Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon
- Published
- Accepted
- Received
- Academic Editor
- Tatiana Tatarinova
- Subject Areas
- Agricultural Science, Genomics, Plant Science
- Keywords
- Watermelon, Dof, Phylogenetic analysis, Expression profile, Abiotic stress
- Copyright
- © 2020 Zhou et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ 8:e8358 https://doi.org/10.7717/peerj.8358
Abstract
The plant DNA-binding with one finger (Dof) gene family is a class of plant-specific transcription factors that play vital roles in many biological processes and stress responses. In the present study, a total of 36 ClDof genes were identified in the watermelon genome, which were unevenly distributed on 10 chromosomes. Phylogenetic analysis showed that the ClDof proteins could be divided into nine groups, and the members in a particular group had similar motif arrangement and exon–intron structure. Synteny analysis indicated the presence of a large number of syntenic relationship events between watermelon and cucumber. In promoter analysis, five kinds of stress-related and nine kinds of hormone-related cis-elements were identified in the promoter regions of ClDof genes. We then analyzed the expression patterns of nine selected ClDof genes in eight specific tissues by qRT-PCR, and the results showed that they have tissue-specific expression patterns. We also evaluated the expression levels of 12 selected ClDof genes under salt stress and ABA treatments using qRT-PCR. As a result, they showed differential expression under these treatments, suggesting their important roles in stress response. Taken together, our results provide a basis for future research on the biological functions of Dof genes in watermelon.
Introduction
DNA binding with one finger (Dof) proteins are a group of plant-specific transcription factors widely present in plants, while there has been no report about them in other eukaryotes such as humans and yeast (Azam et al., 2018; Gupta et al., 2015). Genome-wide surveys showed that the Dof family genes are widely distributed in the genomes of various plant species. For example, as model plants, Arabidopsis and rice include 36 and 30 Dof genes in their genomes, respectively (Lijavetzky, Carbonero & Vicente-Carbajosa, 2003). In addition, it has been reported that there are 25 Dof genes in peach (Prunus persica) (Chen et al., 2017), 29 in eggplant (Solanum melongena) (Wei et al., 2018), 33 in pepper (Capsicum annuum) (Kang et al., 2016; Wu et al., 2016), 34 in tomato (Solanum lycopersicum) (Cai et al., 2013), 36 in cucumber (Cucumis sativus) (Wen et al., 2016), 45 in cassava (Manihot esculenta) (Zou, Zhu & Zhang, 2019), 45 in pear (Pyrus bretschneideri) (Liu et al., 2020), and 60 in apple (Malus domestica) (Zhang et al., 2018). These reports revealed that the Dof proteins are characterized by the highly conserved Dof domain in their N-terminal regions, which is composed of about 52 amino acids with a Cys2/Cys2 zinc finger structure (Umemura et al., 2004; Yanagisawa, 2002). The Dof domain specifically recognizes and combines with a T/AAAAG core sequence in the promoters of target genes (Noguero et al., 2013; Umemura et al., 2004). In addition, the Dof proteins also contain a variable transcriptional activation domain at their C-terminus. The N- and C-terminal regions of the Dof proteins contribute to their bi-functional roles in DNA binding and protein–protein interactions to regulate the expression of the target genes (Gupta et al., 2015; Noguero et al., 2013).
As the first identified Dof gene, ZmDof1 was found to play a role in light-regulated gene expression and affect light response and nitrogen assimilation (Yanagisawa & Izui, 1993; Yanagisawa & Sheen, 1998). Subsequently, a large number of Dof genes were reported to be involved in various plant-specific biological processes, such as seed germination (Boccaccini et al., 2014; Gualberti et al., 2002; Santopolo et al., 2015), fruit ripening (Feng et al., 2016), flowering time control (Li et al., 2009; Liu et al., 2020; Wu et al., 2017), and responses to plant hormones (Boccaccini et al., 2016; Lorrai et al., 2018; Qin et al., 2019; Rymen et al., 2017), as well as various stress responses (Su et al., 2017; Zang et al., 2017). Moreover, some Dof genes can play multifaceted roles in regulating plant development and stress responses. For example, overexpression of Arabidopsis CDF3 could contribute to higher tolerance to drought, cold and osmotic stress and lead to late flowering, suggesting that it is involved in both flowering time control and abiotic stress tolerance (Corrales et al., 2017). In tomato, overexpression of a Dof gene TDDF1 induced early flowering by increasing the expression of flowering-time control genes, and the transgenic plants also displayed higher resistance to drought, salt, and late blight caused by Phytophthora infestans (Ewas et al., 2017). In rice, salt stress repressed the expression of OsDOF15 in roots, and overexpression of OsDOF15 reduced the sensitivity of roots to salt stress via limiting ethylene biosynthesis, suggesting that OsDOF15-mediated ethylene biosynthesis may be involved in the inhibition of primary root elongation by salt stress (Qin et al., 2019). These findings demonstrate that the Dof proteins are involved in diverse biological processes and play important roles in the growth and development of plants.
Although the Dof gene family has been comprehensively analyzed and functionally characterized in a number of plant species, little is known about this gene family in watermelon, an economically important fruit crop cultivated worldwide. In this study, we characterized the Dof family genes in watermelon by analyses of their phylogenetic relationships, conserved motifs, gene structures and chromosomal localizations. In addition, the expression profiles of the selected Dof genes in different tissues and under salt or ABA treatment conditions were also examined. Our findings may lay a foundation for future functional analysis of Dof genes in watermelon.
Materials and Methods
Genome-wide identification and protein properties of Dof family in watermelon
To identify the watermelon Dof family genes, HHM profile of the Dof domain (PF02701) was used as a query to perform an HMMER search against the watermelon proteome, which was downloaded in watermelon (97103) v1 genome from the cucurbit genomics database (CuGenDB; http://cucurbitgenomics.org). A comprehensive search was also performed by using the amino acid sequences of Arabidopsis and rice Dof proteins from a previous study (Lijavetzky, Carbonero & Vicente-Carbajosa, 2003), which were obtained from the TIGR database (https://rice.plantbiology.msu.edu/) and the TAIR database (https://www.arabidopsis.org/), respectively. The putative sequences were submitted to Pfam (http://pfam.sanger.ac.uk/) and SMART (http://smart.embl-heidelberg.de/) for checking the presence of the Dof domain.
Sequence analyses and phylogenetic tree construction
The biochemical features including molecular weight (MW) and isoelectric point (pI) of all Dof proteins were determined by ProtParam server (http://web.expasy.org/protparam/). The subcellular localizations of the watermelon Dof proteins were predicted with CELLO v2.5 tool (http://cello.life.nctu.edu.tw/). The MEME tool (http://meme-suite.org/tools/meme) was used to predict and analyze the conserved motifs of watermelon Dof proteins with the maximum number of motifs being set as 10, and other parameters were set as default. The predicted motifs were further confirmed by searching against InterProScan (http://www.ebi.ac.uk/interpro/search/sequence-search/), and structure schematic diagrams were illustrated using the TBtools software (Chen et al., 2018). The coding region sequences (CDS) and genomic DNA (gDNA) sequences of ClDof genes were downloaded from watermelon (97103) v1 genome database (http://cucurbitgenomics.org/organism/1), and then the exon–intron structures of ClDof genes were displayed by the GSDS tool (Gene Structure Display Server, http://gsds.cbi.pku.edu.cn/) based on the alignment of CDSs with the corresponding gDNA sequences. For gene ontology (GO) analysis, the annotations of ClDof genes were obtained from watermelon (97103) v1 genome database and visualized with the WEGO program (http://wego.genomics.org.cn/). For promoter analysis, we determined the putative promoter sequence for each ClDof gene, which was defined as the upstream 1,500 bp region of the transcription start site (ATG), and analyzed the stress-related and hormone-related cis-elements using the PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). For phylogenetic tree construction, the Dof proteins of watermelon, cucumber, rice and Arabidopsis were aligned by Clustal Omega with default parameters. The Dof protein IDs of above species were listed in Table S1. Then, the MEGA program (v7.0) was used to construct a Neighbor-Joining tree with parameters of 1,000 bootstrap replicates and pairwise deletion.
Chromosomal location, gene duplication, and synteny analysis
The chromosomal location information of watermelon Dof genes was obtained from the watermelon genome database, and MapChart was used to display the physical positions of all ClDof genes along each chromosome. Gene duplication and synteny of Dof genes from watermelon and cucumber were examined using multiple collinear scanning toolkits (MCScanX) software with default parameters as previously reported (You et al., 2018).
Plant materials and treatments
Seeds of the watermelon cultivar “Xinong 8” (Citrullus lanatus L.) were first sterilized and germinated in an incubator (28 °C). Then, the germinated seeds were sown in pots and cultivated under a 12 h day/12 h night cycle (25 °C/19 °C, day/night temperature cycle) until the seedlings developed to four leaves. Uniformly developed four-leaf-stage watermelon plants were then exposed to NaCl (200 mm) and ABA (100 µm) treatments for 0 h, 1 h, 3 h, 9 h and 24 h. All leaves from watermelon plants were collected and rapidly frozen in liquid nitrogen and stored at –80 °C until RNA extraction.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using the total RNA Miniprep Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s protocol. Then, RNase-free DNase I was added in RNA solution to remove any contaminated gDNA. First-strand cDNA synthesis was carried out following the manufacturer’s procedure (ReverTra Ace qPCR-RT Kit, Toyobo, Japan). Primers were designed using Primer Premier 5.0 software (Table S2). The qRT-PCR was performed on an CFX96 instrument (Bio-Rad, Alfred Nobel Drive Hercules, CA, USA) using SYBR Green qPCR kits (TaKaRa, Japan). The watermelon constitutive actin gene (Cla007792) was used as the endogenous control (Zhou et al., 2018b). The PCR amplification conditions included an initial heat-denaturing step at 95 °C for 3 min, followed by 40 cycles of 30 s at 95 °C, 30 s at 58 °C, and 1 min at 72 °C. Relative expression levels were calculated using the 2−ΔΔCt method (Livak & Schmittgen, 2001), and each treatment included three independent biological replicates and three technical replicates. Data were statistically analyzed by one-way ANOVA using SPSS 19.0 software, and Tukey’s multiple range tests were used to detect significant treatment differences (P < 0.05).
Results
Genome-wide identification of Dof family genes in watermelon
A total of 36 Dof genes were identified and named as ClDof1–36 according to their order on the chromosomes. Detailed information including the CDS length, protein length, predicted MW and pI of each gene is listed in Table 1. The amino acid sequences and gene sequences of ClDof members are listed in Tables S3–S5. These genes had CDS lengths ranging from 492 bp (ClDof1) to 1575 bp (ClDof33), and encoded proteins ranging from 163 to 524 amino acid residues, with the predicted MW varying from 17.64 to 56.71 kDa. The pIs of the ClDof proteins ranged from 5.00 (ClDof30) to 9.95 (ClDof13). The CELLO v2.5 tool was used to analyze the subcellular localization of ClDof proteins. The results showed that nearly all ClDof proteins were localized in the nucleus, with the exception of ClDof6, which possibly had a nuclear and extracellular localization (Table 1). The GO annotation results indicated that ClDof proteins were assigned into three major categories and 18 subcategories (Table S6; Fig. S1).
| Gene name | Gene ID | Map position (bp) | CDS length (bp) | Protein length (aa) | MW (kDa) | pI | Subcellular location |
|---|---|---|---|---|---|---|---|
| ClDof1 | Cla000091 | Chr0:12921851–12922342 | 492 | 163 | 17.64 | 8.21 | Nuclear |
| ClDof2 | Cla000604 | Chr0:24087372–24088166 | 795 | 264 | 29.22 | 8.41 | Nuclear |
| ClDof3 | Cla004880 | Chr1:83833–84684 | 852 | 283 | 30.46 | 8.4 | Nuclear |
| ClDof4 | Cla011343 | Chr1:1447591–1449038 | 831 | 276 | 29.57 | 7.72 | Nuclear |
| ClDof5 | Cla000975 | Chr1:10830770–10831984 | 1,011 | 336 | 37.74 | 7.31 | Nuclear |
| ClDof6 | Cla001812 | Chr1:26447800–26448528 | 729 | 242 | 24.73 | 8.34 | Nuclear/extracellular |
| ClDof7 | Cla001818 | Chr1:26513973–26514995 | 1,023 | 340 | 35.45 | 9.21 | Nuclear |
| ClDof8 | Cla014094 | Chr1:28161694–28162635 | 942 | 313 | 33.83 | 8.26 | Nuclear |
| ClDof9 | Cla001373 | Chr1:31447086–31447871 | 786 | 261 | 29.33 | 8.84 | Nuclear |
| ClDof10 | Cla009627 | Chr1:31658215–31659085 | 717 | 238 | 25.84 | 8.84 | Nuclear |
| ClDof11 | Cla009628 | Chr1:31665641–31666539 | 729 | 242 | 26.88 | 9.49 | Nuclear |
| ClDof12 | Cla009692 | Chr1:32112455–32112961 | 507 | 168 | 19.04 | 8.81 | Nuclear |
| ClDof13 | Cla013297 | Chr2:30590643–30592400 | 1,020 | 339 | 37.52 | 9.95 | Nuclear |
| ClDof14 | Cla000540 | Chr2:31118585–31119331 | 747 | 248 | 27.29 | 8.73 | Nuclear |
| ClDof15 | Cla008250 | Chr3:1516113–1517286 | 1,026 | 341 | 37.25 | 9.31 | Nuclear |
| ClDof16 | Cla005059 | Chr3:2677903–2678760 | 858 | 285 | 31.75 | 8.39 | Nuclear |
| ClDof17 | Cla019672 | Chr3:8389380–8389913 | 534 | 177 | 20.21 | 7.13 | Nuclear |
| ClDof18 | Cla019705 | Chr3:8782843–8783751 | 909 | 302 | 33.57 | 7.46 | Nuclear |
| ClDof19 | Cla019706 | Chr3:8791610–8792131 | 522 | 173 | 18.69 | 9.22 | Nuclear |
| ClDof20 | Cla018219 | Chr4:19894774–19896290 | 813 | 270 | 29.93 | 9.9 | Nuclear |
| ClDof21 | Cla018604 | Chr4:23659963–23661769 | 1,308 | 435 | 47.56 | 7.04 | Nuclear |
| ClDof22 | Cla021140 | Chr5:723346–723861 | 516 | 171 | 18.06 | 8.99 | Nuclear |
| ClDof23 | Cla004274 | Chr5:9417748–9418525 | 678 | 225 | 24.96 | 8.32 | Nuclear |
| ClDof24 | Cla010192 | Chr5:31339279–31340779 | 1,296 | 431 | 47.33 | 8.11 | Nuclear |
| ClDof25 | Cla006705 | Chr6:3496040–3496858 | 819 | 272 | 29.94 | 8.26 | Nuclear |
| ClDof26 | Cla019034 | Chr6:24515454–24516428 | 975 | 324 | 34.96 | 8.08 | Nuclear |
| ClDof27 | Cla019107 | Chr6:25139609–25141772 | 1,395 | 464 | 50.63 | 6.19 | Nuclear |
| ClDof28 | Cla004013 | Chr7:3742674–3743851 | 969 | 322 | 34.24 | 9.24 | Nuclear |
| ClDof29 | Cla012621 | Chr7:24693545–24694168 | 624 | 207 | 22.36 | 8.36 | Nuclear |
| ClDof30 | Cla013851 | Chr8:15842719–15843486 | 768 | 255 | 28.77 | 5 | Nuclear |
| ClDof31 | Cla022532 | Chr8:24427298–24428044 | 747 | 248 | 25.77 | 8.12 | Nuclear |
| ClDof32 | Cla004676 | Chr9:32014839–32016085 | 1,077 | 358 | 39.08 | 8.43 | Nuclear |
| ClDof33 | Cla016993 | Chr10:21239053–21241153 | 1,575 | 524 | 56.71 | 5.07 | Nuclear |
| ClDof34 | Cla002907 | Chr10:21961596–21963908 | 1,527 | 508 | 54.74 | 6.06 | Nuclear |
| ClDof35 | Cla017622 | Chr10:24621093–24621851 | 759 | 252 | 27.81 | 6.76 | Nuclear |
| ClDof36 | Cla017890 | Chr10:27032680–27034515 | 1,053 | 350 | 37.19 | 9.85 | Nuclear |
Phylogenetic characterization of the watermelon Dof gene family
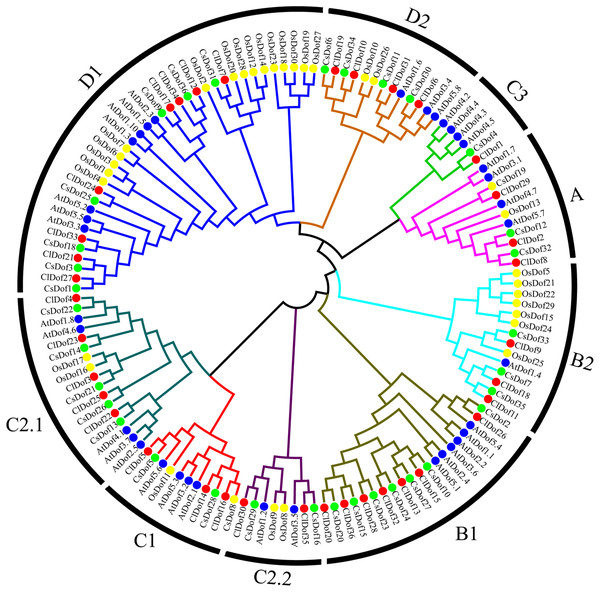
To study the evolutionary relationship of Dof family genes between watermelon and other plants, a phylogenetic tree based on multiple sequence alignment was constructed by using the amino acid sequences of ClDofs together with those from cucumber (CsDofs) (Wen et al., 2016), rice (OsDofs) and Arabidopsis (AtDofs) (Lijavetzky, Carbonero & Vicente-Carbajosa, 2003). The phylogenetic tree showed that these Dof proteins could be classified into nine groups, namely A, B1, B2, C1, C2.1, C2.2, C3, D1 and D2, with well-supported bootstrap values (Fig. 1). Nearly all groups included ClDofs, CsDofs, OsDofs and AtDofs, with the exception of group C3, which comprised only dicotyledonous Dofs (ClDofs, CsDofs, and AtDofs). Besides, the numbers of ClDofs in groups A, B1, B2, C1, C2.1, C2.2, C3, D1 and D2 were 3, 7, 3, 3, 5, 2, 1, 8 and 4, respectively (Fig. 1).
Figure 1: Phylogenetic relationships of Dof family proteins in watermelon, cucumber, rice and Arabidopsis.
Different plant species are represented with different colors: watermelon (red, Cl), cucumber (green, Cs), rice (yellow, Os), and Arabidopsis (blue, At).Conserved motif analysis of ClDofs
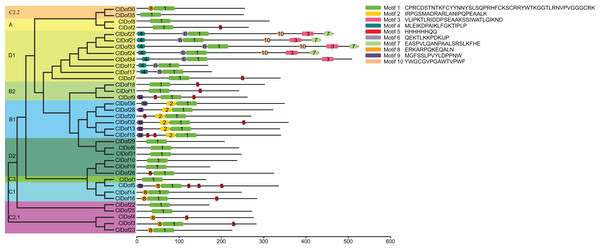
By using the MEME program, a total of 10 conserved motifs were identified (Fig. 2). Amongst them, motif 1 was annotated as a Dof domain, which was widely present in all ClDof proteins, with the exception of ClDof4. Some other motifs were specifically present in individual groups. For example, motif 3, 4, 6, 7 and 10 were exclusively present in the ClDofs in group D1, while motif 2 was present in all ClDofs of group B1. Besides motif 2, nearly all group B1 ClDofs included motif 9 (except for ClDof20). In addition, motif 8 was present in all group C1 ClDofs, as well as some ClDofs in groups C2.1 and C2.2 (Fig. 2). It is worth noting that besides motif 1 and 8, three motif 5 and one motif 9 were also present in ClDof5 of group C1.
Figure 2: Conserved domains of ClDofs based on the evolutionary relationship.
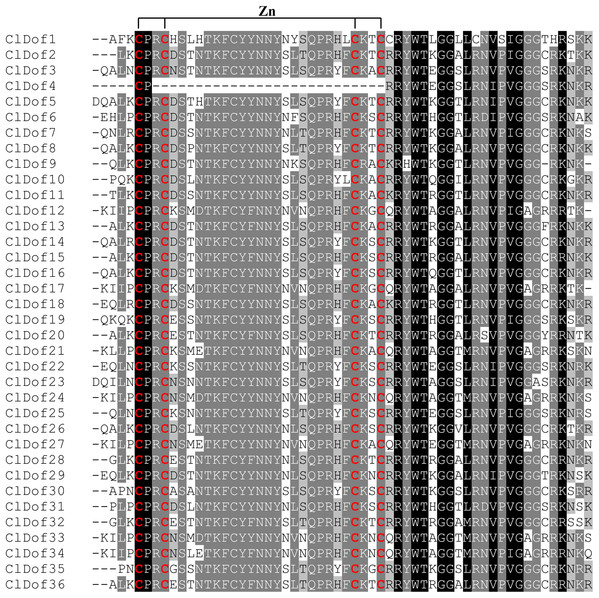
Distribution of conserved motifs in the ClDof proteins.To better understand the structural features of Dof domain, multiple sequence alignment of the Dof domain sequences of ClDofs was carried out. As a result, the Dof domain of ClDofs was highly conserved, and nearly all ClDof proteins harbored the four Cys residues associated with zinc finger structure, with the exception of ClDof4 (Fig. 3), which may result in the divergence of its function from that of other ClDofs.
Figure 3: Dof domain sequence alignment of watermelon Dof proteins.
The Dof DNA-binding domains among watermelon Dof proteins were aligned and the four Cys residues associated with zinc finger structure of the ClDofs are colored in red.Exon–intron arrangement analysis of Dof family genes in watermelon
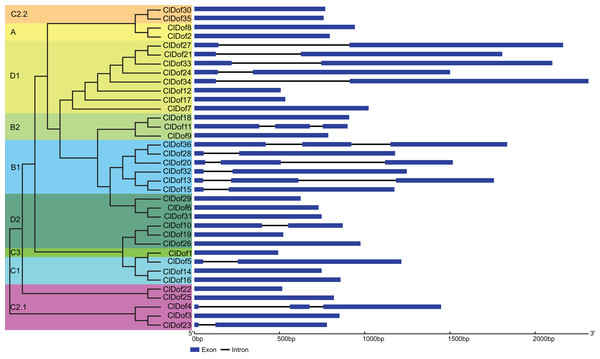
The CDS and gDNA sequences of the 36 ClDof genes were used to examine the distribution of exons and introns. As a result, most of the ClDof genes (20 out of 36) contained no introns, 11 ClDof genes (ClDof5, ClDof10, ClDof15, ClDof23, ClDof27, ClDof28, ClDof21, ClDof24, ClDof32, ClDof33 and ClDof34) had one intron each, whereas five ClDof genes (ClDof4, ClDof11, ClDof13, ClDof20 and ClDof36) possessed two introns (Fig. 4).
Figure 4: Exon–intron structure of ClDof genes based on the evolutionary relationship.
Chromosome distribution, gene duplication and synteny analysis of ClDof genes
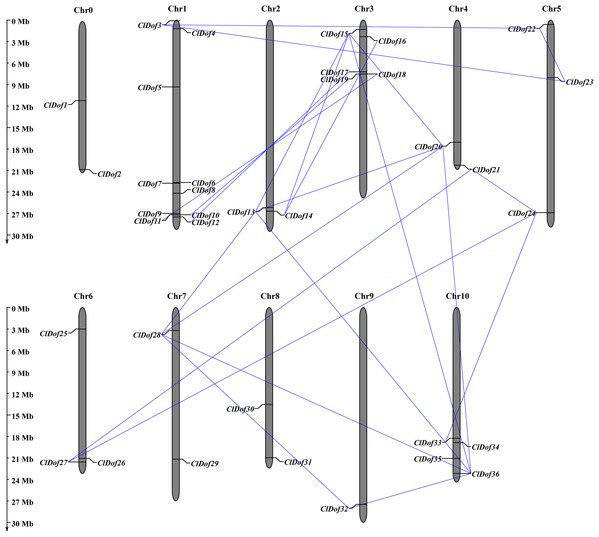
Using the MapInspect program, a total of 34 ClDof genes were mapped on 10 of the 12 chromosomes in watermelon genome, while ClDof1 and ClDof2 were located on chromosome 0 (Fig. 5). In detail, there were 10, 2, 5, 2, 3, 3, 2, 2, 1 and 4 ClDof genes on chromosome 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10, respectively. Moreover, the gene duplication events were analyzed using the MCScanX program, and a total of 20 ClDof genes exhibited segmental duplication, which made up 21 pairs of segmental duplication genes (Fig. 5).
Figure 5: Chromosomal distribution of ClDof genes in watermelon genome.
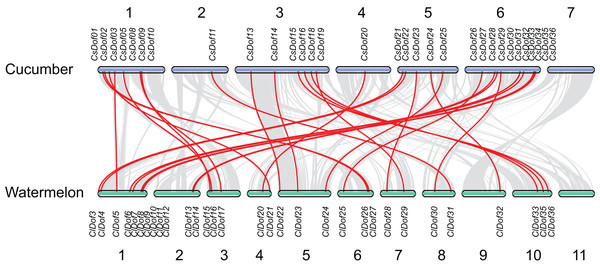
The segmental duplication genes are connected by lines.To reveal the orthologous relationships of Dof genes on chromosomes between watermelon and cucumber genomes, a comparative analysis was performed between Dof genes in watermelon and cucumber by MCScanX. A total of 31 orthologous gene pairs were identified between watermelon and cucumber (Fig. 6), indicating close relationships between ClDof and CsDof genes.
Figure 6: Synteny analysis of Dof genes from watermelon and cucumber genomes.
Gray lines in the background indicate the collinear blocks within watermelon and cucumber genomes. The orthologous gene pairs are linked with red lines.Promoter cis-elements of the ClDof genes
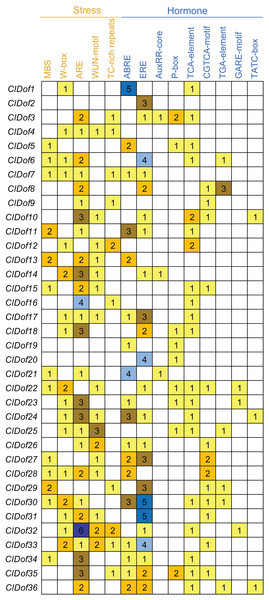
To assess the transcriptional regulation and potential functions of the ClDof genes, the cis-elements in the promoter regions of the ClDof genes were investigated and described in Fig. 7. Five kinds of stress-related and nine kinds of hormone-related cis-elements were identified in this study (Fig. 7). The ARE element was the most abundant among the stress-related cis-elements, and the promoter regions of 28 ClDof genes harbored the ARE element (Fig. 7), which is essential for the anaerobic induction. Other four stress-related cis-elements, including W-box, WUN-motif, MBS and TC-rich repeats, were found in 17, 16, 13 and 10 promoter regions of ClDof genes, respectively. Furthermore, various hormone-related cis-elements were also identified among the promoters of ClDof genes (except for ClDof4), including abscisic acid (ABA)-responsive element (ABRE), ethylene-responsive element (ERE), salicylic acid (SA)-responsive element (TCA-element), methyl jasmonate (MeJA)-responsive element (CGTCA-motif), auxin-responsive elements (AuxRR-core and TGA-element), and gibberellin-responsive elements (P-box, GARE-motif and TATC-box) (Fig. 7). These findings indicated that ClDof genes might play certain roles in various stress responses and hormone signaling.
Figure 7: Distribution of stress- and hormone-related cis-elements in the promoter regions of ClDof genes.
Tissue-specific expression profiles of the ClDof genes
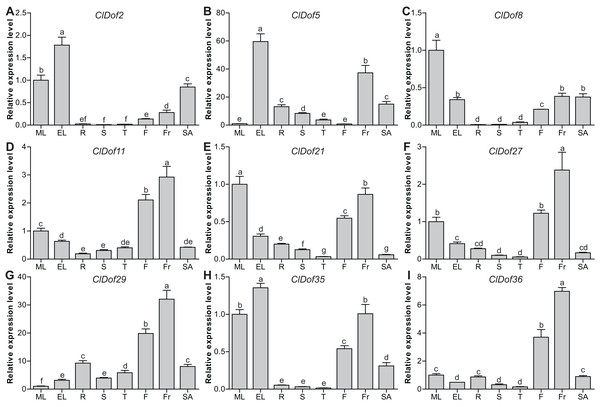
To evaluate the functions of ClDof genes in the growth and development of watermelon, the expression of nine selected ClDof genes in different tissues (mature and expanding leaves, roots, stems, stem apexes, flowers and fruits) was examined with qRT-PCR. Most ClDof genes were highly expressed in flowers and/or fruits, such as ClDof11, ClDof21, ClDof27, ClDof29, ClDof35 and ClDof36 (Fig. 8; Table S7), suggesting that they may function in flower and fruit development of watermelon. In addition, ClDof2, ClDof5, ClDof8, ClDof21 and ClDof35 displayed the highest expression in leaves, and relatively lower expression in other tissues, especially roots, stems, and tendrils (Fig. 8). Besides expanding leaves, ClDof5 also showed relatively higher expression in fruits as compared with other tissues, while its expression was extremely low in flowers. Finally, nearly all ClDof genes exhibited moderate transcript abundance in stem apexes (Fig. 8), implying their possible roles in stem apex development of watermelon.
Figure 8: Expression profiles of nine selected ClDof genes (A–I) in various tissues determined by qRT-PCR.
ML, mature leaves; EL, expanding leaves; R, roots; S, stems; T, tendrils; F, flowers; Fr, fruits; SA, stem apexes. Different letters above the bars stand for significant differences (Tukey’s multiple range tests, P < 0.05) between different treatment times.Expression profiles of ClDof genes in response to salt stress and ABA treatment
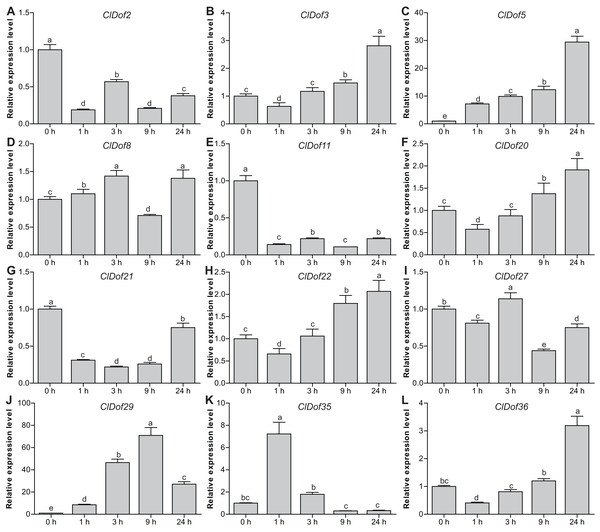
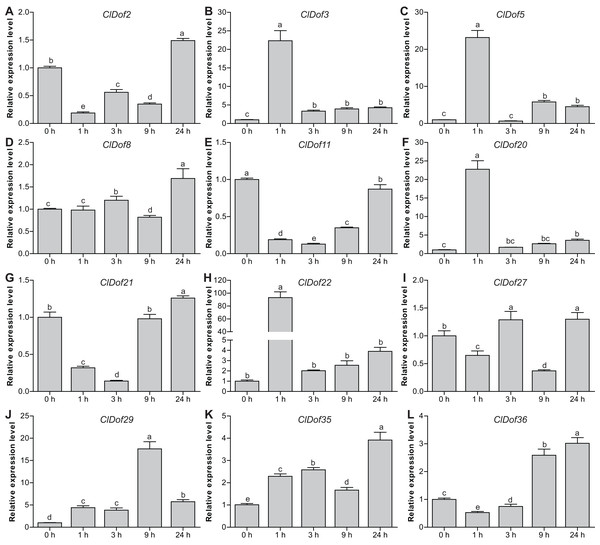
To reveal the possible roles of ClDof genes in response to abiotic stress, we determined the expression levels of 12 selected ClDof genes under salt stress and ABA treatments using qRT-PCR. Under salt stress, nine ClDof genes (ClDof3, ClDof5, ClDof8, ClDof20, ClDof22, ClDof27, ClDof29, ClDof35 and ClDof36) were up-regulated at certain time points (Fig. 9; Table S7). Amongst them, ClDof5 was induced gradually and reached the highest transcript abundance at 24 h, while the expression of ClDof36 showed a decrease at early time point (1 h) and then gradually increased until 24 h (Fig. 9). In addition, three ClDof genes (ClDof2, ClDof11 and ClDof21) were down-regulated across all time points under salt stress, indicating their negative roles in response to salt stress. It should be noted that the expression levels of ClDof3, ClDof8, ClDof20, ClDof22, ClDof27 and ClDof35 were significantly decreased at some time points (Fig. 9). We also determined whether these ClDof genes are regulated by ABA. As shown in Fig. 10 (Table S7), the expression of all detected ClDof genes was significantly altered by ABA treatment, and the expression of five ClDof genes (ClDof2, ClDof11, ClDof21, ClDof27 and ClDof36) showed a decreasing tendency at early time points (1 h and 3 h) and finally increased at the late time points (24 h). It is worth noting that the expression of four ClDof genes (ClDof3, ClDof5, ClDof20 and ClDof22) was dramatically induced at 1 h, followed by sharp decreases thereafter. These results indicated that the ClDof genes may play crucial roles in stress responses.
Figure 9: Expression profiles of twelve selected ClDof genes (A–L) in response to salt stress determined by qRT-PCR.
Different alphabets above the bars indicate significant differences (Tukey’s multiple range tests, P < 0.05) between different treatment times.Figure 10: Expression profiles of twelve selected ClDof genes (A–L) under ABA treatment determined by qRT-PCR.
Different letters above the bars indicate significant differences (Tukey’s multiple range tests, P < 0.05) between different treatment times.Discussion
In the present study, we systematically predicted and identified 36 Dof genes in the watermelon genome (Table 1). The number of ClDof genes is similar to that in many other plant species, such as pepper (33 genes) (Kang et al., 2016; Wu et al., 2016), tomato (34 genes) (Cai et al., 2013), potato (35 genes) (Venkatesh & Park, 2015), foxtail millet (35 genes) (Zhang et al., 2017), cucumber (36 genes) (Wen et al., 2016), chickpea (37 genes) (Nasim et al., 2016), and pigeonpea (38 genes) (Malviya et al., 2015), suggesting that Dof genes usually form multigene families in plants. Duplication events were found to be the primary driving force for the evolution of Dof genes. For example, two pairs of tandemly duplicated genes and six pairs of segmentally duplicated genes were identified in the cucumber genome (Wen et al., 2016). In poplar, up to 49% (20 out of 41) of PtrDof genes were found to be located in both segmental and tandem duplicated regions (Wang et al., 2017). In apple, a total of 57 and 18 MdDof genes were located in segmental and tandem duplicated regions, respectively, and 13 MdDof genes were both segmentally and tandemly duplicated genes (Hong et al., 2019). In this study, more than half of the ClDof genes (20 out of 36) exhibited segmental duplications, while no tandem duplication was identified in watermelon chromosomes, suggesting that segmental duplication has been predominant in the expansion of the Dof genes in watermelon. Similar results have also been reported in other plants such as cotton (Li et al., 2018).
The phylogenetic results revealed that ClDofs could be clearly divided into nine groups: A, B1, B2, C1, C2.1, C2.2, C3, D1 and D2 (Fig. 1), which is consistent with the results in eggplant (Wei et al., 2018), pear (Liu et al., 2020), Arabidopsis and rice (Lijavetzky, Carbonero & Vicente-Carbajosa, 2003). Besides, each of the watermelon Dof genes has at least one homologous gene in Arabidopsis (Fig. 1), implying that Dof genes might play similar roles in watermelon as their homologs in Arabidopsis. In addition, nearly all ClDofs had a common Dof motif (motif 1), but there were also some unique motifs in certain groups with nearly conserved motif compositions (Fig. 2). However, gain or loss of certain motifs was observed between several duplicate pairs, such as ClDof3/ClDof23, ClDof13/ClDof15, ClDof14/ClDof16, ClDof13/ClDof20 and ClDof20/ClDof36 (Figs. 2 and 5), suggesting that these motifs might be involved in the functional divergence of ClDofs. The organization of exon–intron structures can provide insights into the evolutionary relationships within certain gene families (Zhou et al., 2018a). In this study, the number of introns of ClDof genes varied from 0 to a maximum of 2, and most of them contained one intron or no intron at all (Fig. 4). Similar results were obtained in many other plant species, such as cucumber (Wen et al., 2016), poplar (Wang et al., 2017), eggplant (Wei et al., 2018), pear (Liu et al., 2020), Arabidopsis and rice (Lijavetzky, Carbonero & Vicente-Carbajosa, 2003), revealing that the exon–intron structure of Dof genes is highly conserved in plants, which may be related to their similar functions.
The specificity of gene expression in plant tissues and developmental stages can provide important information about the possible functions of genes, and previous reports have revealed that some Dof genes usually have tissue-specific expression patterns (Ma et al., 2015; Venkatesh & Park, 2015; Zou & Yang, 2019). For example, ZmDof3 was found to be exclusively expressed in the endosperm of maize kernel and participate in the regulation of starch accumulation and aleurone development in maize endosperm (Qi et al., 2017). Another maize Dof gene ZmDof36 was also reported to be highly expressed in maize endosperm and function in starch synthesis by regulating the expression of starch synthesis genes (Wu et al., 2019). In this study, ClDof2, ClDof5, ClDof8, ClDof21 and ClDof35 showed much higher expression in leaves than in other tissues, suggesting that they play essential roles in leaf development. Similarly, seven potato Dof genes (StDof15a, StDof22, StDof26, StDof29a, StDof32 and StDof34) exhibited higher expression in leaf tissues than in other tissues (Venkatesh & Park, 2015). In addition, ClDof11, ClDof27, ClDof29 and ClDof36 were predominantly expressed in fruits (Fig. 8), suggesting that they may be associated with fruit development of watermelon. In a previous study, a number of MaDof genes were markedly regulated throughout the fruit development of banana (Dong, Hu & Xie, 2016), and MaDof23 can act as a transcriptional repressor and interacts with MaERF9 to regulate the fruit ripening by controlling specific ripening-related genes (Feng et al., 2016). Besides fruits, the flowers also showed high expression of ClDof11, ClDof21, ClDof27, ClDof29, ClDof35 and ClDof36, which was also observed in other plants. For example, all PheDof genes displayed differential expression patterns during the flower development stage of moso bamboo (Cheng et al., 2018; Wang et al., 2016), and overexpression of PheDof12-1 in Arabidopsis resulted in early flowering under long-day conditions (Liu et al., 2019a). In rubber tree, the HbDof genes in Cluster III and Cluster VI are typically expressed in male and female flowers, respectively (Zou & Yang, 2019). It should be noted that ClDof21 and ClDof27 were segmental duplication genes and they exhibited similar expression patterns in various tissues (Figs. 5 and 8). The qRT-PCR results revealed that both of them were highly expressed in mature leaves and fruits but lowly expressed in tendrils (Fig. 8). Therefore, the tissue-specific expression patterns revealed that ClDof genes play vital and seemingly redundant roles in watermelon growth and development.
Dof genes are known to play a crucial role in stress responses. For example, tomato SlCDF1–5 genes were differentially up-regulated by osmotic, salt, heat and cold stresses, and transgenic Arabidopsis plants overexpressing SlCDF1 or SlCDF3 displayed higher drought and salt tolerance (Corrales et al., 2014). Another Dof gene SlDof22 was shown to control the ascorbate accumulation and salt stress in tomato (Cai et al., 2016). In this study, many stress-related cis-elements were found in the promoter regions of the ClDof genes (Fig. 7), implying their roles in regulating responses to various stresses. In addition, all of the detected ClDof genes showed differential expression under salt stress (Fig. 9), suggesting their regulatory roles in salt stress response. It should be noted that ClDof5 was induced gradually by salt stress (Fig. 9), and had the highest expression in leaves (Fig. 8). Similarly, GhDof1 also showed the highest expression in leaves as compared with in any other tissues, and salt treatment induced its transcript accumulation. Overexpression of GhDof1 in cotton resulted in significantly higher salt and cold tolerance (Su et al., 2017). However, the expression levels of ClDof2, ClDof11 and ClDof21 significantly declined across all time points (Fig. 9), implying that they may play negative regulatory roles in salt stress response. In banana, many MaDof genes were found to be down-regulated under drought and salt stress conditions (Dong, Hu & Xie, 2016). In addition, two pairs of segmental duplication genes, ClDof3/ClDof22 and ClDof20/ClDof36, displayed similar expression patterns under salt treatment, whose expression levels decreased at early time points but increased at the late time points (Fig. 9), suggesting their similar roles in response to salt stress. Notably, since the expression of detected ClDof genes was altered under salt treatment, ClDof29 exhibited much higher expression levels than other detected genes (Fig. 9), indicating that ClDof29 might be the primary regulator of response to salt stress in watermelon. Moreover, a large number of cis-elements associated with stress-related hormonal response, such as ABA, GA, SA, MeJA, ethylene and auxin (Fig. 7), and all of the detected ClDof genes exhibited ABA-dependent expression patterns (Fig. 10). In castor bean, a large number of RcDof genes were regulated (13 up-regulated and 2 down-regulated) in response to ABA treatment (Jin, Chandrasekaran & Liu, 2014). In Arabidopsis, the expression of AtCDF3 was induced by cold, drought, salt, and ABA treatment, and AtCDF3 overexpression could promote tolerance to drought, cold and osmotic stress (Corrales et al., 2017). ABA is thought to participate in the adaptation of plants to various abiotic stresses by regulating the expression of numerous stress-related genes (Zhou et al., 2019). These results indicate that the ClDof genes may play important roles in plant adaptation to salt stress through ABA-dependent pathways.
Conclusions
In this study, we performed a comprehensive analysis of the phylogenetic relationships, conserved motifs, gene structures, chromosome distributions, and gene duplications of 36 Dof genes in watermelon. In addition, qRT-PCR was employed to examine the expression profiles of the ClDof genes in different tissues and in responses to salt and ABA treatments. All of the detected ClDof genes were regulated by salt and ABA treatments. Our findings may help the functional research of ClDof genes for dissecting their roles in the growth, development and stress response of watermelon.
Supplemental Information
The qRT-PCR results of the expression profiles of nine selected ClDof genes in various tissues, under salt and ABA treatment.
GO analysis results for ClD of genes.
Three categories, including cellular component, molecular function, and biological process, were identified and visualized with the WEGO program.