Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant
- Published
- Accepted
- Received
- Academic Editor
- Genlou Sun
- Subject Areas
- Agricultural Science, Biotechnology, Genomics, Molecular Biology
- Keywords
- Eggplant, WRKY transcription factor, Phylogenetic analysis, Inton-exon structure, Conserved motifs, Differentially expressed, Cold stress
- Copyright
- © 2020 Yang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 8:e8777 https://doi.org/10.7717/peerj.8777
Abstract
Background
WRKY proteins play a vital role in the plants response to different stresses, growth and development. Studies of WRKY proteins have been mainly focused on model plant Arabidopsis and a few other vegetable plants. However, the systematical study of eggplant WRKY transcription factor superfamily is scarce.
Methods
Bioinformatics has been used to identify and characterize the eggplant WRKY gene family. For the exploration of the differentially expressed WRKY genes, two cultivars with different cold-tolerance were used. Finally, we performed a virus-induced gene silencing (VIGS) experiment to verify the functions of SmWRKY26 and SmWRKY32.
Results
Fifty eight (58) genes encoding eggplant WRKY proteins were identified through searching the eggplant genome. Eggplant WRKY proteins could be classified into three groups or seven subgroups in accordance with other plants. WRKY variants were identified from the eggplant. Gene structure analysis showed that the number of intron in eggplant WRKY family was from 0 to 11, with an average of 4.4. Conserved motif analysis suggested that WRKY DNA-binding domain was conserved in eggplant WRKY proteins. Furthermore, RNA-seq data showed that WRKY genes were differentially expressed in eggplant response to cold stress. By using VIGS, the two differentially expressed genes-SmWRKY26 and SmWRKY32 were verified in response to cold stress.
Discussions
This study provides a foundation for further exploring the functions of WRKY proteins in eggplant response to stresses and eggplant genetic improvement in stresses.
Introduction
WRKY proteins, a family of transcription factor specific in plants, have been identified for more than twenty years since they were first named as SPF1 in sweet potato (Ipomoea batatas L.) and seldom found in other species (Ishiguro & Nakamura, 1994; Eulgem et al., 2000; Rinerson et al., 2015). They are named as WRKY because of their highly conserved WRKY domain present in the protein sequence. The WRKY domain consists of a conserved core sequence WRKYGQK and a zinc-finger motif (Eulgem et al., 2000). WRKY proteins can be classified into three groups based on the number and amino acid sequences of the conserved WRKY domains. Group I WRKY proteins contain two WRKY domains, while the other two WRKY groups compose of one. The zinc finger motif is different among the three WRKY groups. A C2H2 motif is found in the C-terminal of group I and II WRKY proteins. Instead of C2H2 motif, a C2HC motif is in the C-terminal of group III WRKY proteins (Eulgem et al., 2000). Moreover, group II WRKY proteins can be further divided into five subgroups in accordance with the number and type of amino acids between the C2 in the zinc finger motif (Rushton et al., 2010). WRKY transcription factors can specially bind to the target genes by recognizing the (T)TGAC(C/T) sequences in the promoters of target genes (Yu, Chen & Chen, 2001).
Since its first identification in sweet potato, the functions of WRKY proteins have been focused on biological progresses, including plant responses to biotic stress, abiotic stress and plant development. Several studies have confirmed that WRKY transcription factors are involved in hair development (Verweij et al., 2016), seed size (Luo et al., 2005; Gu et al., 2017), pollen development (Lei et al., 2017), growth types (Yang et al., 2016), flowering time (Yang et al., 2016), fruit ripening (Cheng et al., 2016) and leaf senescence (Ay et al., 2009; Brusslan et al., 2012; Yang et al., 2016). WRKY transcription factors modulates plant hormone signaling pathways (Liu et al., 2017a; Liu et al., 2017b; Jin et al., 2018; Zheng et al., 2018). Despite the important roles in plant growth and development and signal pathways, the most significant function of WRKY proteins studied thoroughly and reported widely is transcription regulation of response to abiotic and biotic stresses. In Arabidopsis thaliana, AtABO3, a WRKY protein, was involved in plant response to drought stress (Ren et al., 2010). In rice (Oryza sativa L.). OsWRKY11 was induced by heat stress and overexpression of the OsWRKY11 enhanced the tolerance to high temperature stress (Wu et al., 2009). In addition, a vast number of studies reported that WRKY transcription factors were correlated with the resistance to salt stress (Xu et al., 2014; Cai et al., 2017), ozone stress (Gottardini et al., 2016; Zhang et al., 2017) and cold stress (He et al., 2016; Zhang et al., 2016). Futhermore, the functions and roles of WRKY proteins in plant immune response are outstanding. For example, Xu et al. (2006) found that AtWRKY18, AtWRKY40 and AtWRKY60 interacted with each other physically and functionally in an overlapping and antagonistic pattern, which played distinct roles in plant response to two types of pathogens-P.syringae and B.cinerea. Molecular complementation and gene silencing have confirmed that WRKY33 homologues in Arabidopsis and tomato (Solanum lycopersicum L.) played a critical role in resistance to botrytis (Zhou et al., 2015). In a recent report, BROWN PLANTHOPPER RESISTANCE 14 (BPH14), the first planthopper resistance gene, mediated the planthopper resistance by interaction with WRKY46 and WRKY72 that could bind to receptor-like cytoplasmic kinase genes and the callose synthase genes in rice. This result threw a light on the role of WRKYs in plantthopper resistance (Hu et al., 2018). Taken together, it is obvious that WRKY proteins can mediate plant defense mechanism in different manners.
Reports about WRKY proteins in some crops including vegetable crops are relatively limited, although all studies about WRKY proteins have focused mainly on model plant Arabidopsis and a few other plants. For instance, there are more than 70 WRKY proteins in Arabidopsis (Eulgem et al., 2000; Dong, Chen & Chen, 2003), 102 WRKY proteins in rice (Ross, Liu & She, 2007), 174 WRKY proteins in soybean (Glycine max L.) (Yang et al., 2017a; Yang et al., 2017b) and 81 WRKY proteins in tomato (Huang et al., 2012a; Huang et al., 2012b). Eggplant, a world-wide cultivated species, is one of the most important vegetable crops (Wang, Sulli & Fu, 2017; Yang et al., 2017a; Yang et al., 2017b). It is especially popular in Asia, the Middle and Near East, Southern Europe and Africa (Daunay & Lester, 1988). The information of eggplant genome has been released in 2014 (Hirakawa et al., 2014). Several gene families were identified, such as CBL and CIPK genes (Li et al., 2016), R2R3MYB transcription factor superfamily (Wang et al., 2016), calcium-dependent protein kinases (CDPKs) (Kumari et al., 2017) and CBF genes (Zhou et al., 2018a; Zhou et al., 2018b). Previously, Yang et al. (2015) have identified the WRKY proteins in Solanum melongena L. and Solanum torvum Sw. using the RNA-sequencing data, respectively. Herein, we identified 58 WRKY transcription factors in eggplant through genome-wide analysis. Subsequently, a comprehensive analysis of eggplant WRKY genes was carried out based on their sequences and structures, phylogenetic relationship and expression. Meanwhile, several WRKY genes were found to actively respond to cold stress. SmWRKY26 and SmWRKY32 were preliminarily verified involved in regulating eggplant tolerance to cold stress through VIGS. Therefore, cold-responsive genes in eggplant genome were preliminarily validated and the transcript level of these responsive genes may influence eggplant response to cold stress. The results threw light on their probable functions in eggplant response to abiotic stresses and provided a reference for genetic improvement in the stress tolerance.
Materials & Methods
Plant treatment
In the present study, the seedlings of ‘sanyueqie’ eggplant variety were germinated and grown in a growth chamber at 28 °C with a 16/8 h light/dark photoperiod. The cotyledons of plants were inoculated with the cultures at the cotyledon stage and transferred to 22 °C to induce gene-silencing. When the fourth leaves were fully expanded, the seedlings were transferred into 4 °C for five days, followed by three days of recovery at 28 °C.
Identification of WRKY family in eggplant
To identify the WRKY proteins in eggplant, two methods were employed. We searched the eggplant genome database (http://eggplant.kazusa.or.jp/) for WRKY proteins based on the sequences of conserved WRKY domains in Arabidopsis (Dong, Chen & Chen, 2003). All peptide sequences of eggplant were download from the eggplant database. The Hidden Markov Model (HMM) of WRKY was used to BLASTP. The candidate WRKY transcription factors were obtained subsequently. The Pfam databases (http://pfam.xfam.org/) and SMART databases (http://smart.embl-heidelberg.de/) were used to validate the candidate WRKY proteins. ProtParam tool (https://web.expasy.org/protparam/) was used to predict the molecular weights (MWs) and isoelectric points (pIs) of WRKY proteins.
Phylogenetic analysis of SmWRKY proteins
To understand the evolutionary relationship between the eggplant and other plants, multiple sequence alignment was constructed using CLUSTAL W based on the conserved WRKY domain sequences of WRKY proteins from eggplant, tomato, Arabidopsis and rice (Thompson, Higgins & Gibson, 1994; Ross, Liu & She, 2007). To know the relationship among eggplant WRKY proteins, the phylogenetic tree of eggplant WRKY proteins was conducted with the conserved WRKY domain sequences of WRKY proteins from eggplant. Phylogenetic analysis was performed with MEGA v5.1 (Tamura et al., 2011; Hall, 2013). A phylogenetic tree was produced following the neighbor-joining method.
Intron-exon structure and conserved motifs analysis
The intron-exon structures of SmWRKY genes were determined by submitting a txt file of gene features with gff3 format to Gene Structure Displayer Server (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015) (Data S1). The conserved motifs in the WRKY proteins were predicted using the MutipleEm for Motif Elicitation (MEME) program (http://alternate.meme-suite.org/tools/meme). The parameters in this study were set as follows: 10 motifs should be found, minimum width was 15, maximum width was 50. Interpro Scan was used to annotate the ten motifs.
Virus-induced gene silencing (VIGS)
Differentially expressed WRKYs were identified from the RNA-seq data conducted previously. SmWRKY26 and SmWRKY32 were up-regulated in both cultivars after 4 °C treatment and the proteins were encoded by two genes with a high sequence similarity. We selected these two genes to performed a functional verification through VIGS. The fragments of SmWRKY26 and SmWRKY32 were amplified using the specific primers (Data S2), verified using VIGS tool in SGN (http://vigs.solgenomics.net/) and cloned to TRV (tobacco rattle virus) RNA2 vector (Fu et al., 2005; Huang et al., 2012a; Huang et al., 2012b). The empty vector TRV RNA2 was used as the control. The pTRV1, pTRV2, pTRV2-SmWRKY26 and pTRV2-SmWRKY32 were introduced into agrobacterium GV3101 with electroporation method, respectively. The mixture of Agrobacterium cultures containing pTRV1 and pTRV2 (1:1, v/v), pTRV1 and pTRV2-SmWRKY26 (1:1, v/v) and pTRV1 and pTRV2-SmWRKY32 (1:1, v/v) at OD600 = 1 were incubated for 4 h in the dark at room temperature before inoculation. Three independent replicates were performed.
Quantative real time-polymerase chain reaction (qRT-PCR)
Specific primers were designed in the website (http://primer3.ut.ee/) (Data S2) (Koressaar & Remm, 2007). Total RNA was isolated from the tissue samples using the Trizol reagent according to the manufacturer’s instruction. Subsequently, RNA was reverse transcribed using the HiScript II Q Select RT SuperMix for qPCR kit (Vazyme, China). qRT-PCR was performed on an LightCycler 480 II (Roche, Switzerland) using the AceQ®qPCRSYBR® Green Master Mix (Vazyme, China). The PCR conditions consisted of denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing and extension at 60 °C for 30 s. Melt curve analysis was performed to determine the specificity of reactions. The transcript level was calculated according to the ΔΔCt method (Livak & Schmittgen, 2001). The eggplant Actin gene (Sme2.5_01462.1) was used as internal control. Three biological and technical repetitions were performed during the whole experiment.
| Protein name | Gene ID | Group | Amino acid | Molecular weight | pI |
|---|---|---|---|---|---|
| SmWRKY1 | Sme2.5_00009.1_g00017.1 | 3 | 340 | 38.90 | 5.14 |
| SmWRKY2 | Sme2.5_00013.1_g00024.1 | 1 | 511 | 57.35 | 6.58 |
| SmWRKY3 | Sme2.5_00013.1_g00025.1 | 3 | 360 | 40.79 | 5.70 |
| SmWRKY4 | Sme2.5_00016.1_g00020.1/pseudo | 2c | 338 | 37.43 | 5.58 |
| SmWRKY5 | Sme2.5_00029.1_g00022.1 | 2d | 324 | 35.87 | 9.57 |
| SmWRKY6 | Sme2.5_00038.1_g00016.1/TE | 2d | 367 | 40.39 | 9.72 |
| SmWRKY7 | Sme2.5_00100.1_g00012.1 | 1 | 595 | 64.89 | 6.56 |
| SmWRKY8 | Sme2.5_00161.1_g00022.1 | 1 | 490 | 54.66 | 5.71 |
| SmWRKY9 | Sme2.5_00196.1_g00004.1 | 1 | 989 | 109.90 | 6.58 |
| SmWRKY10 | Sme2.5_00232.1_g00014.1 | 2d | 239 | 27.26 | 5.55 |
| SmWRKY11 | Sme2.5_00232.1_g00015.1 | 2d | 285 | 30.87 | 5.53 |
| SmWRKY12 | Sme2.5_00264.1_g00017.1 | 2d | 306 | 33.62 | 9.64 |
| SmWRKY13 | Sme2.5_00281.1_g00010.1 | 2b | 454 | 50.55 | 5.92 |
| SmWRKY14 | Sme2.5_00386.1_g00007.1 | 2b | 561 | 61.09 | 6.89 |
| SmWRKY15 | Sme2.5_00423.1_g00013.1 | 2e | 277 | 30.16 | 5.51 |
| SmWRKY16 | Sme2.5_00556.1_g00018.1 | 2a | 224 | 25.48 | 6.45 |
| SmWRKY17 | Sme2.5_00556.1_g00019.1 | 2a | 261 | 29.75 | 5.75 |
| SmWRKY18 | Sme2.5_00574.1_g00007.1 | 2c | 171 | 19.67 | 5.40 |
| SmWRKY19 | Sme2.5_01030.1_g00008.1 | 1 | 486 | 54.38 | 6.50 |
| SmWRKY20 | Sme2.5_01060.1_g00010.1 | 2d | 402 | 45.04 | 9.43 |
| SmWRKY21 | Sme2.5_01071.1_g00003.1 | 2e | 297 | 33.64 | 6.45 |
| SmWRKY22 | Sme2.5_01077.1_g00010.1 | 1 | 649 | 71.02 | 6.46 |
| SmWRKY23 | Sme2.5_01130.1_g00003.1 | 2b | 334 | 37.60 | 7.01 |
| SmWRKY24 | Sme2.5_01183.1_g00010.1 | 2d | 337 | 36.62 | 9.55 |
| SmWRKY25 | Sme2.5_01372.1_g00013.1/pseudo | 2a | 353 | 38.71 | 8.55 |
| SmWRKY26 | Sme2.5_01585.1_g00006.1 | 1 | 499 | 55.49 | 7.64 |
| SmWRKY27 | Sme2.5_01670.1_g00011.1 | 2e | 353 | 39.38 | 5.64 |
| SmWRKY28 | Sme2.5_01689.1_g00004.1 | 2e | 332 | 36.60 | 5.13 |
| SmWRKY29 | Sme2.5_02107.1_g00005.1 | 1 | 615 | 66.36 | 6.12 |
| SmWRKY30 | Sme2.5_02381.1_g00007.1 | 2b | 534 | 58.28 | 6.44 |
| SmWRKY31 | Sme2.5_02389.1_g00002.1 | 2c | 134 | 15.75 | 9.64 |
| SmWRKY32 | Sme2.5_02587.1_g00015.1 | 1 | 529 | 58.74 | 7.17 |
| SmWRKY33 | Sme2.5_02680.1_g00006.1 | 1 | 450 | 49.73 | 9.51 |
| SmWRKY34 | Sme2.5_02752.1_g00007.1 | 3 | 296 | 33.91 | 5.94 |
| SmWRKY35 | Sme2.5_02954.1_g00006.1 | 1 | 389 | 43.05 | 7.69 |
| SmWRKY36 | Sme2.5_03205.1_g00005.1 | 2b | 434 | 47.31 | 6.90 |
| SmWRKY37 | Sme2.5_03353.1_g00002.1 | 2c | 288 | 32.62 | 6.18 |
| SmWRKY38 | Sme2.5_03471.1_g00002.1 | 2a | 182 | 20.83 | 9.45 |
| SmWRKY41 | Sme2.5_03997.1_g00004.1 | 2c | 406 | 44.18 | 5.83 |
| SmWRKY39 | Sme2.5_03533.1_g00001.1 | 2e | 299 | 33.37 | 5.70 |
| SmWRKY40 | Sme2.5_03980.1_g00003.1 | 2c | 319 | 36.33 | 6.49 |
| SmWRKY42 | Sme2.5_03997.1_g00007.1 | 2c | 448 | 48.81 | 8.75 |
| SmWRKY43 | Sme2.5_04027.1_g00002.1 | 3 | 356 | 39.06 | 6.32 |
| SmWRKY44 | Sme2.5_04190.1_g00001.1 | 2a | 339 | 37.64 | 8.84 |
| SmWRKY45 | Sme2.5_04253.1_g00002.1 | 2c | 227 | 25.60 | 8.20 |
| SmWRKY46 | Sme2.5_04516.1_g00005.1 | 2c | 301 | 33.88 | 6.46 |
| SmWRKY47 | Sme2.5_04517.1_g00003.1/TE | 2c | 1046 | 116.54 | 6.30 |
| SmWRKY48 | Sme2.5_05222.1_g00004.1 | 1 | 505 | 54.91 | 7.60 |
| SmWRKY49 | Sme2.5_06310.1_g00004.1 | 2a | 586 | 66.70 | 8.48 |
| SmWRKY50 | Sme2.5_06988.1_g00004.1 | 1 | 492 | 55.21 | 6.65 |
| SmWRKY51 | Sme2.5_07339.1_g00001.1 | 3 | 341 | 38.36 | 7.01 |
| SmWRKY52 | Sme2.5_08092.1_g00001.1 | 2b | 505 | 55.81 | 6.58 |
| SmWRKY53 | Sme2.5_11773.1_g00001.1/partial | 3 | 355 | 40.35 | 5.78 |
| SmWRKY54 | Sme2.5_14019.1_g00001.1/partial | 2c | 241 | 27.65 | 7.67 |
| SmWRKY55 | Sme2.5_14251.1_g00004.1/partial | 2c | 202 | 23.01 | 7.63 |
| SmWRKY56 | Sme2.5_15021.1_g00001.1 | 2d | 358 | 40.55 | 9.58 |
| SmWRKY57 | Sme2.5_17732.1_g00001.1 | 2b | 362 | 39.30 | 9.17 |
| SmWRKY58 | Sme2.5_18444.1_g00001.1/partial | 2c | 298 | 33.79 | 9.51 |
Results
Identification of WRKY protein family in eggplant
To identify the WRKY proteins in eggplant, BLASTP search was performed against the eggplant genome database (http://eggplant.kazusa.or.jp/) using the conserved WRKY domains in Arabidopsis. About 79 putative WRKY proteins were identified. Meanwhile, 68 putative WRKY proteins were identified using HMM (Hidden Markov Model) search program with a Hidden Markov Model (PF03106). By removing the redundant proteins and proteins without WRKY domain verified by Pfam and SMART programs, a total of 58 WRKY proteins were finally confirmed in eggplant. Obviously, the WRKY family of eggplant was smaller than those of other Solanaceae plants such as tomato (81 members) (Huang et al., 2012a; Huang et al., 2012b), pepper (Capsicum annuum L.) (61 members) (Cheng et al., 2016) and potato (82 members) (Liu et al., 2017a; Liu et al., 2017b).
The length of WRKY proteins in eggplant ranged from 134 to 1046 aa, with an average length of approximately 396 aa. The proteins with minimum length and maximum length were SmWRKY31 and SmWRKY47, respectively. Correspondingly, the molecular weights ranged from 15.75 kDa (SmWRKY31) to 116.54 kDa (SmWRKY47). The pIs of 58 SmWRKY proteins ranged from 5.13 to 9.72, among which 34 pIs<7 and 24 pIs>7 (Table 1) (Data S3).
Classification and phylogenetic analysis of SmWRKY proteins
Based on the number and amino acid sequences of the conserved WRKY domains, eggplant WRKY family can be classified into three main groups or seven subgroups, which is consistent with other plants. The group I contained 13 WRKY members which had two conserved WRKY domains, while 37 members were classed into group II (6 group II a, 7 group II b, 13 group II c, 8 group II d and 5 group II e). The remaining six WRKY proteins belonged to group III (Table 1).
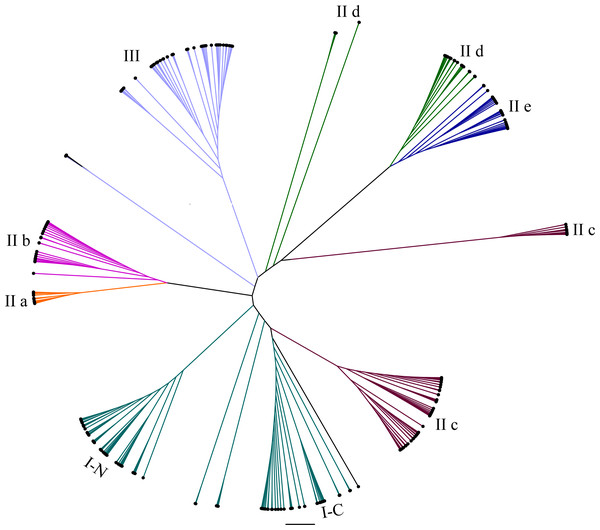
To study the evolutionary relationships of WRKY proteins between eggplant and other plants, a phylogenetic tree was constructed based on the conserved sequences of WRKY domains from eggplant, tomato, Arabidopsis and rice. All the WRKY proteins were divided into seven subgroups (Fig. 1). Phylogenetic analysis indicated that WRKY proteins in eggplant and tomato were closely related, compared with those in Arabidopsis and rice, such as SlWRKY54 and SmWRKY1, SlWRKY53 and SmWRKY3, SlWRKY31 and SmWRKY26, SlWRKY33 and SmWRKY32, SlWRKY3 and SmWRKY8 and so on (Data S4).
Figure 1: Radial phylogenetic tree of WRKY family in eggplant, tomato, Arabidopsis and rice.
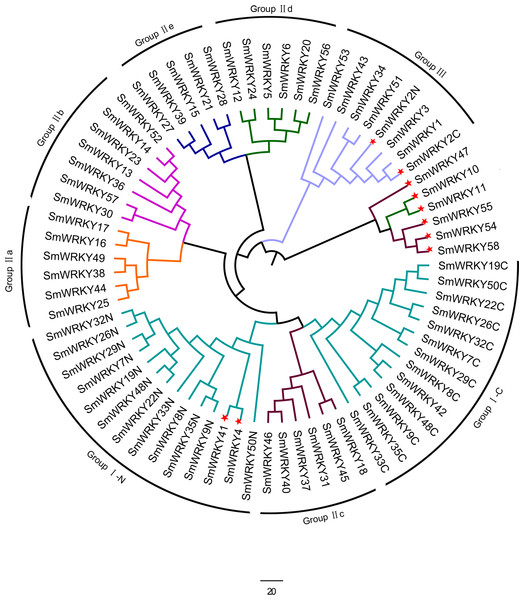
The conserved WRKY domain sequences of eggplant, tomato, Arabidopsis and rice were used to construct the phylogenetic tree by MEGA v5.1. The suffix ‘N’ and ‘C’ indicated the N-terminal and C-terminal WRKY domain of group I, respectively. The different groups were indicated with colors. The conserved WRKY domain sequences of eggplant, tomato, Arabidopsis and rice were used to construct the phylogenetic tree by MEGA v5.1. All the WRKY proteins were divided into three groups (I, II and III) and five subgroups (II a, II b, II c, II d and II e). The suffix ‘N’ and ‘C’ indicated the N-terminal and C-terminal WRKY domain of group I, respectively. The different groups were indicated with colors.To determine the phylogenetic relationships among the SmWRKY proteins, a phylogenetic tree was also constructed with WRKY conserved sequences using MEGAv5.1. As shown in Fig. 2, most of the SmWRKY members clustered in accordance with the classification, while SmWRKY2, SmWRKY4, SmWRKY10, SmWRKY11, SmWRKY41, SmWRKY42, SmWRKY47, SmWRKY54, SmWRKY55 and SmWRKY58 were not clustered into the corresponding branches.
Figure 2: Phylogenetic tree of WRKY family in eggplant.
The phylogenetic tree was constructed using the neighbor-joining method in MEGA v5.1 with the conserved WRKY domains of eggplant WRKY proteins. The eggplant WRKY proteins were grouped into three groups (I, II and III) and five subgroups (II a, II b, II c, II d and II e). The suffix ‘N’ and ‘C’ indicated the N-terminal and C-terminal WRKY domain of group I, respectively. The WRKY proteins which were not clustered into the groups they belong to were indicated with red stars. The different groups were indicated with colors.Structural analysis of WRKY variants
The WRKY domain consists of a WRKY amino acid sequence at the N-terminal and a zinc-finger structure that is either Cx4–5Cx22–23HxH or Cx7Cx23HxC at the C-terminal. Some variants in WRKY domain were reported in the earlier studies. For example, the core sequence of WRKY domain was replaced by WRRY, WSKY, WKRY, WVKY or WKKY (Eulgem et al., 2000; Xie et al., 2005; Wei et al., 2016). The amino acids of GQK were substituted by GKK or EIG. Interestingly, WHKF and WDKF were found in eggplant WRKY proteins and GEK replaced GQK following the WRKY sequence (Fig. 3). Furthermore, it existed that the sequence of the zinc-finger motif at the C-terminal was Cx23RxH (SmWRKY58) or Cx23QxH (SmWRKY42) in eggplant WRKY proteins (Fig. 3). In SmWRKY11 protein, the sequence between the core domain (WRKYGQK) and the first cysteine of zinc-finger was two amino acids less than those normal sequences (Fig. 3). SmWRKY42 and SmWRKY58 were lack of the first histidine of conserved zinc-finger (Fig. 3). It may result in the failure of zinc-finger formation.
Figure 3: Identification of WRKY variants in eggplant.
The sequences of WRKY variants were aligned. The conserved WRKYGQK and zinc-finger residues were marked in red.Intron-exon structure and conserved motifs of SmWRKY genes
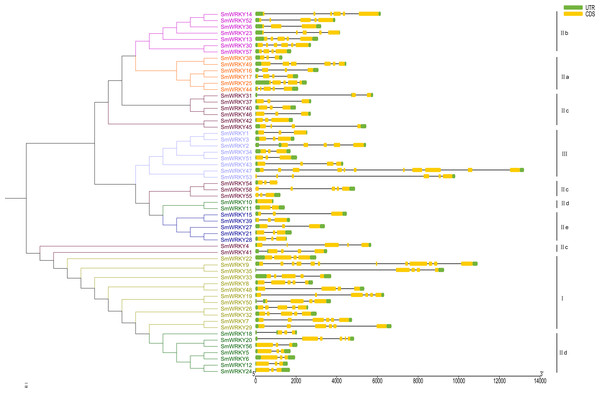
To understand the intron-exon structure of SmWRKY genes, we submitted the gff3 file of SmWRKY genes to Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) (Data S1). As shown in Fig. 4, the scope of intron number in eggplant WRKY family was from 0 (SmWRKY10) to 11 (SmWRKY9), with an average number of 4.4. In rice and cassava, the number of introns varied from 0 to 8 and 1 to 5, respectively (Xie et al., 2005; Wei et al., 2016). Group I contained 3-5 introns with the exception of SmWRKY9 (11 introns). Group II a contained 3-5 introns, except for SmWRKY38 (2 introns). Group II b contained 2–5 introns. Although several WRKY proteins were not clustered together in group II c, most of WRKYs contained 2 introns. The number of intron in group II d WRKYs was almost 2, excluding SmWRKY10 (0 intron) and SmWRKY20 (5 introns). All of the group II e SmWRKYs consisted of 2 introns. The majority of group III WRKY genes also included 2 introns.
Figure 4: The intron-exon structure of eggplant WRKY genes according to the phylogenetic relationship.
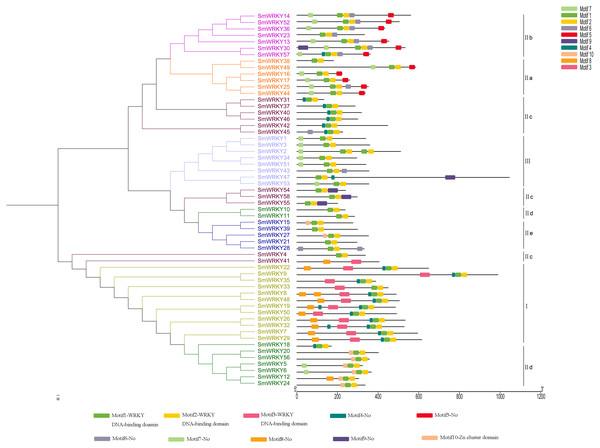
The phylogenetic tree was constructed with the full length sequences of eggplant WRKY proteins using MEGA v5.1. The intron-exon structure analysis was performed in Gene Structure Display Server. Lengths of intron and exon of each WRKY gene were showed in proportion. Every group was marked with different colors.To further know the structure of eggplant WRKYs and their phylogenetic relationships, conserved motifs were analyzed using MEME tool and annotated with InterPro Scan. As we set, ten motifs were found in the sequences (Fig. 5). The first three motifs were annotated as WRKY DNA-binding domain. All the SmWRKY proteins contained two WRKY motifs at least. These results suggested that the features were conserved in the WRKY family identified from eggplant. Notably, almost the members in each group contained the same motifs. However, Yang et al. (2019) mentioned that the function of family members largely varied, even if these members were from the same clade. Therefore, these WRKY proteins classified into the same group may differentially function. Although SmWRKY18 belonged to group II c, it had the similar conserved motifs with the group II d.
Figure 5: Conserved motifs of eggplant WRKY proteins according to the phylogenetic relationship.
The phylogenetic tree was constructed with the full length sequences of eggplant WRKY proteins using MEGA v5.1. The conserved motifs were predicted using the MEME program. The gray line indicated the non-conserved sequences. Each conserved motif was represented by a colored box. The length of each motif was showed in proportion.The differentially expressed SmWRKYs in response to cold stress
Eggplant is sensitive to cold stress and often undergoes the cold stress during its growth and development, especially in the early spring (Zhou et al., 2018a; Zhou et al., 2018b). Therefore, it is important to explore the mechanism for cold tolerance and how to improve this tolerance. Previous studies have shown that WRKY transcription factors regulated the plant defense against cold stress (He et al., 2016; Zhang et al., 2016). In order to study the function of WRKY gene family in eggplant response to cold stress, the leaf samples from a cold-tolerant genotype and a cold-sensitive genotype were collected with the three biological replicates when the seedlings were treated for twelve hours. Then the differentially expressed WRKY genes in eggplant cultivars to cold stress were investigated from RNA-seq data which has been not published yet (Data S5). As shown in Fig. 6, twenty SmWRKY genes were identified in each cultivar. Interestingly, sixteen WRKY genes were common in both cultivars. Meanwhile, the sixteen WRKY genes showed similar expression pattern in these two cultivars when exposed to cold stress. Only SmWRKY1 and SmWRKY8 were blocked in the sensitive cultivar (Fig. 6A). SmWRKY4, SmWRKY7, SmWRKY29 and SmWRKY39 were also down-regulated in the tolerant cultivar except these two WRKY genes, (Fig. 6B). The transcript level of the rest of WRKY genes in both cultivars were enhanced. These results indicated that WRKY transcription factors could respond to cold stress and constituted a complicated network to regulate the cold tolerance in eggplant.
Figure 6: Expression profiles of the differentially expressed eggplant WRKY genes under cold stress.
(A) The differentially expressed WRKY genes in sensitive cultivar after 4 °C treatment for 12 h. (B) The differentially expressed WRKY genes in tolerant cultivar after 4 °C treatment for 12 h. The units of the scale was Log Fold change. Green, black and red indicated down-regulated, no change and up-regualted, respectively.Silencing SmWRKY26 and SmWRKY32 increased the sensitivity to cold
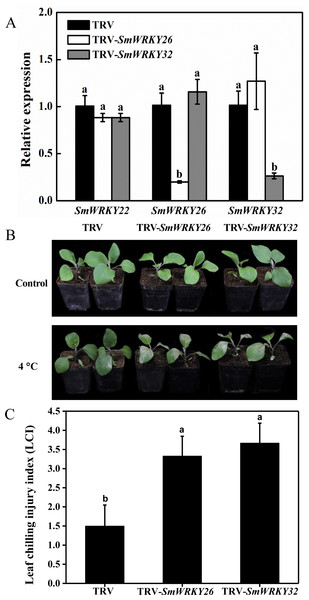
VIGS was an effective tool to study the gene functions (Zhang et al., 2018; Xu et al., 2019). VIGS mediated by TRV has been successfully employed in the functional analysis of eggplant genes (Zhou et al., 2018a; Zhou et al., 2018b). We conducted a functional validation for SmWRKY26 and SmWRKY32 using VIGS. The fragments of SmWRKY26 and SmWRKY32 were cloned into the TRV silencing vector. Subsequently, these vectors were introduced into the eggplant. Using the online VIGS tool, we found that the transcript level of SmWRKY22 was likely affected. To detect the efficiency of silencing, the transcript level of these two WRKY genes was determined with qRT-PCR. As shown in Fig. 7A, the transcript level of SmWRKY26 and SmWRKY32 in the leaves with target gene silenced displayed a reduction of approximately 80%, compared with the leaves inoculated with the control bacterium. However, the expression of SmWRKY22 showed no alter in any one kind of the silenced seedlings. The seedlings were transferred into 4 °C for 5 days when the fourth leaf was fully expanded. The gene-silenced seedlings showed more severe wilting and shrink, while the control seedlings presented a little chlorosis (Fig. 7B). Whereafter, the leaf chilling injury index (LCI) was calculated after treatment. The LCI of SmWRKY26- and SmWRKY32- silenced seedlings were significantly higher than that of control seedlings (Fig. 7C). The phenotypes and index upon cold stress suggested that SmWRKY26 and SmWRKY32 positively regulated the tolerance to cold stress in eggplant.
Figure 7: Silencing SmWRKY26 and SmWRKY32 enhanced the sensitivity to cold stress in eggplant.
(A) The transcript level of SmWRKY22, SmWRKY26 and SmWRKY32 tested by qRT-PCR. The black, white and shaded columns represent the empty TRV control plants, SmWRKY26-silenced plants and SmWRKY32-silenced plants, respectively. Data were means ± SE calculated from three biological replicates. Values with the same letter were not significantly different according to Duncan’s multiple range tests (P < 0.05, n = 3). (B) The phenotypes of silenced plants after cold stress for five days followed by three days of recovery at 28 °C. (C) The leaf chilling injury index of VIGS-plants after cold stress. Values with the same letter were not significantly different according to Duncan’s multiple range tests (P < 0.05, n = 6). The inoculations were conducted for three independent times.Discussion
Identification of WRKY proteins in eggplant
Characterization of gene families from eggplant genome is more and more important and imperative due to its importance for understanding the roles in plant responses to environmental stresses. In this study, we have identified 58 WRKY genes through searching the sequenced genome (Table 1). The WRKY gene family of eggplant was smaller than those of other plants such as the representative of Solanaceae plant tomato (81 WRKY genes). The WRKY members identified in this study was fewer than that reported in the PlantTFDB website (http://planttfdb.cbi.pku.edu.cn/family.php?sp=Sme&fam=WRKY#family_intro). 65 eggplant WRKY proteins were recorded in the plant transcription factor database. However, nine proteins either contained no WRKY feature structures, or had incomplete WRKY domains. Moreover, another two WRKY proteins were found in our study. 74 hits were obtained when “WRKY” was set as the keyword in eggplant genome. Among these, 55 hits were consistent with the WRKY proteins in our study. It lacked three WRKY proteins (Sme2.5_06310.1_g00004.1, Sme2.5_04517.1_g00003.1 and Sme2.5_00196.1_g00004.1) which were found in our study. The rest 19 predicted proteins were misannotated and listed in Data S6.
Interestingly, it was larger than that reported in Yang’s study (50 SmWRKY genes) (Yang et al., 2015), which might be due to that Yang identified the WRKY genes from transcriptome data.
Phylogenetic tree and variant analysis of eggplant WRKY
The phylogenetic tree among the eggplant WRKY proteins has shown their relatively conserved evolutionary relationships (Fig. 2). As shown in Fig. 2, some WRKY proteins were not clustered into the corresponding branches. These WRKY proteins were identified as WRKY variants through analyzing the amino acid sequence of these proteins (Fig. 3). For example, the C-terminal WRKY domain of WRKY2 protein had the sequence similarity with group III WRKY proteins and the last amino acid in the zinc-finger of the N-terminal WRKY domain is cysteine. These results suggested that the substitutions, addition and subtraction of amino acids of WRKY domain might be responsible for the changes in clustering. Furthermore, these WRKY variants probably lose the DNA-binding activity and the zinc-finger motif can not be formed, which lead to new molecular activities, novel biology functions and a non-functional protein. We found this phenomenon in our earlier study performed in soybean (Yang et al., 2017a; Yang et al., 2017b). The phylogenetic relationship between eggplant and other plants confirmed that WRKY proteins in eggplant contained the same subgroups with other reported plants (Data S4). In addition, eggplant WRKY proteins had a highly close relationship to tomato WRKY proteins, on account of being the Solanaceae.
Intron-exon structure and conserved motifs in eggplant WRKY
Analysis of the distribution of intron and exon is a better way to understand the phylogenetic relationship, because intron-exon structural diversity is an important part in the evolution of gene families (Wei et al., 2016). The WRKY genes in group I, group II a and group II b have more introns, whereas a vast majority of the rest WRKY genes have only two introns. It seems that eggplant WRKY proteins originate from the first group WRKY. The conserved motifs of eggplant WRKY proteins were identified using MEME tool and InterPro Scan. All the eggplant WRKY proteins contained at least two WRKY motifs with the exception of SmWRKY41. Unlike SmWRKY4, another WRKY variant in which the Cx22HxH replaced for Cx23HxH, the core sequence of the WRKY domain in SmWRKY41 was replaced by WRKYGQN. This specific variation was potentially responsible for one WRKY motif in SmWRKY41. Motif 10 was mainly included in group II d proteins except SmWRKY10 and SmWRKY11 (Fig. 5). Using InterPro Scan, motif 10 was identified as Zn-cluster domain (IPR018872) which was found associated with the WRKY domain (Babu et al., 2006). The types of conserved motifs contained in SmWRKY10 and SmWRKY11 more closely resembled those of group II e proteins, which might be one of the reasons for their clustering (Fig. 5).
SmWRKY genes in response to cold stress
Cold stress is an important factor affecting the growth and development of eggplant. To a certain extent, the response of WRKY genes to cold stress reflects the roles of WRKY genes in cold tolerance of plants (He et al., 2016). For example, Zhou et al. (2008) found that GmWRKY21 was induced by cold treatment and the transgenic plants over-expressing GmWRKY21 exhibited enhanced tolerance. In this study, we identified 24 differentially expressed SmWRKY genes in the two investigated materials from the RNA-seq data (Fig. 6). Four eggplant WRKY genes were specially differentially expressed in any one cultivar. This indicated that the differentially expression of eight WRKY genes might lead to difference in cold tolerance. In the common sixteen genes, WRKY26 and WRKY32 which shared a high sequence-similarity displayed up-regulated under cold stress. We conducted a VIGS experiment to verify the function of these two genes in response to cold. The different phenotypes and the chilling injury index between the silenced and the control plants illuminated that SmWRKY26 and SmWRKY32 could positively regulate the response to cold stress (Fig. 7).
Zhou et al. (2016) identified that several genes in response against Verticillium dahlia were annotated as WRKY genes, such as Unigene21521, Unigene11753, Unigene6502 and Unigene20920. SmWRKY26 and SmWRKY32 were the orthologous of Unigene20920 and Unigene6502 in cultivated eggplant, respectively. In addition, the proteins encoded by WRKY26 and WRKY32 had a 93% and 85% degree of similarity with SlWRKY31 and SlWRKY33 in tomato, respectively. SlWRKY31 and SlWRKY33 were reported as SlWRKY33A and SlWRKY33B to function in disease resistance and heat tolerance in tomato (Zhou et al., 2015). These results implied that orthologous could perform variant functions in different plants.
Conclusions
WRKY proteins play a vital role in response to different stresses, plant growth and development. In order to understand the WRKY proteins family in eggplant, a genome-wide identification of WRKY proteins was performed in this study. 58 genes encoding eggplant WRKY proteins were identified through searching the eggplant genome, which could be classified into three groups or seven subgroups in accordance with other plants. WRKY variants were identified in eggplant. Gene structure analysis showed that the number of intron in eggplant WRKY family was from 0 to 11, with an average number of 4.4. Based on conserved motif analysis, the WRKY DNA-binding domain was found conserved in eggplant WRKY proteins. Furthermore, RNA-seq data showed that WRKY genes were differentially expressed in eggplant response to cold stress. Structurally related SmWRKY26 and SmWRKY32 were confirmed to positively regulate the response to cold stress in eggplant. Therefore, the two preliminarily verified genes can be used as target genes for genetic improvement in cold stress. This study provides a foundation and practical reference for the functional analysis of the WRKY gene family in eggplant and the genetic improvement in cold-tolerance.
Supplemental Information
The rectangular phylogenetic tree of WRKY family in eggplant, tomato, Arabidopsis and rice
The differentially expressed WRKY genes in sensitive and tolerant cultivars under cold stress
The sheet “S0 vs S12” and “T0 vs T12” indicated the differentially expressed WRKY genes in sensitive and tolerant cultivars after cold stress for 12 h from the RNA-seq raw data, respectively.