5 mean xs = div (sum xs) (size xs)
6 mean xs = div (fold add 0 xs) (size xs)Here we source an implementation directly from C++ and also write two local definitions. Which definition is used at a given place in a program will depend on context. In a C++ context, the C++ sourced definition will be used. In other cases, the smaller definition that uses sum will be chosen if sum is implemented for the contextually chosen language. Otherwise, the larger expression that sums explicitly by folding will be chosen.
The compiler is responsible for selecting the implementation that maximize desired qualities of the final program. This is a complex optimization problem that will be a major focus of future work. For now we use a simple scoring system that penalizes between-language calls and the use of “slower” languages. When terms have equal scores, the term with fewer elements in its abstract syntax tree is chosen. When terms have the same size and score, an error is raised stating that there is no rule to resolve the definitions.
All implementations for a given term must have the same general type; this is enforced by the typechecker. Type equivalence theoretically guarantees that the functions may be substituted and yield programs that can still be compiled and run, but it does not require functional equivalence. For control functions like map, all implementations should be equivalent (testing can confirm this). However, other functions, such as heuristic algorithms and machine learning models, may differ systematically. The compiler cannot yet model performance of non-equivalent functions, so programmers should avoid equating them.
Support for multiple definitions alters the meaning of equality in morloc. The “=” operator in morloc implies that the right-hand expression is being added as one of the implementations of the left-hand term. Thus one may write:
x = 1
x = 2morloc has a rudimentary value checker that will raise an error for this class of primitive contradictions. Every pair of implementations for a given term are recursively evaluated to check for such contradictions. The value checker cannot currently check past source call boundaries, however, so contradictions such as the following will not be caught:
x = div 1 (add 1 1)
x = div 2 1Without specific knowledge about div, morloc cannot know that the functions are not equivalent. So the contradiction is missed and the simpler second definition is ultimately selected.
Terms may be overloaded through typeclasses
As discussed above, the equals operator can bind a term to multiple instances of the same type. Through typeclasses, a term may also be associated with instances of different types. A typeclass defines a set of generic terms with type-specific instances. They were inspired by Haskell typeclasses and are similar to interfaces in Java, traits in Rust, and concepts in C++20.
Below are example definitions of Addable and Foldable classes:
1 class Addable a where
2 zero a :: a
3 add a :: a -> a -> a
4
5 instance Addable Int where
6 source Py "arithmetic.py" ("add")
7 source Cpp "arithmetic.hpp" ("add")
8 zero = 0
9
10 instance Addable Real where
11 source Py "arithmetic.py" ("add")
12 source Cpp "arithmetic.hpp" ("add")
13 zero = 0.0
14
15 class Foldable f where
16 foldr a b :: (a -> b -> b) -> b -> f a -> b
17
18 instance Foldable List where
19 source Py "foldable.py" ("foldr")
20 source Cpp "foldable.hpp" ("foldr")
21
22 sum = foldr add zeroLines 1–3 define the typeclass Addable with two terms: zero and add. Lines 5–13 define integer and real instances for the Addable typeclass. The native functions may themselves be polymorphic, as is the case with add, which may be implemented in Python as:
def add(x, y)
return x + yAnd in C++ as
template <class A>
A add(A x, A y){
return(x + y);
}Lines 15–16 define the Foldable typeclass. Here is a container of generic elements that can be iteratively reduced to a single value. Lines 18–20 define a Foldable instance for the List type.
With the Addable and Foldable classes, we can define the polymorphic sum function (Line 22) that folds the add operator over a list of values with the initial accumulator of zero. The instances will be chosen statically after the types have been inferred by the typechecker.
Types may be defined and passed between languages
A core principle of morloc is that cross-language interoperability should be invisible. All terms in morloc have both a native type and a general type. The native type specifies how the data is represented in a given language. The general type specifies a common memory layout. The morloc compiler can automatically cast data in each native type to this common binary form. This is the foundation of morloc interoperability.
morloc supports several fixed-width primitives and two collection types. The primitives include a unit type, a boolean type, signed and unsigned integers of 8, 16, 32 and 64 bit widths, and 32 and 64 bit floats. Next morloc offers a list type which is represented by a 64-bit integer storing the container size and a pointer to a vector of contiguous, fixed-size morloc values. Finally, morloc offers a tuple type that contains a fixed number of fixed-size values in contiguous memory. The primitives and the two types of collections are sufficient to represent all forms of data. The morloc compiler translates general types into schemas that are used by language-binding libraries to cast these common binary forms to/from native types.
Records, such as structs in C or dictionaries in Python, are represented as tuples in memory. The field names are stored only in the type schema. Tabular data can be specified in the same way as records, but with field types describing column types. Records and tables can be defined and instantiated as shown below:
1 record Person = Person { name :: Str, age :: UInt8 }
2 alice = { name = "Alice", age = 27 }
3
4 table People = People { name :: Str, age :: Int }
5 students = { name = ["Alice", "Bob"], age = [27, 25] }We may also define new types from these base types, for example we can define a Pair type as a tuple:
type Pair x y = (x, y)Many types, though, have different structures in different languages. Suppose we want to use the parameterized type (Map k v). This type represents a data structure that associates keys of generic type with values of generic type . It may be structured in many ways, including a hashmap, binary tree, two-column table, or list of pairs. Before being converted to the morloc binary form, these structures must be reformatted into a common form. This common form for Map may be a list of pairs (row form) or a pair of equal-length lists (column form). Transforming native types to the common form requires knowledge about the native data structure that the morloc compiler does not possess, so a pair of functions must be given that converts the type to and from a more basic form. These functions are provided as methods of the Packable typeclass. The class is defined as follows:
class Packable a b where
pack a b :: a -> b
unpack a b :: b -> aand refer to the unpacked and packed forms of the type, respectively. The packed type is the type used by morloc functions, such as Map k v. The unpacked type is a reduced form that eliminates the top type term (in this case Map). The unpack functions may be recursively applied until a data structure is reduced entirely to basic types (primitives, lists, and tuples). This final structure may then be transformed to the common binary form by the language-binders and passed to a foreign language. Reversing this process in the foreign language constructs the corresponding foreign type. The morloc compiler generates the code to perform these transformations; the morloc user does not need to directly use the pack and unpack functions. This framework provides a general template for unifying data structures across languages.
The Packable instance for the column-based representation of the Map type may be written as follows:
1 type Py => Map key val = "dict" key val
2 type Cpp => Map key val = "std::map<$1,$2>" key val
3
4 instance Packable ([a],[b]) (Map a b) where
5 source Cpp from "map-packing.hpp" ("pack", "unpack")
6 source Py from "map-packing.py" ("pack", "unpack")Some languages may not support the fully general parametric form. This can be expressed by implementing a more specific instance of Packable. For example:
type R => Map key val = "list" key val
instance Packable ([Str],[b]) (Map Str b) where
source R from "map-packing.R" ("pack", "unpack")Here we define an instance of Map for R that is defined only when keys are strings. In cases where non-string keys are required, R solutions will be eliminated. If no solutions remain, a compile-time error will be raised.
Modules may be defined and compiled into executables
The organizational unit of morloc is the module. A module defines a set of terms and types and specifies what is exported. Below is a simple example:
1 module foo (mean, add)
2 import types
3 source Cpp from "foo.hpp" ("mean", "add")
4 mean :: [Real] -> Real
5 add :: Real -> Real -> Realmorloc has no special “main” function. Instead, a module may be directly compiled into an executable if all exports are non-generic (see Fig. 3). The functions exported from this module are translated into an inventory of commands. Each command takes one positional parameter for each argument of the original function. Each positional parameter expects user input with format corresponding it the argument’s general type. Help messages are generated based on this type. Input may be supplied as either literal JSON strings or files containing JSON, MessagePack or morloc binary data. These user arguments will be translated automatically to native data types before being passed to the wrapped native functions.
Figure 3: The compiler typechecks the program, selects instances, and generates polyglot executables.
The Front End reads and checks morloc scripts. It produces a typed polyglot plural tree where every term has a single general type and where functional terms may have many definitions in many languages. The Middle End prunes the tree down to a forest of monoglot singular trees. The Back End organizes the many monoglot trees and generates the source code that will be compiled into the final product. Starting at the first step in the Front End, Parse builds a syntax tree from the morloc code and resolves inheritance across modules. Typecheck infers a general type for every term in the tree and resolves typeclass instances. Valuecheck searches for conflicts between the implementations of each term. Prune is a major optimization step that chooses a single implementation for each functional node. Segment breaks the pruned tree into subtrees by language and derives language-specific types for each term. Serialize takes each monoglot subtree and determines where (de)serialization is needed, generates serialization strategies, and determines how serial and native forms are passed through a subtree. Pool partitions subtrees by language and gathers their dependencies. Generate makes the source code for each language that handles interop and implements the compositions specified in the morloc script. Build interacts with language-specific compilers as needed to generate binaries. Dispatch makes the command line user interface.We can compile, print usage, and run an executable as follows:
$ morloc make -o foo mean.loc
$ ./foo -h
The following commands are exported:
mean
param 1: [Real]
return: Real
add
param 1: Real
param 2: Real
return: Real
$ ./foo mean "[1,2,3]"
2.0This simple interface serves as a toolbox of functions that can be used interactively on the command line. Future releases of morloc will support within-code documentation that is propagated to the generated interface. We may also explore alternative backend generators that make REST APIs, documentation pages, and basic graphical interfaces.
When a morloc program is compiled, the compiler writes language-specific code to “pools” (one for each required language) and writes a “nexus” executable that accepts arguments from the user (see Fig. 3). When the user passes a command to the nexus, the nexus starts a pool for each language used by the specified command. Each pool contains wrappers for all functions that are used in the pool language. When initialized, the pools listen over UNIX domain sockets for commands from the nexus or from other pools. When a command arrives, the pool spawns a new job in the background. The job executes a composition of native functions and handles transformations to native data types and foreign calls as needed.
All communications between pools and the nexus are mediated through binary packets that each consist of a 32-byte header, a metadata block, and a data block. The header specifies version info, metadata length, data length, and packet type. The main packet types are “data” packets and “call” packets. A data packet describes a unit of morloc data and specifies how it is represented. The packet may contain a type schema in its metadata section. A call packet specifies the command that will be executed in the receiving pool and its data block is a contiguous vector of arguments formatted as morloc data packets.
When a pool makes a foreign call, all arguments are translated to the common binary form and written to a memory volume shared between the nexus and all pools. Then a call packet is generated where each argument is written to the call packet data block as a data packet that stores a relative pointer to the argument data in the shared memory volume. The call packet is then sent to the foreign pool over a socket. The foreign pool reads the packet, translates the data in shared memory into native data structures (when needed), and executes the code. On success, the foreign pool writes the result to shared memory and returns a packet containing the relative pointer to the result. On failure, the foreign pool will return a data packet with the failing bit set and a message containing an error message. This message will be propagated back to the nexus and printed to the user.
An analysis of performance
The runtime of a pipeline of identical components passing data of equal size can be modeled as:
(1)
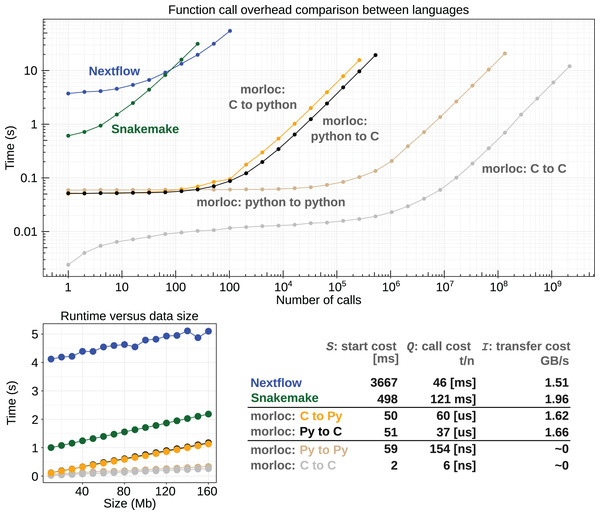
Where is the runtime of the pipeline as a function of the number of nodes in the pipeline ( ) and the amount of data passing between each node ( ). The runtime is equal to the constant cost of starting the pipeline (S seconds), plus the variable cost of loading initial data (L seconds per GB), plus the cost of running nodes. Each node’s cost is equal to the constant node startup cost (Q seconds), plus the cost of reading and writing data (I seconds per GB), plus the cost of running the program of interest (R seconds per GB, linear for our benchmark case).
In a single-language functional program running locally, “nodes” are function calls and the runtime will typically be dominated by the cost of loading data ( ) and applying it to each function in the pipeline ( ). Q and I will be low since function calls are fast and data can be passed in memory.
In conventional workflow languages, the costs of starting nodes and transferring data between them (Q and I, respectively) are high. Invoking a node requires a system call to a program, container, or web service. Transferring data between nodes requires a serialization cycle and the cost of sending data over a connection or moving it to and from files on the disk.
morloc is a hybrid between these two approaches. Nodes within the same language can communicate as they would natively, with Q and I both nearly zero. Communication between languages is slower since a message must be passed over a socket and data may need to be refactored and possibly copied. The best possible performance in this architecture is limited to a few microseconds by the speed of transmission over a UNIX domain socket. The current morloc implementation generates code with modest additional overhead for processing the packets and starting workers (see Fig. 4).
Figure 4: Runtime comparison between morloc and conventional workflow programs.
(Top) Log-log plot comparing runtimes for a linear pipeline of nodes acting on data of zero-length. All tests involve sequential calls between languages in morloc or to a Python script in Nextflow and Snakemake. (Bottom Left) Comparison between runtimes for linear pipelines of constant length with varying data input size. Each node copies the data and performs a constant time modification. (Bottom right) A table comparing start costs (S), call costs (Q), and data transfer costs (I) (see Eq. (1)). These values were statistically inferred from the benchmark data. The call cost column of the table is split into three cases by time units: conventional workflow languages (milliseconds), morloc programs with foreign interop (microseconds), and morloc programs with no foreign interop (nanoseconds). Code and documentation are available at https://github.com/morloc-project/examples. Benchmarks were run on an i7-10510U CPU and Samsung PM981A 512 GB SSD.Call overhead in morloc ranges from nanoseconds for native calls (depending on the cost of a function call in the native language) to tens of microseconds for foreign calls. For conventional workflows, call overhead is lower-bound by the cost of invoking an external resource. In this study, that resource is a light Python program with a roughly 100 ms startup time. The cost of passing data between nodes is nearly zero for native calls in morloc. For foreign morloc calls, data transfer rates measured in this benchmark were similar to Snakemake/Nextflow.
These benchmarks were done on a local machine and without parallelism. All morloc language pools are multi-threaded and naturally support local parallelism. Work may be parallelized using generic control functions, such as parallel versions of the basic map function. This might be implemented in Python as follows:
import multiprocessing
def pmap(f, xs):
with multiprocessing.Pool() as pool:
results = pool.map(f, xs)
return resultsThe morloc type signature of pmap is the same as its non-parallel cousin, so it is a drop-in replacement. In contrast, conventional workflow managers delegate fine-grained parallelism to the application.
Where workflow managers excel is in distributed computing where many large applications are run in parallel on different data. morloc has experimental support for remote job submission as well. In morloc, a remote job is not unlike a local foreign call. In both cases, data must be sent from one language pool to another. The data reformatting steps are the same for both. Within the compiler, the main difference is that the function tree needs to be segmented by locallity, not just language. On a shared file system, the data may be “sent” to the remote machine by serializing the call packet and associated data from the shared memory volume to the disc. Then a job is submitted that starts the remote morloc nexus with the call packet. The remote nexus sends the job to the proper pool and writes the returned result to a binary output file. This output is then read by the local calling pool after the remote worker closes. Caching of intermediate results is also supported via functional memoization.
morloc terms may be given labels that target them for remote execution. For example:
foo = merge . map big:myJobThe big label is a hook we can use to provide annotations such as where and how myJob is run. The annotations are included in a YAML file associated with the morloc program. In this example, merge and map are both local and each myJob function is run on a remote node. myJob may be an arbitrarily complex morloc composition and may submit its own jobs recursively. While still experimental, this general approach allows fine, unobtrusive control over execution.
Case study: influenza strain classification with three languages
This case study shows how multiple languages are interwoven and how complex programs and types are defined in morloc. We reproduce an influenza virus classification pipeline developed by Chang et al. (2019). In this workflow, influenza strains within a given time range are retrieved from an online database, a phylogenetic tree is constructed, clades (biologically distinct subtrees) are identified using a set of reference strains, and the final labeled tree is plotted (see Fig. 5). The pipeline applies Python for data retrieval, C++ for algorithms, and R for visualization.
Figure 5: Influenza classification case study overview.
The case study consists of four major steps (top). Retrieve takes a configuration record, FluConfig, and prepares a list of sequences and their annotations (represented as the generic , for simplicity). Make tree builds a phylogenetic tree from the retrieved sequences, returning a RootedTree object. The tree type has three parameters representing the node type, edge type, and leaf type. Classify determines the clade of each unlabeled leaf based on the labeled reference leaves. Visualize makes a plot of the tree.The main morloc script is shown below:
1 module flucase (plot)
2 import types
3 import lib.flutypes
4 import lib.retrieve (retrieve, setLeafName, FluConfig)
5 import lib.classify (classify)
6 import lib.treeplot (plotTree)
7 import bio.algo (upgma)
8 import bio.tree (treeBy, mapLeaf)
9 plot :: FluConfig -> ()
10 plot config =
11 ( plotTree config@treefile
12 . mapLeaf setLeafName
13 . classify
14 . treeBy upgma
15 . retrieve
16 ) configThis script defines the module flucase and exports the function plot. It imports required types and functions (lines 2–8) and defines the exported plot function (lines 9–16) as a composition of five functions that takes a FluConfig record as input. The following sections will outline the four steps in Fig. 5 and describe how the tree type is defined.
Retrieve and clean data
The first step in the pipeline is the retrieval of data from the Entrez database (Schuler et al., 1996). This task is performed by the retrieve function with the following signature:
retrieve :: FluConfig -> [((JsonObj, Clade), Sequence)]
The input is a FluConfig record that is defined in lib.flutypes as:
record FluConfig = FluConfig
{ mindate :: Date
, maxdate :: Date
, reffile :: Filename
, treefile :: Filename
, query :: Str
, email :: Str }The record contains the query data range, the query string, an email (reqired by the remote database), and reference and output filenames. morloc does not, and never will, have keyword arguments. When many arguments are needed, they may be organized into records, allowing full sets of parameters to be clearly defined and transported.
The output of retrieve is a list of annotated sequences. The annotation is a pair of values, first a JSON object storing the full metadata record and second the clade assignment (either an empty string or the clade stored in the reference map). The JsonObj type specifies the JSON formatted metadata that is retrieved from the remote database. This type is defined in the json module as follows:
type Py => JsonObj = "dict"
type R => JsonObj = "list"
-- from Niels Lohmann’s json package (https://github.com/nlohmann)
type Cpp => JsonObj = "ordered_json"
instance Packable (Str) JsonObj where
…The Packable instance for JsonObj defines functions for translating JSON strings to and from the different native data structures, such as dictionaries in Python.
The Clade, Date, and Filename types are all aliases for the string type. While the specialized names clarify type signatures, they do not provide additional type safety. We could alternatively define them as unique types by specifying Packable instances that map them to strings with identity functions for the pack and unpack methods. These types would then raise helpful errors at compile time when misused.
The retrieve function is defined as follows:
1 retrieve config =
2 ( map (onFst (labelRef refmap)) -- add reference clades
3 . concat -- flatten list of lists
4 . map ( map parseRecord -- parse wanted data from XML records
5 . sleep 1.0 -- pause between calls to not overuse the API
6 . fetchRecords fetchConfig -- retrieve XML records for chunk
7 )
8 . shard 30 -- split list into chunks
9 . join (keys refmap) -- add the reference IDs to list
10 . fetchIds searchConfig -- send query to get strain IDs
11 ) config@query -- access the query in the config record
12 where
13 refmap = readMap config@reffile -- open the reference to clade map
14 searchConfig = -- set parameters for the search
15 { email = config@email
16 , db = "nuccore"
17 , mindate = config@mindate
18 , maxdate = config@maxdate
19 , retmax = 1000
20 }
21 fetchConfig = { email = config@email } -- set parameters for fetchRecordsLines 1–11 define the function composition that runs a query, retrieves full data records on all returned ids in chunks of 30, flattens the list of chunks to a list, and finally adds in clade labels from the table of references. Lines 12–21 is a where block that defines terms that are available within the scope of the retrieve function.
Define tree types
Phylogenetic trees are represented with the (RootedTree n e l) type. The parameters represent node type ( ), edge type ( ), and leaf type ( ). In a phylogenetic tree, the node often contains a metric of confidence in the inference of its children, though we will use it later to store clade names. The edge type usually represents branch length. The leaf may contain the taxon name and other metadata.
The RootedTree type is unpacked as a tuple of three elements: a node list, an edge list, and a leaf list. The node and leaf lists are ordered sets of data for each node and leaf. The edge list is a list of tuples of three elements: parent index, child index, and generic edge data. Node indices range from 0 to , where N is the number of nodes. Leaf indices range from N to , where L is the number of leaves. This convention is adapted from the R phylogenetic representation of trees used in phylo objects.
The RootedTree type is declared in bio.tree as:
type Cpp => (RootedTree n e l) = "RootedTree<$1,$2,$3>" n e l
type R => (RootedTree n e l) = "phylo" n e l
instance Packable ([n], [(Int, Int, e)], [l]) (RootedTree n e l) where
source Cpp from "rooted_tree.hpp" (
"pack_tree" as pack, "unpack_tree" as unpack)
instance Packable ([Str], [(Int, Int, Real)], [Str])
(RootedTree Str Real Str)
where
source R from "tree.R" ("pack_tree" as pack, "unpack_tree"
as unpack)In C++, we map the RootedTree type to a custom recursive structure. In R, we map it to the pre-existing R phylo class. Unlike the morloc RootedTree type, R phylo objects are not generic; nodes and leaves are always strings and edges are always numeric. This type limitation is reflected in the R instance above. Attempts to use the phylo object more generically will fail at compile time.
Build trees and classify strains
The next steps in the pipeline are to build phylogenetic trees and then classify the strains. We implement these computationally expensive steps in C++.
In the main plot function, the tree is built with the command (treeBy upgma). treeBy is a generic control function that applies a tree building algorithm to an annotated list of sequences and returns an annotated tree. It has the following signature:
treeBy n e l b :: ([b] -> RootedTree n e Int)
-> [(l, b)] -> RootedTree n e ltreeBy accepts two arguments: a tree building algorithm and a list of annotated sequences. It unzips the list of annotation/sequence pairs into two lists and feeds the list of sequences to the tree algorithm. This algorithm creates a tree from just the sequences and stores sequence indices in the leaves. treeBy then weaves the original annotations back into the new tree using the indices in the leaves. This allows the tree algorithm to be a pure function of the sequences and guarantees that the sequence annotations are not altered by the tree builder.
In this case study, we use a simple UPGMA algorithm that builds a tree from a distance matrix (see Fig. 6). This is implemented in the bio module as follows:
Figure 6: Algorithm for building a tree from sequence.
The phylogenetic tree building starts with unaligned DNA sequences, counts the k-mers in each sequence (2-mers in this figure), creates a distance matrix from the counts, and then creates a tree from the distance matrix using the UPGMA algorithm.source Cpp from "algo.hpp"
("countKmers", "kmerDistance", "upgmaFromDist")
countKmers :: Int -> Str -> Map Str Int
kmerDistance :: Map Str Int -> Map Str Int -> Real
upgmaFromDist :: Matrix Real -> RootedTree () Real Int
makeDist :: Int -> [Str] -> Matrix Real
makeDist k = selfcmp kmerDistance . map (countKmers k)
upgma :: [Str] -> RootedTree () Real Int
upgma = upgmaFromDist . makeDist 8In our implementation, the distance matrix is made by comparing the number of occurences of each k-mer in each sequence. This is done in makeDist by composing countKmers and selfCmp. The latter function creates a square matrix from a vector by calling a distance function on each pair-wise value in the input vector. The distance metric is the square root of the sum of squared k-mer frequency differences (as defined in kmerDistance). A general implementation of the UPGMA algorithm is provided by the upgmaFromDist function. This is written as a simple C++ function that takes a matrix of doubles (using the matrix type from the eigen library) and returns a RootedTree structure. The upgma function is a composition of a function that builds a distance matrix and a pure implementation of the tree-building UPGMA algorithm.
The input to the upgma function is a list of unaligned sequences and the output is a rooted tree with null node labels, numeric edge values (branch lengths), and integers on the leaves representing the sequence indices in the input list. This signature is shared by all functions in the family of phylogenetic algorithms that create rooted trees from sequence alone. There are other families. Algorithms that estimate uncertainty at each node would replace the node parameter with a numeric type. Algorithms that produce an ensemble of trees would return a list of trees. The type system here provides a succinct, machine-checked method for logically organizing families of algorithms.
Traverse the tree to assign clade labels to leaves
After building the tree, the reference strains with labeled clades are used to infer the clades of unlabeled strains. This is done by the classify function:
1 classify n e a :: RootedTree n e (a, Clade) -> RootedTree Str e (a, Clade)
2 classify
3 = push id passClade setLeaf
4 . pullNode snd pullClade
5 where
6 passClade parent edge child =
7 (edge, ifelse (eq 0 (size child)) parent child)
8 setLeaf parent edge leaf = (edge, (fst leaf, parent))
9 pullClade xs
10 = branch (eq 1 . size) head (const "") seenClades
11 where
12 seenClades = ( unique
13 . filter (ne 0 . size)
14 ) xsThis function relies on two general tree traversal algorithms. The first, pullNode, makes distal nodes from leaves and then makes parent nodes from child nodes all the way down to root. The second, push, creates new child nodes based on old parent and old child nodes. In this case, it pushes the parent label into unlabeled children.
The pullNode function is a specialization of the pull function:
1 pull n e l n’ e’
2 :: (l -> n’) -- create a new node from a leaf
3 -> (n -> e -> n’ -> e’) -- synthesize a new edge
4 -> (n -> [(e’, n’)] -> n’) -- synthesize a new node
5 -> RootedTree n e l -- input tree
6 -> RootedTree n’ e’ l -- output treepull is a highly general function defined in the bio.tree morloc library for altering trees from leaf to root. It takes three functional arguments. The first extracts an initial value from a leaf. The second makes a new edge from the old parent node, the old edge, and the new child node. The third makes a new parent node from the old parent node and the new child nodes and edges. pullNode is defined in terms of pull as follows:
1 pullNode n e l n’
2 :: (l -> n’) -> ([n’] -> n’) -> RootedTree n e l -> RootedTree n’ e l
3 pullNode f g = pull
4 f -- generate a new node using f
5 (\n e n’ -> e) -- return the original edge
6 (\n es -> g (map snd es)) -- create new node from child nodesThis specialized function requires only two functional arguments, one for creating initial values from leaf values and one for creating new parent nodes from new child nodes. In the classify definition, pullNode is passed the functional arguments snd and pullClade. snd determines how the new node is created from a leaf at the tip of the tree: it selects the second element in a tuple. Based on the type signature of classify, the input tree has type: (RootedTree n e (a, Clade)). So snd sets the new terminal nodes to the strain clade. pullClade, from Lines 9–14 of the classify definition, specifies how the children of a node determine the new node value. It sets a parent node to the clade of its children if all child nodes either share the same clade or are undefined. Otherwise it sets the parent node as undefined (an empty string).
The generic branch function used in pullClade is a variant of an if-else with the signature:
branch a b :: (a -> Bool) -> (a -> b) -> (a -> b) -> a -> b
If the first argument to branch, a predicate of the input , returns true, the second functional argument is called on the input . Otherwise the third function is called. In our context, if children are from different clades, no parent clade is inferred. seenClades is a unique list of non-empty child clades. If its size is exactly 1, then children share a common clade and the parent clade is set to the first (and only) element in the list using the head function. Otherwise, the parent clade is set to an empty string using the const function. const has type a->b->a; here it is given an empty string and will ignore the empty list it is passed.
A simpler alternative to branch would be an ifelse function of type:
ifelse a :: Bool -> a -> a -> aHowever, this function would evaluate both the “if” and “else” blocks in non-lazy languages, leading to an error when head tries to take the first element from an empty list.
Returning to the classify implementation, the generic push function rewrites a tree from root to leaf. It takes three functional arguments as shown in the signature below:
1 push n e l n’ e’ l’
2 :: (n -> n’) -- initialize new root
3 -> (n’ -> e -> n -> (e’, n’)) -- alter child nodes
4 -> (n’ -> e -> l -> (e’, l’)) -- alter leaves
5 -> RootedTree n e l -- input tree
6 -> RootedTree n’ e’ l’ -- output treeIn classify, the first argument is the identity function since we are not changing the node type. The second function, passClade, passes the parent clade to the child if the child clade is undefined (empty string). The third function, setLeaf, sets the child leaf to the parent leaf while preserving the original leaf annotation. The final effect is to label all leaves that descend from labeled nodes.
We opted for a fine-grained implementation of classify. We could instead have sourced pullClade, push or classify directly from C++ rather than composing them in morloc. Such granularity decisions are common when writing morloc programs. A fine-grained representation exposes more workflow logic to the reader, expands the code that is typechecked, allows more code reuse, and increases the modularity of the program. A coarse-grained representation, in contrast, grants more control over implementation.
Visualize the tree
The final step in the pipeline is to plot the tree. The tree object returned from the classify function contains a metadata record for each leaf. From these records, we can synthesize informative leaf labels that will be used in the plotted tree. Recalling the definition of plot from the main script and adding types in comments:
plot config =
( plotTree config@treefile -- ()
. mapLeaf setLeafName -- RootedTree () Real Str
. classify -- RootedTree Str Real(JsonObj, Clade)
. treeBy upgma -- RootedTree () Real (JsonObj, Clade)
. retrieve -- [((JsonObj, Clade), Sequence)]
) config -- FluConfigThe output of plotTree is the Unit type () indicating nothing is returned. As a side-effect, the function creates a tree file with the name given in the configuration object (config@treefile). The final tree is shown in Fig. 7.
Figure 7: The final tree.
Leaves are labeled with the strain clade, accession, and sequence length.We use mapLeaf to apply setLeafName to each leaf in the tree. setLeafName reads a leaf’s metadata and generates the label that will appear in the final phylogenetic tree. Since the metadata was originally retrieved and parsed using Python code, it reasonable to write setLeafName in Python as well and include it in the “entrez.py” file. This encapsulates all Entez-related code in one place.
But if setLeafName is in Python and tree-handling algorithms, including mapLeaf, are all in C++, then a foreign call from C to Python will be required for every leaf. Any overhead in foreign function calls will be multiplied by the number of leaves in the tree. In morloc, this overhead is around 60 microseconds per call (Fig. 4). If better performance is needed, we can translate setLeafName to C++ and add a one-line type signature to the morloc source statement. The compiler will automatically choose the new C++ implementation since it reduces the number of foreign calls.
After naming the leaves, the next step is to plot the tree. We could implement plotting at a fine scale in morloc, but most plotting libraries do not lend themselves well to functional composition since they rely on unique grammars (e.g., ggplot) or mutability (e.g., matplotlib). So we source the plotting function from R with the type:
plotTree n :: Filename -> RootedTree n Real Str -> ()
We require the edges be parameterized as real numbers since they represent branch lengths. The nodes may be left generic since clade labels have been pushed into the leaves. plotTree returns the null type. It is run for its side effect of writing a plot of the given tree to a file.
Comparison to conventional pipeline
This case study demonstrates several features of morloc that are lacking in conventional pipelines. Together, these features solve each of the problems presented in the introduction. Many of the features are common in general programming languages but are lost in workflow languages where functions are replaced with applications.
morloc simplifies testing, benchmarking, maintenance, and composability. morloc programs compose functions with well-defined types rather than command-line tools that read arbitrary files. morloc functions can be easily unit tested and benchmarked across languages without the added complexity of mocking files and system state. Benchmarking is more precise since the algorithmic action is measured directly without the confounding cost of system calls and reading/writing files. Removing UI elements and file handling reduces the amount of code that needs to be maintained. Functions with the same type signatures can be safely composed without considering subtle variations in file formats and interfaces.
morloc allows data to be represented in its most natural form. Functions in morloc align neatly to conceptual algorithmic forms. The UPGMA tree building algorithm, for instance, is fundamentally a function from a distance matrix to a tree. The morloc type exactly matches this form by sourcing an idiomatic C++ function of a numeric matrix that returns a simple tree object. Traditional bioinformatics pipelines replace pure functions like this with applications. Rather than a distance matrix, the application must choose a file format to carry the distance matrix and must decide how to parse and propagate any associated metadata. More likely, an application would merge the distance matrix creation step and the tree building step into one function. This merge within the application prevents the two individual functions from being reused in other contexts.
morloc supports unconstrained modularity. Most morloc functions are pure functions. Complex behavior is built through composition. These compositions are checked and data is passed in well-defined structures. There is little or no overhead to morloc function calls and no formatting limitations. The programmer is free to organize functions to match the layout of the algorithm. Further, as seen in the implementation of upgma, all functions used in the algorithm can be exported and reused independently. In contrast, traditional pipelines must force enough work into each node to justify their high overhead and input/output must conform to accepted file formats. So modularity is limited to large operations over a sparse set of intermediate data types. Even this limited modularity is corrupted by format ambiguities and side effects that, for every new use context, necessitate careful testing and often new glue code.
morloc supports generic and compound data. In the case study, both genetic sequences and their metadata are passed as a compound data structure of type [(a, Sequence)]. Functions may be mapped across the sequence values without the possibility of altering the metadata (and vice versa). The metadata may be passed cleanly into completely new structures, such as the tree type in the case study. Such flexibility and well-defined coupling and transport are not possible in traditional bioinformatics pipelines where metadata is strongly limited by file format and where transformations between formats is usually lossy and ambiguous.
morloc supports higher-order functions. In morloc, functions can be passed as arguments, enabling functions to control the flow of operations. For example, onFst applies a given function exclusively to the first element of a tuple. map applies a function to every element in a list. foldTree recursively applies provided functions to reduce a tree into a single value. In each case, structural control logic is cleanly separated from application logic. This increases the reuse of complex control logic which improves consistency and reduces the likelihood of bugs. In conventional workflows, this finer logic would have to be handled within the applications themselves. Each application would have to independently solve problems like tree traversal. This limitation of traditional workflow languages restricts programmer freedom and forces more work onto application developers.
morloc nodes are simple functions rather than scripts. Each node in a morloc program is implemented as an independent, idiomatic function in the chosen language. morloc imposes no constraints on these functions beyond the expected type signature (see Table 1). In a conventional workflow, a node is instead an application that is saddled with all the complexity described in the introduction (see Fig. 1). Some workflow managers reduce the complexity through specialized code evaluation that bypasses the need to give the scripts full command line interfaces. Nextflow can pre-process a script template before execution to expand workflow variables to arbitrary strings of code. Snakemake can evaluate a script in a special context where a Snakemake object that stores workflow variables is added to scope. In both cases, the target source code uses workflow-specific syntax to access the workflow namespace. This complicates the code, interferes with testing and linting, and prevents reuse outside the workflow (see Table 1).
| main.loc—Morloc | sqr.py |
|
module sqr (val) source Py from "sqr.py" ("sqr") sqr :: Int -> Int type Py => Int = "int" val = sqr 2 |
def sqr(x): return (x * x) |
| main.nf — Nextflow | templates/sqr.py |
|
process SQR { input: val x output: path "result" script: template "sqr.py" } workflow { SQR( 2 ) } |
#!/usr/bin/env python3 with open("result", "w") as fh: print( { x } * { x }, file=fh) |
| Snakefile — Snakemake | scripts/sqr.py |
|
rule add: output: "result" params: x = 1, script: "sqr.py" |
with open(snakemake.output[0], "w") as fh: print ( snakemake.params.x * snakemake.params.x, file = fh) |
morloc nodes are not containerized by default. In Nextflow, all nodes are run in containers. This approach binds code to its environment, which improves reproducibility and portability. In contrast, morloc programs are normally built in a single environment. When unified builds are insufficient, containerized morloc components may be composed (see Fig. 8D). Alternatively, morloc functions may directly invoke containers. Containerization, though, is not built into morloc.
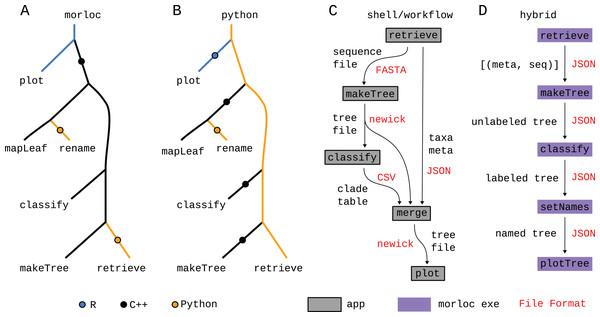
Figure 8: Comparison of paradigms.
(A–D) Call trees for four possible implementations of this case study. In the morloc implementation (A), the R plot function is evaluated first from an R context. A foreign call is made to the C++ pool requesting a tree from mapLeafs rename. The last C++ function, makeTree, makes a foreign call to retrieve in Python. The Python implementations (B) differs in that Python is the main language from root to tip. The Python programmer is responsible for designing the Python/C++ and Python/R interfaces. The Bash/workflow implementation (C) replaces each function with a standalone application (represented by a box). Each edge in the tree represents data passed as files in a specialized format (in red). Since free annotations cannot easily be added to file formats like FASTA and newick (for sequences and trees, respectively), the annotation metadata and clade predictions must be sent to separate files and then woven together with a dedicated tool (merge). Hybrid systems are possible (D), where a conventional workflow system uses morloc executables as nodes. In this case, the files between nodes are JSON representations of morloc data structures. Six implementations—morloc, Python, Bash, Snakemake, Nextflow, and hybrid (Snakemake and morloc)—are available at https://github.com/morloc-project/examples.morloc reduces function call overhead. Traditional workflow managers support mapping inputs (usually files) over applications, but high overhead costs (Fig. 4) make these programs inefficient at running many small functions. To compensate, they pack more work into each node. Individual nodes often operate on many values (e.g., sequences in multi-entry FASTA files) and implement their own particular parallelization schemes with accompanying dependencies and architectural requirements. morloc’s overhead, in contrast, is nearly zero in the best case and orders of magnitude lower in the worst case (Fig. 4). So the morloc programmer can efficiently work with concise functions and exercise fine control over their execution and parallelization. Further, they can reuse high-performance parallelization code, thus reducing the number of dependencies and improving performance.
morloc programs are typechecked and support type driven design. In the conventional bioinformatics pipeline, the form of intermediate data is only vaguely specified by the file type. Few errors can be caught before runtime. In contrast, morloc offers type checking and inference that allow the entire pipeline to be checked before it is run. This is of practical significance since scientific pipelines are often computationally expensive and the cost of late failure can be high. The typed functions also simplify reasoning and improve readability both for humans and machines.
Comparison to programming with one main language and foreign function interfaces
The influenza case study shows how morloc can specify a program that spans three languages: Python, C++, and R. For comparison, we implemented the same case study using a single primary language, Python, and making foreign function calls to C++ and R (see Fig. 8B). For interop we used the rpy2 library for R interop and pybind11 for C++. The program retrieves data in Python, calls three sequential C++ functions (to make the tree, classify it, and rename the leaves) and then translates the final C++ tree object to an R phylo object and sends it to the R interpreter to create the final tree.
The translation of the C++ tree object uses the same basic steps as morloc’s automatically generated interop code. It first calls the C++ unpack function to extract the tree data as a tuple (leaf list, edge list, and node list), then it uses rpy2 to cast the data into Python-wrapped R types. Next, it calls the R pack function to create a Python object that holds the Python-wrapped R phylo type. This final phylo object is passed to the R plotting function. This manual process achieves the same transformation—from C++ tree object to R phylo object — as the code automatically generated by morloc. While using rpy2 and pybind11 produces a clean and efficient solution, morloc offers several advantages.
morloc interoperability is declarative and generative. In traditional foreign function interface (FFI) workflows, developers must research, select, and integrate a different interoperability library for each language pair, often learning new APIs and manually writing complex data marshaling code. This can lead to code that is tightly coupled to specific interop tools (such as rpy2 and pybind11), which adds dependencies and complicates future refactoring and language substitution. In morloc, this work is shifted from the programmer to the compiler. The morloc programmer does not need to add any interop-specific modifications to their code. They only need to declare the morloc type signature for each imported function and then all interop code will be generated by the compiler. This simplifies code, reduces dependencies, and allows improvements in the compiler to benefit all morloc programs in parallel.
morloc symmetrically organizes program logic. In the morloc case study, we present a project with functions evenly partitioned between data retrieval steps in Python, algorithmic steps in C++, and visualization in R. With morloc, each partition can be presented as a composition of typed, modular functions and these may be transparently substituted and extended by the morloc programmer. If instead we choose one focal language, say Python, then the data retrieval steps would be accessible to the Python programmer, but the algorithmic and statistical logic would be hidden in external codebases written in foreign languages.
morloc enforces no central runtime language. In the Python case study, Python handles the user interface and the coordination of foreign function calls. Switching the base language to R or C++ would require very different implementations and dependencies. Because the core language manages the call sequence, data must repeatedly return to it. In our Python version, three sequential C++ functions are called. For each, Python stores the result and passes it to the next function without using it, adding unnecessary overhead and complexity. In the penultimate step, a tree object is returned from C++ to Python and then sent from Python to R for plotting—Python acts only as a conduit. With morloc, these redundant Python steps and their unused type representations disappear.
morloc allows easy polyglot prototyping. The leaf renaming step in the morloc case study starts in R and passes a Python renaming function to a C++ tree traversal algorithm. Each leaf renaming step requires a foreign call to Python. While not ideal for production code (tens of microseconds of overhead per loop), this freedom of mixing functions is valuable in a fast prototyping context. Importantly, the prototype is not a dead end. It may be optimized by adding a C++ implementation of the Python function and sourcing it into the morloc code. The morloc programmer can focus on quickly building the function composition that best describes the problem. In conventional systems, the extra complexity and maintenance costs incurred by manually written interoperability code often outweighs the advantage of reusing the foreign code.
morloc provides a framework for modular library development. Most work on language interoperability focuses on calling one language from another. This allows reuse of code written in foreign libraries. Most commonly, slower languages like Python and R call functions in fast C libraries. morloc offers a more symmetrical language-agnostic approach. Libraries can be built in the abstract on the foundational common type system. Implementations can be imported for defined functions and can share common benchmarking and test suites. From these libraries, we can build databases of verified, composable functions.
Related work
To the best of my knowledge, morloc is unique as a language based on multi-lingual native function composition under a common type system. However, morloc shares the idea of multi-lingual composition with workflow managers; the idea of a common type system with data serialization systems; and the idea of native composition with language runtimes and foreign function interfaces. In the following sections, we will discuss these relations.
Languages that separate scripts from components
The separation between script and component is the defining principle of all workflow approaches (Schneider, 1999). The script specifies the connectivity of the components and the components perform the actual data transformations. In morloc, the script is a functional programming language and the components are native functions from other languages. We will discuss the most common types of scripts below.
Domain specific languages (DSLs) are languages designed for a special purpose, in our case the implementation of workflows. Cuneiform is a functional, Erlang-based language focusing on automatic parallelization (Brandt, Reisig & Leser, 2017). BioShake is a DSL embedded in Haskell that allows metadata for files across a workflow to be expressed and checked by the Haskell type system (Bedő, 2019). BioNix implements purely functional workflows using the Nix language and package manager (Bedő, Di Stefano & Papenfuss, 2020). Nextflow is a Groovy-based DSL popular in bioinformatics (Di Tommaso et al., 2017).
Scripting languages are languages focused on automating tasks. These include shell languages such as Bash. While Bash may be used as a general programming language, it is more often used to manage files and the execution of components (programs) written in other languages. In addition to Bash, there are specialized scientific scripting languages, for example BPipe (Sadedin, Pope & Oshlack, 2012) and BigDataScript (Cingolani, Sladek & Blanchette, 2015), which may automate job submission and reuse past results.
Rule-based languages are declarative languages that use a recipe file (e.g., Makefile) to coordinate the execution of commands and caching of intermediate results for efficient building of projects. GNU Make is the most common of these. While Make is primarily used for software compilation, it has also been applied to scientific analysis (e.g., Askren et al., 2016). Many workflow languages have descended from it, including the popular Snakemake (Mölder et al., 2021) manager.
Specification languages are declarative languages for describing the behavior and requirements of components and their connectivity. Two popular examples are the Common Workflow Language (CWL) (Crusoe et al., 2022) and the Workflow Description Language (WDL) (Voss, Auwera & Gentry, 2017). These workflow specifications may be run by external execution engines such as Arvados (Amstutz, 2015), Cromwell (Voss, Auwera & Gentry, 2017) or Toil (Vivian et al., 2017).
Language-specific workflow managers are packages in a given language that manage the execution of functions in the same language. Examples of these include Parsl (Babuji et al., 2018) in Python and targets in R (Landau, 2021). Though all composed functions are in one language, functions may make system calls to external applications or have foreign function interfaces to external languages.
Frameworks for interoperability and serialization
Interoperability through serialization has been heavily explored and effectively used in practice. Many data serialization frameworks use common type systems to generate serialization code. These frameworks include Google Protocol Buffers (https://protobuf.dev), Apache Thrift (Slee, Agarwal & Kwiatkowski, 2007), and Apache Avro (Vohra & Vohra, 2016). Each of these has a means of specifying type schemas that direct the generation of serialization code in supported languages. Remote Procedure Call (RPC) systems build on these serialization frameworks to allow calls between systems in a language and platform independent manner.
Interoperability runtimes circumvent the need for serialization by allowing shared memory between languages. The Common Language Runtime (CLR) in the .NET framework is one such system that allows typed communication between supported languages (Gough & Gough, 2001). Interoperability is based on a common binary layout specified by the Common Language Infrastructure (CLI). Languages designed for this infrastructure can share objects without any special boilerplate. Similarly, GraalVM allows interoperability between supported languages by executing them in a common runtime through the TruffleVM framework (Grimmer et al., 2015).
Beyond these general purpose runtimes, many tools have been developed to enable pairs of languages to interoperate seamlessly. The Simplified Wrapper and Interface Generator (SWIG) (Beazley, 1996) allows C and C++ code to be called from many high-level programming languages including Python, Perl, Ruby, and Tcl. MetaCall goes even further, offering binary interfaces between a wide range of languages (https://metacall.io/). Other tools are specialized for one pair of languages. These include the ctypes and pybind11 modules for calling C++ from Python; Rcpp for calling C++ from R (Eddelbuettel & François, 2011), and PypeR (Xia, McClelland & Wang, 2010) and rpy2 for calling R from Python.
While these projects overlap with morloc, their focus differs. Their purpose is to provide interoperability laterally between languages. They are a glue between languages. Dedicated code is written in the target languages to allow them to communicate. morloc, in contrast, is a system under languages that works in the background to tie the polyglot components together. There is no morloc-specific code written in the component source.
Future work
The current implementation of morloc (v0.53.7) demonstrates a strongly-typed solution to high-performance, multi-lingual function composition. morloc is still being actively developed. A few of the major goals for future work are listed below:
Improve workflow features. Though morloc has experimental support for remote job execution and caching, it lacks the full feature set of standard workflow programs. Until these features are mature, hybrid solutions using a conventional workflow manager and morloc-generated components may be a helpful compromise (see Fig. 8D). Future work includes improved error recovery, monitoring, and broader support for cloud computing.
Increase language support. Currently only C++, Python, and R are supported. An obvious goal is to add more languages and streamline the onboarding process. A deeper challenge is improving language feature support. Polyglot morloc programs are currently limited to composition of eager functions of immutable and non-streaming data. Further generalizing morloc will require additional work on the backend generators and the typesystem.
Expand the type system. We plan to add sum types, refinement types, contracts, extensible records, and effect handling. Sum types, though absent in many languages, greatly improve data modeling. Refinement types will allow the expected behavior of a function to be described more clearly. Contracts allow the specification of pre- and post- conditions to a function. Extensible records will improve reuse and specialization of records and tables. Effect handling will allow behavior such as mutability, console printing, randomness, and exceptions to be modeled by the user and correctly handled by the compiler.
Develop the ecosystem. Usability can be improved by generating executables with richer usage statements, error messages, profiling options, debugging support, dependency handling, and documentation integration. The generators could also be extended to make REST APIs or simple graphical user interfaces in addition to command line executables. We also intend to develop a searchable database of functions that can integrate with IDEs and AI code generators.
Open the black boxes. Currently the value checker cannot determine if foreign source code is what morloc expects. The source code is a black box. But we may be able to peak inside with LLMs and other static analysis tools. We may similarly be able to automatically generate candidate type signatures for foreign libraries, reducing integration effort and improving safety.
Conclusion
We present morloc as an alternative to the file and application based paradigm that now dominates bioinformatics. Replacing the old paradigm is an ambitious but necessary goal. It will require stripping existing monolithic applications down to their algorithmic cores and exposing them as simple programmatic libraries. These libraries may then be raised into the morloc ecosystem by adding type signatures to exported functions. Functions may be shared through functional databases searchable by type. These may be easily benchmarked and integrated into new pipelines. Novel algorithmic work may be shared as ageless functions rather than heavy, idiosyncratic, high-maintenance applications. Conventional bioinformatics file formats may be replaced with simple, well-defined, generic data structures that map cleanly to native types in memory and storage types on the disk or in databases. This transition will require refactoring most bioinformatics code and developing infrastructure for reproducible polyglot builds. We hope to free future scientists from floundering in software engineering and infrastructure minutiae and instead focus on designing and using elegant functions and solving logical problems using the languages of their choice.
Availability
The morloc source code is published under a GPL license and is freely available on GitHub (https://github.com/morloc-project/morloc). The present work describes morloc version 0.53.7. We make no guarantees of backwards compatibility for this version. The influenza case study, the five alternative implementations, and all benchmarking code are available at https://github.com/morloc-project/examples (release v1.1). Archived versions of the code and examples are also available through Zenodo: code and examples.