Evaluation of leaf rust resistance in the Chinese wheat cultivar ‘Een1’
- Published
- Accepted
- Received
- Academic Editor
- Guorong Zhang
- Subject Areas
- Agricultural Science, Plant Science
- Keywords
- Wheat, Leaf rust disease, Leaf rust resistance gene, Puccinia triticina, Polymorphism, Molecular mapping, SSR markers, Resistance identification, Inoculation, Gene postulation
- Copyright
- © 2020 Zhang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Evaluation of leaf rust resistance in the Chinese wheat cultivar ‘Een1’. PeerJ 8:e8993 https://doi.org/10.7717/peerj.8993
Abstract
Wheat cultivar Een1, 34 near isogenic lines (NILs), and two cultivars were used as plant materials to evaluate the resistance of Een1 to leaf rust disease. Infection type identification and gene postulation were carried out by inoculation of 12 Chinese Puccinia triticina (Pt) pathotypes. Based on the unique phenotype of Een1, we speculated that Een1 might carry Lr gene(s) different from the tested ones. The chromosomal locations for resistance gene to leaf rust disease was employed using SSR primers mapping the populations derived from the cross between Een1 and susceptible Thatcher. A total of 285 plants in the F2 population were tested by inoculating Pt pathotype FHNQ during the seedling stage. Results from the segregation analysis fits a ratio of 3:1 (, P = 0.12), indicating the presence of a single dominant gene in Een1 conferring resistance to FHNQ. A total of 1,255 simple sequence repeat (SSR) primers were first used to identify the likely linked markers based on bulk segregation analysis (BSA), and then those likely linked markers were further genotyped in the F2 population for linkage analysis. Our linkage analysis found that the resistance gene (LrE1) was distal to seven SSR loci on the long arm of chromosome 7B, with distances from 2.6 cM (Xgwm344) to 27.1 cM (Xgwm131). The closest marker Xgwm344 was further verified with F3 lines.
Introduction
Wheat leaf rust, caused by Puccinia triticina (Pt), is one of the most devastating fungal diseases affecting wheat, causing severe yield losses globally. It has caused serious epidemics in North America and South America, and is a major seasonal disease in India. Destructive epidemics of leaf rust disease occurred in the 1970s in China (Dong, 2001), and in recent years, leaf rust pathogens caused epidemics in major wheat production regions in China including Gansu, Sichuan, Shaanxi, Henan, Anhui, Hebei and Shandong Provinces (Zhou et al., 2013a). These epidemics could be related to global warming and continuous intensive crop production in the same fields. Utilization of resistant cultivars would still be the most effective, economic and eco-friendly way to control wheat leaf rust disease (Chen et al., 1998).
To date, more than 100 leaf rust resistance (Lr) genes/alleles have been identified in wheat and its relatives (Singla et al., 2017). Only a few designated genes (Lr9, Lr19, Lr24, Lr25, Lr28, Lr29, Lr38 and Lr47) are effective at the seedlings stage against the prevalent Chinese Pt pathotypes (Zhang et al., 2020). The fact that most of these effective genes have not been detected in the Chinese wheat cultivars, means that most cultivars from the Chinese wheat germplasms could be rapidly overcome by these pathogens. It is therefore risky to release cultivars with limited or single gene resistance (also termed as major, seedling or race specific resistance). To cope with the dynamic and rapid evolution of Pt populations, it is necessary to identify new and effective resistance genes in different germplasms, so as to enlarge the Chinese wheat gene pool, thereby pyramiding multiple genes in wheat breeding programs.
Due to the advantages of high polymorphism and known chromosome location, simple sequence repeat (SSR) markers have been widely used in genetic studies during the last two decades (Röder et al., 1995). Many Lr genes such as Lr3, Lr12, Lr37, Lr39/41, Lr51 and Lr52 have been mapped on wheat chromosomes using SSR markers (Singh & Bowden, 2011; McIntosh et al., 2013). We have mapped several Lr genes in our previous studies including Lr19 on chromosome 7D (Li et al., 2005), Lr45 on chromosome 2A (Zhang et al., 2007), LrZH84, LrBi16 and LrXi on chromosome 1BL (Zhou et al., 2013b; Zhang et al., 2011; Li et al., 2010a), LrNJ97 on 2BL (Zhou et al., 2013a), and LrFun on chromosome 7BL (Xing et al., 2014). Several adult resistance loci have also been mapped in our previous studies (Qi et al., 2016; Zhang et al., 2017).
Released by Hongmiao State Agricultural Science Research Institute in Hubei province (China), the wheat cultivar Een1 showed high resistance to multiple fungal diseases including leaf rust, stripe rust, stem rust and powdery mildew. This cultivar also has moderate resistance to wheat scab and lodging resistance (https://baike.baidu.com/item/%E9%84%82%E6%81%A91%E5%8F%B7). The objectives of the present study were to (1) investigate the resistance of Een1 to 12 Chinese P. triticina pathotypes, and (2) explore and map the leaf rust resistance gene in Een1.
| Cultivars/lines | Pathotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| THST | FHNQ | PHGS | THTQ | THPS | FHTR | SHKN | PHST | THGR | THTS | PHTT | THPS | |
| RL6003 (Lr1) | 3 | ;1 | 3 | 3 | 3 | ;1 | 4 | 4 | 3 | 3 | 4 | 4 |
| RL6016 (Lr2a) | 3 | 1 | 1 | 3 | 3 | 2 | 4 | 2 | 3 | 3 | 2 | 4 |
| RL6047 (Lr2c) | 3 | 4 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 3 | 4 | 4 |
| RL6002 (Lr3) | 3 | 4 | 3 | 3 | 3 | 3 | ; | 4 | 4 | 3 | 4 | 4 |
| RL6010 (Lr9) | ; | ; | ; | ; | ;1 | ; | 0 | 0 | ; | 1 | 0 | 0 |
| RL6005 (Lr16) | 3 | 4 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 |
| RL6040 (Lr24) | ;1 | ; | ;1 | ; | ; | ;1 | ; | ;1 | ;1 | ; | ; | ; |
| RL6078 (Lr26) | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 3 | 4 | 4 |
| RL6007 (Lr3ka) | 3 | 3 | ;1 | 3 | 3 | 3 | ; | 4 | ; | 3 | 4 | 4 |
| RL6053 (Lr11) | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 4 | 2 |
| RL6008 (Lr17) | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 2 | 4 | 4 | 4 |
| RL6049 (Lr30) | 1 | ;1 | ;1 | 3 | 3 | 3 | 3 | 2 | ; | 3 | 3 | 3 |
| RL6051 (LrB) | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 3 | 3 | 4 | 4 |
| RL6004 (Lr10) | 3 | 3 | 3 | 3 | 3 | 3 | ; | 4 | 4 | 3 | 4 | 4 |
| RL6013 (Lr14a) | 3 | 2 | 4 | 1 | 3 | 2 | 3 | 3 | 2 | 4 | 4 | 4 |
| RL6009 (Lr18) | 3 | 2 | 1 | 1 | 2 | 3 | ; | 3 | 4 | 3 | 3 | 2 |
| RL6019 (Lr2b) | 2 | 3 | 1 | 3 | 3 | 3 | 4 | 3 | 4 | 3 | 4 | 3 |
| RL6042 (Lr3bg) | 3 | 4 | 3 | 3 | 3 | 3 | ; | 3 | 4 | 3 | 4 | 3 |
| RL6006 (Lr14b) | 3 | 3 | 4 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 4 |
| RL6039 (Lr14ab) | 3 | ;1 | 1 | 1 | 3 | 2 | 3 | 3 | 2 | 3 | 4 | 4 |
| RL6052 (Lr15) | 3 | ;1 | 3 | 3 | 3 | 3 | ; | 4 | 3 | 3 | ; | 4 |
| RL6040 (Lr19) | ; | ; | ; | 0 | ; | ; | ; | ; | ; | 1 | 0 | ; |
| RL6043 (Lr21) | 3 | 2 | 3 | 3 | 2 | 2 | 4 | 3 | ; | 3 | 4 | 4 |
| RL6012 (Lr23) | 3 | 3 | 4 | 3 | 3 | 3 | 4 | 3 | 4 | 4 | 4 | 4 |
| RL6084 (Lr25) | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 4 |
| RL6079 (Lr28) | ; | ; | ; | ; | ; | ; | ; | ;1 | ;1 | ; | ; | ; |
| RL6080 (Lr29) | 3 | ; | 3 | 3 | 3 | 3 | ; | ; | ; | ; | ; | ; |
| RL6057 (Lr33) | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 4 | 4 |
| E84018 (Lr36) | 0 | ; | 3 | 1 | ; | ;1 | 4 | 1 | 3 | 3 | 4 | 4 |
| RL6097 (Lr38) | ;1 | ; | ;1 | ; | ; | ; | ; | ;1 | 0 | ; | ; | ; |
| KS86WGRC02 (Lr39) | 3 | ; | 3 | 3 | 3 | 1 | 2 | 2 | 2 | ; | ; | ; |
| KS91WGRC11 (Lr42) | ;1 | 3 | ; | ; | ; | ; | ; | ; | 2 | ; | 3 | ; |
| RL7147 (Lr44) | 4 | 3 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 2 | 4 | 4 |
| KS96WGRC36 (Lr50) | 4 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 3 | 4 |
| Een1 | ; | ; | ; | ; | ; | ; | ; | 1 | 2 | 3 | 3 | 4 |
| Bimai 16 | 1 | 1 | 1 | ;1 | 1 | 2 | ; | 3 | ;1 | 0; | 3 | ; |
| Fundulea 900 | 0; | ;1 | ; | 0; | ; | ;1 | 1 | ;1 | 0; | 4 | 0; | 0; |
| Thatcher | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
Materials & Methods
Plant materials and leaf rust evaluation in the greenhouse
Experimental plant materials including 34 near-isogenic lines (NILs) with Thatcher background, susceptible line Thatcher, cultivar Een1, Bimai16 and Fundulea900 were used for gene allelic polymorphism evaluations (Table 1). Five to seven seedling plants for each tested line were grown in a growth chamber and the inoculations were performed by spraying urediniospores of the tested 12 Pt pathotypes when the first leaves appeared fully expanded. Inoculated seedlings were placed in a chamber and incubated at 20 °C with 100% relative humidity in dark for 16 h. They were then transferred to a greenhouse with 12 h light/12 h darkness at 22 ± 3 °C with 70% relative humidity (RH). Infection types (ITs) were scored at 14 days post inoculation according to the Stakman scale modified by Roelfs, Singh & Saari (1992). The standards applied in this scale were: “0” representing immunity (no sign of infection), “;” for necrotic flecks, “1” for small uredinia surrounded by a necrosis, “2” for small to medium uredinia surrounded by chlorosis, “3” for medium uredinia without chlorosis or necrosis, and “4” for large uredinia without chlorosis or necrosis. Plants that scored IT 3 or higher were considered susceptible. The F1 population (15 plants), F2 population (285 plants) and F3 population (30 seedlings from each of the 155 plants selected from the F2 population), derived from Een 1 ×Thatcher, were used for mapping to identify SSR molecular markers associated with the leaf rust resistance genes. Pt pathotype FHNQ was used to phenotype all the plants tested in the F1, F2 and F3 populations.
Simple Sequence Repeat (SSR) marker analysis
Genomic DNAs were extracted from seedlings using the CTAB method. A total of 1255 SSR primers (Zhang et al., 2011) were selected randomly from the GWM, WMC, CFA, CFD, and BARC primer series covering each wheat chromosome (https://wheat.pw.usda.gov/). All the primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. Bulked segregant analysis described by Michelmore, Paran & Kesseli (1991) (equal amounts of genomic DNA from 10 resistant (Br) and 10 susceptible (Bs) from F2 plants, along with the two parents) was performed in a preliminary screen to identify molecular markers likely to be linked to the resistance gene in Een1. Polymorphic primers for the parents and the bulked pools were then genotyped across the individual lines in the F2 and F3 populations. PCR amplification was carried out as described by Xing et al. (2014). Mixed with 8 µL formamide loading buffer, all the denatured PCR products were separated on 6% denaturing polyacrylamide gels for approximately 1.5 h at 100 W and viewed by the silver staining method.
Construction of the linkage map
To evaluate the deviations of the observed and expected segregation ratios, Chi-squared (χ2) tests for goodness-of-fit were calculated using Microsoft Excel 2010 software. Linkage analysis between SSR markers genotyping and the phenotyping results were performed by MapManager QTXb20 software (LOD = 3) (Manly, Cudmore & Meer, 2001). And Kosambi mapping function (Kosambi, 1944) was used to calculate the genetic distances between the markers and the resistance gene. The linkage map was drawn using Mapdraw Version 2.1 software (Liu & Meng, 2003).
Results
The gene postulation for Een1
Twelve Chinese Pt pathotypes (THST, FHNQ, PHGS, THTQ, THPS, FHTR, SHKN, PHST, THGR, THTS, PHTT, THPS) were used to phenotype the wheat cultivar Een1, susceptible line Thatcher and 34 near isogenic lines (NILs) carrying different Lr genes. Two other wheat cultivars, Fundulea900 and Bimai16, were also tested. Results for all the phenotypic infection types (IT) are listed in Table 1. Een1 showed high resistance to nine of the tested Pt races (THST, FHNQ, PHGS, THTQ, THPS, FHTR, SHKN, PHST, THGR) whilst three Pt races (THTS, PHTT and THPS) showing virulence. An overall analysis showed that Pt race THTS had low IT records for the near-isogenic lines including TcLr9, TcLr24, TcLr19, TcLr28, TcLr29, TcLr38, TcLr39, TcLr42 and TcLr44. The Pt race PHTT produced low ITs with TcLr2a, TcLr9, TcLr24, TcLr15, TcLr19, TcLr28, TcLr29, TcLr38, and TcLr39 lines. Pathotype THPS recorded a low infection type on TcLr9, TcLr24, TcLr11, TcLr18, TcLr19, TcLr28, TcLr29, TcLr38, TcLr39, and TcLr42. The combined results from these three Pt races enable us to conclude that none of the corresponding genes including Lr2a, Lr9, Lr11, Lr15, Lr18, Lr19, Lr24, Lr28, Lr29, Lr38, Lr39, Lr42 and Lr44 existed in our tested wheat cultivar Een1.
With a different infection type (“;”) on TcLr3ka, TcLr30 and TcLr21 lines, the Pt race THGR showed infection type “2” on Een1, we can conclude that cultivar Een1 carried no Lr3ka, Lr30 and Lr21 genes. The general results in Table 1 reveal that the phenotype of Een1, in relation to the pathotypes, had different result patterns for TcLr1, TcLr2c, TcLr2b, TcLr3, TcLr3bg, TcLr10, TcLr17, TcLr14ab, TcLr14a, TcLr36 and TcLr50 lines. These results indicate that Een1 carried none of the genes mentioned above. In this study, final conclusions for genes in Een1 with high IT records on corresponding NILs, Lr2c, LrB, Lr16, Lr26, Lr14b, Lr23, Lr25 and Lr33 were not possible. Based on the combined phenotyping results, it could be concluded that Een1 may carry new Lr gene(s) besides one or several of Lr2c, LrB, Lr16, Lr26, Lr14b, Lr23, Lr25 or Lr33. In addition, depending on the unique phenotypes of Een1 responding to the 12 tested Pt pathotypes, we questioned whether wheat cultivar Een1 may carry unknown leaf rust resistance gene(s) other than the tested Lr genes, or there may be a combined action of more than one Lr gene.
Inheritance of leaf rust resistance in Een1
All the plants in the F2 and F3 populations, along with the parents, were inoculated with Pt pathotype FHNQ (avirulent on Een1 and virulent on Thatcher) at the seedling stage and the test results were presented in Table 2 and Table 3. Of all the 285 plants tested in F2 population, 225 individuals showed resistance phenotype and 60 were susceptible, giving a suitable 3:1 ratio (χ23:1 = 2.37, P = 0.12) (Table 2). Among the tested 155 families in the F3 population, 40 were homozygous resistant, 80 heterozygous and 35 homozygous susceptible, fitting an expected ratio of 1:2:1 (χ21:2:1 = 0.48, P = 0.78) (Table 3). Results from both the F2 and F3 populations indicated that leaf rust resistance to Pt FHNQ in wheat cultivar Een1 was conferred by a single dominant gene, tentatively designated as LrE1.
| Material | Total plant | Resistance | Susceptible | Goodness of fit test |
|---|---|---|---|---|
| Een1 | 15 | 15 | ||
| Thatcher | 15 | 15 | ||
| F1 | 15 | 15 | ||
| F2 | 285 | 225 | 60 | χ23:1 = 2.37, P = 0.12 |
| F3 phenotype | F3 genotype | Goodness of fit test | Allele | |
|---|---|---|---|---|
| D | B | |||
| Resistance 120 | RR 40 | 39 | 1 | |
| Rr 80 | χ21:2:1 = 0.48, P = 0.78 | 78 | 2 | |
| Susceptible 35 | rr 35 | 4 | 31 | |
Notes:
- RR
-
homozygous resistant
- Rr
-
segregating
- rr
-
homozygous susceptible
- D
-
homozygous for Een1 allele or heterozygous
- B
-
homozygous for Thatcher allele
Linkage analysis and genetic map
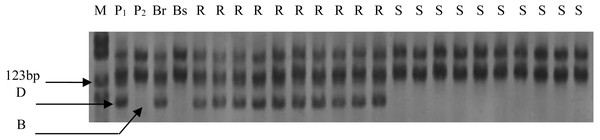
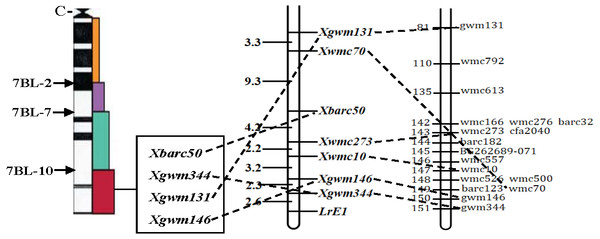
Out of the 1255 randomly selected wheat SSR primers, seven SSR primers (Xgwm344, Xgwm146, Xwmc10, Xwmc70, Xwmc273, Xbarc50, Xgwm131) located on chromosome 7BL showed polymorphism between the resistance and the susceptible bulks as well as the parents (Table 4). All the seven polymorphic SSR primers were further used to genotype the DNA samples from each of the 285 F2 plants (Fig. 1). The linkage analysis using Mapmanager QTXb20 software showed that Xgwm344 was linked to LrE1 with a genetic distance of 2.6 cM as the closest SSR marker (Fig. 2). The SSR marker Xgwm146 had a value of 4.9 cM, thus scoring the second genetic distance. And the furthest marker was Xgwm131 with 27.1 cM from the gene LrE1. In the 155 families in the F3 population, Xgwm344 proved to be 4.8 cM to gene LrE1.
| Marker loci | F2 phenotype | Allele | ||||
|---|---|---|---|---|---|---|
| A | H | B | D | B | ||
| Xgwm344 | Resistance | – | – | – | 222 | 3 |
| Susceptible | – | – | – | 4 | 56 | |
| Xgwm146 | Resistance | 76 | 143 | 6 | – | – |
| Susceptible | 1 | 6 | 53 | – | – | |
| Xwmc10 | Resistance | 70 | 138 | 17 | – | – |
| Susceptible | 1 | 7 | 52 | – | – | |
| Xwmc273 | Resistance | – | – | – | 208 | 17 |
| Susceptible | – | – | – | 14 | 46 | |
| Xbarc50 | Resistance | 53 | 154 | 18 | – | – |
| Susceptible | 1 | 24 | 35 | – | – | |
| Xwmc70 | Resistance | 78 | 126 | 21 | – | – |
| Susceptible | 1 | 46 | 13 | – | – | |
| Xgwm131 | Resistance | – | – | – | 197 | 28 |
| Susceptible | – | – | – | 46 | 14 | |
Notes:
- D
-
homozygous for Een1 allele or heterozygous
- B
-
homozygous for Thatcher allele
- A
-
homozygous for Een1 allele
- H
-
heterozygous
Figure 1: Electrophoresis of PCR products amplified with Xgwm344 on polyacrylamide gels.
M: PBR322/Msp I Marker; P1: resistance parental line Een1; P2: susceptible parental line Thatcher; Br: resistance bulk; Bs: susceptible bulk; R: resistance F2 plants; S: susceptible F2 plants; B: homozygous for Thatcher allele; D: homozygous for Een1 allele or heterozygous.Figure 2: Linkage map of the resistance gene LrE1 using SSR markers on chromosome 7BL.
Deletion bin map on wheat chromosome 7BL (Sourdille et al., 2004) (left); linkage map of leaf rust resistance gene LrE1 generated using data from F2 population of Een1 × Thatcher (centre). Locus names and corresponding locations are indicated on the right, genetic distances are labeled on the left in centiMorgans; and compared with previously published wheat chromosome 7BL map (Somers, Isaac & Edwards, 2004) (right).Discussion
Seedlings of the winter wheat cultivar Een1 showed high resistance to multiple fungal diseases. With good agronomic traits, this cultivar has significant potential for the future genetic improvement in wheat. Results from this study indicated that Een1 has a high resistance to several epidemic Pt pathotypes including THST, FHNQ, PHGS, THTQ, THTS, FHTR and SHKN. Specific genetic analysis on the F2 and F3 populations indicated that the resistance of Een1 against FHNQ was conferred by a single dominant gene, provisionally designated as LrE1. This resistance locus was distal to the seven SSR markers (Xgwm344, Xgwm146, Xwmc10, Xwmc273, barc50, Xwmc70, Xgwm131) on the long arm of chromosome 7B. The closest SSR marker, Xgwm344, was linked to LrE1 with a genetic distance of 2.6 cM. The linear order for these markers in the genetic map drawn in this study was similar to the high-density consensus map developed by Somers, Isaac & Edwards (2004), with the exception that Xwmc70 was proximal to Xgwm344 and Xgwm146. The difference between these results could be due to the specific populations analyzed or the molecular markers employed in the tests.
Currently, there are five designated Lr genes on wheat chromosome 7B (Lr14a, Lr14b, Lr68, LrBi16, and LrFun) (Herrera-Foessel et al., 2008; Dyck & Samborski, 1970; Herrera-Foessel et al., 2012; Zhang et al., 2011; Xing et al., 2014). LrE1 is a resistance gene activated at the seedling stage, since Lr68 has been previously reported as an adult resistance gene (Herrera-Foessel et al., 2012), this indicates that LrE1 is different from Lr68. Since wheat near isogenic lines (NILs) carrying Lr14a or Lr14b showed different phenotypes than Een1 when inoculated with the FHNQ, LrE1 could be different from Lr14a and Lr14b. Een1 had similar resistant reaction to FHNQ as Bimai16 and Fundulea900. However, Zhang et al. (2011) reported that the LrBi16 in Bimai16 was flanked between molecular markers Xcfa2257 (2.8 cM) and Xgwm344 (2.9 cM), with Xgwm146 at the same side of the chromosome. The LrFun gene in wheat cultivar Fundulea900 was located between molecular markers Xgwm344 (4.4 cM) and Xwmc70 (5.7 cM) (Xing et al., 2014). It seems like the LrE1 gene has a different chromosome position, which is outside the region of Xgwm344 and Xwmc70. Therefore, LrE1 might be different from both LiBi16 and LrFun. Future studies are needed to further clarify the genetic relationship between these three genes, which would include phenotyping and fine mapping on the populations derived from the crosses between Een1 and the lines with single Lr gene including LrBi16 and LrFun as what has been done by Zhang et al. (2015). Since only the Pt race FHNQ was tested in this study, other low virulence Pt pathotypes such as THST, PHGS, THTQ should also be the focus in the future.
Based on previous research findings, only a few Lr genes, including Lr1, Lr3, Lr3bg, Lr10, Lr13, Lr14a, Lr16, Lr23, Lr26, Lr34 and Lr35, were detected in Chinese cultivars. Most of the Lr genes are likely to lose their resistance function due to the rapid evolution of Pt pathotypes in China (Li et al., 2010b). The pedigree of Een1 are Lvorin10/761//Sumai3. No Lr gene has been found in 761. Sumai3 contained Lr1 and Lr34, and showed slow-rusting resistance at the adult stage with a disease index (DI) of 0.8 in the field (Ding et al., 2010), Een1 showed slow-rusting resistance to mixed Pt pathotypes at the adult stage (Fig. S2), so Een1 may have inherited Lr34 (on chromosome 7D) from Sumai3. Previous research showed that Een1 has a 1BL.1RS and carries another leaf rust resistance gene Lr26 on chromosome 1B (Li et al., 2010b; Yan et al., 2017). According to Hu & Chen (1992), there were/was more Lr gene(s) in Lovrin10 besides Lr26 and Lr2c, so the resistance to FHNQ in Een1 may be derived from Lovrin10. Generally, utilization of wheat cultivars carrying multiple resistance genes is an effective way to improve both wide-spectrum resistance against various pathogens and durability of such resistance. The cultivar Een1, with its characteristic of multi-resistance (with the known genes Lr26 and LrE1 identified in this paper) and other good agronomic traits, could be widely distributed in China to delay the “loss of resistance”.
Conclusions
A seedling leaf rust resistance gene (provisionally named LrE1) was identified in Een1, which showed high resistance to nine Puccinia triticina (Pt) pathotypes prevalent in China. With multi-resistance traits and slow-rusting resistance, the cultivar could become important in delaying loss of disease resistance if widely distributed.