Molecular taxonomy of endemic coastal Ligia isopods from the Hawaiian Islands: re-description of L. hawaiensis and description of seven novel cryptic species
- Published
- Accepted
- Received
- Academic Editor
- Rita Castilho
- Subject Areas
- Marine Biology, Taxonomy, Zoology
- Keywords
- Oniscidea, Intertidal, Species description, Ligiidae, Pacific biodiversity, Cryptic species
- Copyright
- © 2019 Santamaria
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Molecular taxonomy of endemic coastal Ligia isopods from the Hawaiian Islands: re-description of L. hawaiensis and description of seven novel cryptic species. PeerJ 7:e7531 https://doi.org/10.7717/peerj.7531
Abstract
Past phylogeographic work has shown Ligia hawaiensis, a coastal isopod species endemic to the Hawaiian Islands, to be a paraphyletic complex of several highly genetically divergent yet morphologically cryptic lineages. Despite the need for a taxonomic revision of this species, the lack of morphological differentiation has proven an impediment to formally describe new Ligia species in the region. Molecular characters and species delimitation approaches have been successfully used to formally describe cryptic species in other crustacean taxa, suggesting they may aid taxonomic revisions of L. hawaiensis. Herein, various distance- and tree-based molecular species delimitation approaches are applied on a concatenated dataset comprised of both mitochondrial and nuclear gene sequences of L. hawaiensis and L. perkinsi, a terrestrial species endemic to the Hawaiian archipelago. Results of these analyses informed a taxonomic revision leading to the redescription of L. hawaiensis and the description of seven new cryptic species on the basis of molecular characters: L. dante, L. eleluensis, L. honu, L. kamehameha, L. mauinuiensis, L. pele, and L. rolliensis. These coastal Ligia species from the Hawaiian archipelago appear to be largely limited to single islands, where they appear largely constrained to volcanic rift zones suggesting allopatric events at local scales may drive diversification for poorly dispersing organisms in the Hawaiian coastlines. Additional work remains needed to fully assess the role of said events; however, the description of these novel species underscore their potential to aid in studies of local diversification of marine organisms in Hawai‘i. Lastly, this represents the first application of molecular taxonomic approaches to formally describe genetic lineages found in Ligia isopods as species, underscoring the promise these methods hold to taxonomic revisions in other species in the genus shown to harbor cryptic genetic lineages.
Introduction
The isopod genus Ligia (Fabricius 1798) consists of ∼40 currently valid species, most of which inhabit rocky intertidal habitats (Schmalfuss, 2003). The genus is known to exhibit several biological traits that severely limit their dispersal potential: Ligia isopods are direct developers who carry their embryos in a brood pouch (i.e., marsupium) until their emergence as fully formed juveniles, have poor desiccation resistance (Barnes, 1934; Barnes, 1935; Tsai, Dai & Chen, 1998), avoid open water (Barnes, 1932), and exhibit poor locomotion outside their rocky habitats (Santamaria personal observation). Such low vagility and the patchiness of Ligia habitats (i.e., rocky intertidal coastlines) have been suggested to restrict gene flow, leading to long-term isolation and deep genetic divergence between populations even across small geographic distances (e.g., Jung et al., 2008; Hurtado, Mateos & Santamaria, 2010; Eberl et al., 2013; Santamaria et al., 2013). Not surprisingly, molecular characterizations have uncovered highly divergent genetic lineages in several Ligia species around the world suggesting that some species may represent cryptic species complexes in need of formal taxonomic description (Jung et al., 2008; Hurtado, Mateos & Santamaria, 2010; Santamaria et al., 2013; Raupach et al., 2014; Santamaria, Mateos & Hurtado, 2014; Santamaria et al., 2017; Greenan, Griffiths & Santamaria, 2018; Hurtado et al., 2018). One such example is that of Ligia from the coastlines of the Hawaiian archipelago.
Two coastal species have been described from the Hawaiian islands to date: L. hawaiensis (Dana, 1853) and L. kauaiensis (Edmondson, 1931). The former was first described by Dana (1853) from specimens collected in Kaua‘i and O‘ahu and later confirmed as a valid species by Jackson (1922). It is currently thought to occur solely in the Hawaiian archipelago where it is widespread (Taiti & Ferrara, 1991; Taiti & Howarth, 1996; Schmalfuss, 2003). Ligia kauaiensis was described by Edmondson (1931) from individuals collected from Kalihiwai Bay, Kaua‘i. He differentiated L. kauaiensis from L. hawaiensis based on differences in inter-eye distance, number of segments in the flagellum of the antenna, and body size; however, posterior authors did not find such differences and suggested L. kauaiensis to be a junior synonym of L. hawaiensis (Arcangeli, 1954). The current taxonomy of the Ligia genus reflects this synonymy, recognizing L. hawaiensis as the sole coastal Ligia species endemic to the Hawaiian archipelago (Schmalfuss, 2003). Recent molecular analyses of coastal Ligia from the region; however, suggest L. hawaiensis may be a cryptic species complex in need of taxonomic revision.
Phylogeographic studies completed in the past two decades suggest L. hawaiensis to be a paraphyletic taxon composed of several deeply divergent and cryptic lineages (Taiti et al., 2003; Santamaria et al., 2013). Maximum Parsimony phylogenetic reconstructions carried out in 2003 and based on a mitochondrial dataset including L. hawaiensis and L. perkinsi, a terrestrial species from the Hawaiian archipelago, from Kaua‘i and O‘ahu uncovered three divergent lineages within L. hawaiensis (Taiti et al., 2003). A decade later, Santamaria et al. (2013) expanded on this work by including samples from previously unsampled islands, using additional genetic markers, and applying model-based phylogenetic reconstruction approaches. They found L. hawaiensis to be a paraphyletic taxon composed of several highly divergent and geographically disjunct lineages, including a clade comprised of coastal Ligia from Maui and Hawai‘i (Clade A), another comprised of Kaua‘i individuals (Clade D), one containing Ligia from O‘ahu and the Maui-Nui complex (Clade E), and lastly one comprised of individuals from the islands of O‘ahu, Maui, and Hawai‘i (Clade F). Given the paraphyly of L. hawaiensis and that levels of divergence between said lineages matched or exceeded those observed between other Ligia species pairs, Santamaria et al. (2013) concluded L. hawaiensis may represent a cryptic species complex in need of taxonomic revision. Unfortunately, such taxonomic revisions have been hindered by an apparent lack of morphological differentiation amongst highly divergent genetic lineages.
Taiti et al. (2003) evaluated eight characters used in Ligia taxonomy for both male and female L. hawaiensis of what are now known to be three highly divergent lineages (i.e., D, E, F) and failed to find any differences between them. More recently, geometric-morphometric comparisons capturing characters used in Ligia taxonomy (e.g., inter-eye distance, shape of telson) were carried out by Santamaria et al. (2013) to determine whether statistically significant shape differences amongst genetic lineages existed. Although their analyses did recover statistically significant differences amongst lineages, cross-validated discriminant function analyses indicated these to be of no taxonomic value as correct classification rates were as low as 26.67%. Similar results have been since reported for other Ligia species. High degrees of overlap in overall body shapes and low classification rates have also been reported for L. occidentalis lineages (Santamaria et al., 2016), while taxonomic examination of highly divergent L. natalensis lineages uncovered by Greenan, Griffiths & Santamaria (2018) failed to uncover taxonomically diagnostic characters amongst in the lineages found in this southern African species (C.L. Griffiths personal communication). Hence, the totality of these findings indicates molecular approaches may be the best-suited approach to formally describe coastal Ligia lineages from the Hawaiian archipelago as species.
Molecular-based species approaches have been successfully used in detecting, delineating, and describing cryptic species in other peracarids such as Gammarus amphipods (Grabowski, Wysocka & Mamos, 2017), munnopsid isopods (Schnurr et al., 2018), and Atlantoscia isopods (Zimmermann et al., 2018). In this study, phylogenetic reconstructions as well as distance- and phylogeny-based molecular species delimitation methods are applied on a multi-locus dataset comprised of L. hawaiensis and L. perkinsi individuals collected throughout the Hawaiian archipelago to inform the revision of the taxonomy of L. hawaiensis. Results reported herein indicate the need to narrowly re-describe L. hawaiensis and to describe seven new species distinguished on the basis of molecular characters. The formal description of these cryptic species not only further highlights Ligia as a rare example of in-situ speciation in a Hawaiian marine taxon bird (Kay & Palumbi, 1987, but see Bird et al., 2011), but may also be of importance to conservation efforts (Bickford et al., 2007; Delić et al., 2017).
Materials and Methods
Sample collection
Ligia specimens were collected from 24 rocky intertidal habitats across the Hawaiian islands of Kaua‘i, O‘ahu, Maui, and Hawai‘i in the summer of 2016. Of these, nine are localities previously sampled by Santamaria et al. (2013) with the remaining fifteen being localities previously not characterized by either Santamaria et al. (2013) or Taiti et al. (2003). Detail locality information is provided in Table 1. All individuals were caught by hand during the day and field-preserved in 70% ethanol. Once in the laboratory, all specimens were identified as L. hawaiensis by comparing the endopod of the 2nd pleopod to the morphology illustrated by Taiti et al. (2003) prior to molecular characterizations.
Molecular laboratory methods
Total genomic DNA was extracted from pereopods and/or pleopods using the Quick g-DNA MiniPrep Kit (Zymo Research) for 1–5 individuals per locality. Afterwards, four mitochondrial and three nuclear gene fragments were amplified using previously published primers and conditions: (a) a 658-bp segment of the Cytochrome Oxidase I gene (hereafter COI, primers LCO1490/HCO2198; Folmer et al., 1994), (b) a ∼490-bp segment of the 16S rRNA gene (primers 16Sar/16Sbr; Palumbi, 1996), (c) a ∼495-bp segment of the 12S rDNA gene (primers crust-12Sf/crust-12Sr; Podsiadlowski & Bartolomaeus, 2005), (d) a 361-bp fragment of the Cytochrome-b gene (hereafter Cytb, primers 144F/151F and 270R/272R; Merritt et al., 1998), (e) a ∼1,000-bp segment of the 28S rDNA gene (primers 28SA/28SB Whiting, 2002) (f) 664-bp region of the alpha-subunit of the Sodium Potassium ATPase (hereafter NaK, primers NaK-forb/NaK-rev2; Tsang et al., 2008), and (g) a ∼328-bp fragment of the Histone H3 gene (primers H3AF/H3AR; Colgan et al., 1998).
Genomic DNA was also obtained for the syntype of L. hawaiensis deposited in the Harvard Museum of Comparative Zoology (MCZ CRU-1543) using a modified version of the protocol of Shokralla, Singer & Hajibabaei (2010): one mL of the specimen’s preservative ethanol was evaporated at 56 °C for 30 min, reconstituted in 250 µL of molecular water, with DNA then extracted using the Quick g-DNA MiniPrep Kit (Zymo Research). Two mitochondrial genes fragments were PCR amplified for this specimen using internal primers designed in Geneious R8.1.9 based on publicly available Ligia sequences: a 122-bp fragment of the 16S rDNA gene (16S-LigiaF: 5′-CGCAGTATCCTGACTGTGCT-3′, 16S-LigiaR: 5′-AGCTTTTAGGGTCTTATCGTCCC-3′) and a 212-bp fragment of the COI gene (COI-LigiaF 5′-CTWGGDCAGCCTGGWAGRTTT-3′; COI-LigiaR 5′-MCCTGTTCCTACTCCTCTTTCA-3′). All PCR products were visualized on a 1% agarose gel stained using SYBR-Safe (Invitrogen) prior to sequencing at the University of Arizona Genetics Core (UAGC).
Sequence alignment and model testing
Sequences produced in this study, with the exception of those for the L. hawaiensis syntype, were combined with those produced by Santamaria et al. (2013) and those publicly available in GenBank. The syntype sequences were excluded from the dataset due to their relative short length. Ribosomal genes (16S rDNA, 12S rDNA) were aligned using the MAFFT algorithm (Katoh & Standley, 2013) as implemented in the GUIDANCE2 server (Sela et al., 2015) using standard settings. Poorly aligned positions in these alignments were removed automatically by masking all positions with a confidence alignment score below 1.00. Protein coding genes were aligned using the online MAFFT server (Katoh, Rozewicki & Yamada, 2017) using default settings. No evidence suggestive of pseudo-genes was observed in any of the protein coding genes alignments. Pairwise genetic distances were estimated with the Kimura-2-Parameter (K2P) correction (excluding ambiguous sites) in MEGA v7.0.18 for the COI dataset (Kumar, Stecher & Tamura, 2016).
For each aligned gene dataset, the most appropriate model of nucleotide evolution was selected from 1,624 models by evaluating their likelihoods on a fixed BioNJ-JC tree under the Bayesian Information Criterion (BIC) in jModeltest v2.1 (Darriba et al., 2012). Afterwards, individual gene alignments were concatenated using SequenceMatrix v.1.7.8.1 (Vaidya, Lohman & Meier, 2011) and the most appropriate model of nucleotide evolution selected for the concatenated alignment as described above.
Molecular species delimitation analyses
Species hypothesis were obtained using several molecular species delimitation analyses (hereafter MSDAs) approaches, including both tree and distance based approaches. Two tree-based MSDA approaches were implemented: the Poisson Tree Processes model used in the PTP server ( http://species.h-its.org/), an approach that delineates species based on branching patterns (Zhang et al., 2013), and the General Mixed Yule Coalescent model (hereafter GMYC; Fujisawa & Barraclough, 2013), an approach that uses branch lengths to determine the transition from intraspecific to interspecific relationships.
As PTP analyses require phylogenetic trees as input, phylogenetic searches on the concatenated alignment of all six genes were carried out under Maximum Likelihood inference as implemented in RAxML v8.0.0 (Stamatakis, 2014). Searches were repeated for three partitioning approaches: unpartitioned, by gene, and as determined by the BIC implemented in PartitionFinder v1.0.0 (Lanfear et al., 2012) using settings as Santamaria et al. (2013). RAxML searches consisted of 1,000 bootstrap replicates followed by a thorough ML search under the GTR + Γ model run under the Thorough Bootstrap Algorithm with all other settings as default. Majority-rule consensus trees were estimated for each analysis using the SumTrees command of DendroPy v3.10.1 (Sukumaran & Holder, 2010). PTP analyses were then carried under both the Maximum Likelihood and Bayesian implementations on both the ML tree and majority consensus bootstrap tree produced by each search. Settings used were as follows: 500,000 MCMC iterations; a burn-in of 0.10; and a thinning value of 100.
In contrast with PTP, GMYC delineations require ultrametric trees as input. Thus, BEAST v2.1.3 (Bouckaert et al., 2014) was used to estimate ultrametric trees for the unpartitioned concatenated mitochondrial dataset using two different approaches: assuming a constant rate of evolution and speciation assuming a Yule process (i.e., constant speciation rate; Yule, 1925; Gernhard, 2008), and under a coalescent model of speciation assuming a constant population size (Kingman, 1982). Both searches were used using the most appropriate model of nucleotide evolution. All BEAST runs were carried out for 50 million generations with sampling every 1,000th. Resulting trees were summarized using the SumTrees command with burn-in discarded and with edges set as per the mean-age option. Resulting ultrametric trees were analyzed using the GMYC approach as implemented by the ‘splits’ package (http://r-forge.r-project.org/projects/splits/) in R using default settings.
Two distance-based approaches were applied on the COI gene dataset alone: the ABGD software (Puillandre et al., 2012) and the BIN system applied in BOLD v3 (Ratnasingham & Hebert, 2013). ABGD analyses were carried out on the entire COI dataset and after masking ambiguous sites using the online server (http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html) under the Kimura 2-Parameter (K2P) nucleotide evolution model, a Pmin value of 0.01, Pmax of 0.20, and a relative width of 1. All other settings were as default. BIN searches in BOLD were carried out using the “Cluster Sequences” option under the K2P distance model and the BOLD aligner option. Sequences shorter than 200-bp, with evidence of contaminants, possibly misidentified, and with stop codons were filtered out. All other parameters were as default.
Candidate species were then identified by comparing results of phylogenetic reconstructions, pairwise COI K2P distances, and MSDAs patterns. In general terms, candidate species were chosen so that all the following criteria were met: (1) all members of the putative species constituted a well-supported (BS >90%) monophyletic clade recovered in all phylogenetic reconstructions; (2) within-group average pairwise COI K2P distances in the clade were <3.0%; and (3) a majority of MSDAs assigned individuals as belonging to the same candidate species. Exceptions to the third criterion were made for those instances where analyses identified several species consisting of a single individual within a well-supported monophyletic group identified as a single species by other analyses. These criteria were chosen to avoid over-splitting.
Candidate species were validated using BPP v4.0 (Yang, 2015) by completing species delimitations under two different partition schemes (i.e., unpartitioned, by gene) and several combinations of θ and τ priors. Diagnostic nucleotide positions were then determined for the validated candidate species using the “Diagnostic character” function of BOLD. Analyses were carried out on all genes independently assuming a K2P distance model, all quality filters, grouping of sequences according to species, and alignments as submitted. All other settings were as default. Diagnostic and partially diagnostic characters for each species were recorded, with others ignored.
Lastly, the identity of the name-carrying L. hawaiensis syntype deposited in the Harvard Museum of Comparative Zoology was established using three approaches. First, the COI sequence obtained from this specimen was queried against all barcode records available in the Barcode of Life Database (BOLD) in May of 2019 (Ratnasingham & Hebert, 2007). Second, the 16S rDNA and COI sequences were each queried against all published sequences in GenBank using BLAST in May 2019. Lastly, 16S rDNA and COI sequences were combined with the corresponding gene dataset produced in this study, and aligned as described above. Neighbor-joining trees were produced for each aligned gene dataset using Geneious R8.1.9.
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/. The LSID for this publication is: urn:lsid:zoobank.org:pub:7FF280B9-13B0-43F9-A9D6-1C2CDF29C0CC. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central and CLOCKSS.
Results
The combination of molecular data produced in this study with that previously published by Santamaria et al. (2013) produced a concatenated dataset 3,889-bp long after to the removal of poorly aligned positions in the 16S, 12S, and 28S rDNA genes (43, 17, and 49 respectively). This alignment included four mitochondrial and three nuclear genes from 193 individuals across 39 localities in the Hawaiian archipelago (Fig. 1, Table 1). The final alignment included 543 parsimony informative sites (COI = 185; Cyt-b: 120; 12S rDNA = 99; 16S rDNA = 91; 28S rDNA = 39; NaK = 6; H3A = 3). All new sequences produced in this study have been deposited in GenBank under accession numbers MK032482 –MK032590, MK032592 –MK032638, MK034474 –MK034685, MK940864 –MK940896 (Table 1) while an annotated alignment is provided as Dataset S1. The sequences produced in this study are also deposited in the BOLD database.
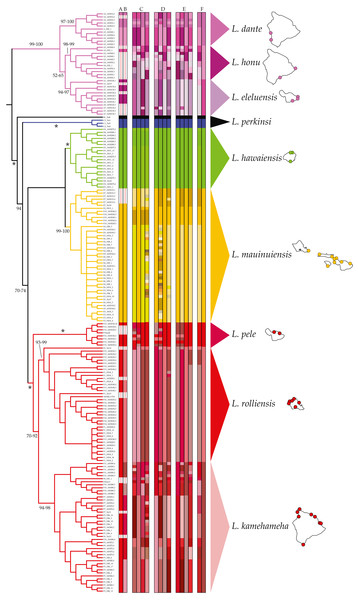
Figure 1: Ligia localities included in this study.
Labels and colors correspond with other figures and tables in this study and that of Santamaria et al. (2013). Detailed information for each locality is presented in Table 1. Localities of the suppralittoral L. hawaiensis included: Kaua‘i: D1-Kalihiwai Beach, D2-Kauapea Beach, D6-Hoai Bay (D6); O‘ahu: E10-Wawamalu Beach Park, F1-Pupukea, F2-Pouhala Marsh, F13-Kahaluu (F13), F14-Kaena Point (North), F15-Kaiaka Bay Beach Park, F16-Kaena Point (South); Moloka‘i: E2-Papohaku Beach Park, E4-Manele Bay; Lana‘i: E3-North of Puko’o; Maui: A1-Wai‘Ōpae; A6-Waianapanapa State Park, A7-Koki Beach Park,E5-Poelua Bay, E6-Spreckelsville, E7-Keanae, E8-DT Fleming Beach Park, E9-Hanakao’o Park, F3-Honomanu Bay, F12-Baby Beach Spreckelsville Area; Hawai‘i: A2-Kealakukea Bay, A3-Pu’unalu Beach Park, A4-Isaac Hale Beach Park, A5-Miloli Beach Park, F4-Keokea Beach, F5-Onekahakaha Beach Park, F6-Leleiwi Beach, F7-South Point, F8-Kapa’a State Park, F9-Kolekole Beach Park, F10-Laupahoehoe Beach Park, F11-Spencer Beach Park. Localities of the terrestrial L. perkinsi included are Kaua‘i: C1-Mt Kahili, C2-Makaleha Mts, C3-Haupu Range; O‘ahu: B1-Nu’uanu Pali. Boldfaced labels indicate type localities.All phylogenetic reconstructions completed in this study were congruent and match those reported by Santamaria et al. (2013). Four highly divergent lineages comprised of coastal Ligia were identified: (a) Clade A (lavenders and purples in all figures) which contained all individuals from three localities in Maui (A1, 6–7) and Hawai‘i (A2–5) each; (b) Clade D (green in all figures) which included all coastal Ligia sampled from Kaua‘i (D1–2, 6); (c) Clade E (oranges and yellows in all figures) from O‘ahu (E10), Moloka‘i (E2, E3), Lana‘i (E4), and Maui (E5–E9); and lastly (d) Clade F (reds in all figures) from O‘ahu (F1–2, 13–16), Maui (F3, 12), and Hawai‘i (F4–F11). No locality was shown to harbor more than one of these lineages. Two highly divergent lineages comprised of L. perkinsi individuals were recovered.
MSDAs assigned individuals identified as L. hawaiensis to 5–57 putative species. Results across analyses were largely congruent, with higher putative species counts produced by some analyses over-splitting larger groups into species with two or less members. No contradictory assignments (e.g., individuals assigned to different species containing three or more species) were observed. ABGD analyses of the COI dataset identified between 5 and 13 species within L. hawaiensis, with 13 and 7 being reported for four partitions each, the former at lower P values (0.010–0.018) and the latter at higher P values (0.022–0.035). BIN analyses in BOLD produced similar results to those of ABGD at higher P values, with the sole difference being the split of Clade A individuals into three rather than two putative species. Tree-based MSDAs recognized 8–57 putative species from specimens identified as L. hawaiensis, with 3–15 species for Clade A, 1–2 for Clade D, 1–22 for Clade E, and 1–22 for Clade F. A detailed breakdown of all MSDA results is presented in Fig. 2.
Comparisons amongst MSDAs, phylogenetic reconstructions, and pairwise COI K2P distances (Table 2) based on the criteria previously led to the identification of eight candidate species of coastal Ligia from the Hawaiian archipelago. Three candidate species were identified from Clade A: (a) one comprising all individuals collected in localities A2 and A5, (b) one from those collected in A3–4, and (c) another containing all specimens from A1, A6–7. Only one candidate species was identified in Clade D (all individuals from localities D1–2, D6) and another in Clade E (all individuals from E2–10). Lastly, three candidate species were identified in Clade F: (a) one comprised of all individuals collected in Maui (F3, 12), (b) one from O‘ahu specimens (F1–2, F13–16), and (c) another composed of all Clade F individuals from Hawai‘i (F4–11). BPP analyses provided high support for this partitioning scheme, as high posterior probabilities (>0.99) were observed for all eight candidate coastal Ligia species regardless of priors used. No analysis clustered two or more candidate species with posterior probabilities >0.01.
Figure 2: Results of molecular species delimitation analyses (MSDAs).
Results are projected on the majority rule consensus tree produced by analyzing the concatenated mitochondrial and nuclear dataset of Ligia samples from the Hawaiian Islands in RAxML under the GTR +Γ under a “by gene” partitioning scheme. Branches are transformed for clarity and are colored per clade as per Santamaria et al. (2013). Vertical bars represent assignments to putative species (identified by colors) under various MSDA methods. Values by nodes correspond with bootstrap support values, with * denoting 100% across all analyses. Bars A–B represent assignments by ABGD and BOLD respectively, two distance-based methods on the COI dataset alone. All other bars are results from tree-based approaches. Results for PTP and bPTP (Bayesian implementation of PTP) based on phylogenetic searches carried out on RAxML under various partitioning schemes are presented in C (unpartitioned), D (by gene), and E (according to PartitionFinder). The first two vertical bars within each of these correspond to PTP and bPTP results based on the most likely tree produced by RAxML, with the last two corresponding to PTP and bPTP results on majority consensus bootstrap trees. The bars denoted by F correspond with GMYC assignments based on Coalescent and Yule speciation models respectively. Consensus species as well as the localities where they have been identified at are shown.Of these, the candidate species comprised of Clade D individuals (green in all Figures) appears to correspond with Dana’s L. hawaiensis description, as BOLD identifications (100% match to a Clade D haplotype), BLAST searches (16S rDNA: 98.36% match, 100% coverage; COI: 100% match, 100% coverage), and NJ analyses (not shown) indicate the name-carrying syntype of L. hawaiensis to be a member of this candidate species. These results thus indicate the need to narrowly redescribe L. hawaiensis and to describe seven novel species of coastal Ligia from the Hawaiian archipelago.
| L. dante | L. eleluensis | L. rolliensis | L. honu | L. kamehameha | L. hawaiensis | L. mauinuiensis | L. pele | L. perkinsi | |
|---|---|---|---|---|---|---|---|---|---|
| L. dante | 0.0–4.6 (2.4) |
||||||||
| L. eleluensis | 9.7–11.2 (10.7) |
0.0–0.9 (0.5) |
|||||||
| L. rolliensis | 14.0–16.5 (15.2) |
13.4–15.4 (14.3) |
0.0–2.0 (0.5) |
||||||
| L. honu | 5.8–7.5 (6.8) |
10.9–11.3 (11.0) |
14.9–15.7 (15.2) |
N/A | |||||
| L. kamehameha | 13.8–16.4 (14.8) |
13.8–15.4 (14.6) |
4.0–6.4 (5.0) |
13.6–15.5 (14.6) |
0.0–5.4 (2.5) |
||||
| L. hawaiensis | 12.8–15.4 (14.4) |
14.7–16.4 (15.8) |
12.5–14.4 (13.6) |
15.0–16.9 (16.4) |
11.6–14.6 (13.0) |
0.0–2.2 (0.9) |
|||
| L. mauinuiensis | 13.6–15.9 (14.9) |
14.4–16.4 (15.4) |
10.9–12.6 (11.6) |
14.5–15.3 (15.0) |
8.7–13.1 (10.6) |
10.3–12.7 (11.5) |
0.0–2.4 (0.7) |
||
| L. pele | 15.0–16.6 (15.8) |
15.4–16.2 (15.7) |
6.1–7.1 (6. 6) |
15.9–16.2 (15.0) |
6.4–8.7 (7.3) |
12.6–14.8 (13.9) |
11.1–12.8 (12.0) |
0.0–0.2 (0.9) |
|
| L. perkinsi | 11.9–15.0 (13. 6) |
12.5–14.8 (13.6) |
12.9–14.7 (13.9) |
13.7–15.1 (14.1) |
11.1–15.0 (13.2) |
13.8–15.8 (15.2) |
12.5–16.6 (13.8) |
13.9–16.1 (14.9) |
1.0–15.3 (8.4) |
Taxonomy
MSDAs results, as well as those of phylogenetic reconstructions, COI K2P pairwise distances, and the geographic distribution of lineages informed the re-description of L. hawaiensis as well as the description of seven novel coastal Ligia species from the Hawaiian archipelago. All type specimens, paratypes, and additional lots have been deposited at the Florida Museum of Natural History (FLMNH) in Gainesville, FL, USA. Descriptions below focus on molecular characters, as past morphological inspections have shown L. hawaiensis lineages to lack diagnostic morphological differences (Taiti et al., 2003; Santamaria et al., 2013). Nonetheless, descriptions briefly touch on some overall body characteristics that may help distinguish Ligia species (e.g., eye size/distance, body ratio) and photographs of all type specimens are provided (Fig. 3). All other traits (e.g., pereopods) are as described and/or illustrated by Taiti et al. (2003), Taiti, Ferrara & Kwon (1992), and Jackson (1933).
Figure 3: Figure 3: Paratypes of novel and re-described Hawaiian Ligia species.
(A) L. honu (UFID 49319); (B) L. eleluensis (UFID: 49329); (C) L. hawaiensis (UFID 49341); (D) L. pele (UFID 49326); (E) L. dante (UFID 49316); (F) L. kamehameha (UFID 49322); (G) L. rolliensis (UFID 49336); (H) L. mauinuiensis (UF 49331).| Ligia hawaiensis species re-description |
| LSID: urn:lsid:marinespecies.org:taxname:257550. |
| BOLD BINs : AAD0842. |
Materials examined: the L. hawaiensis syntype deposited in the Harvard Museum of Comparative Zoology (MCZ:IZ:CRU-1543) as well as twenty individuals, both male and female, from 6 coastal localities across the island of Kaua‘i (D1, D2, and 6). A neotype (UFID 49339), paratype (UFID 49341) and a lot of five individuals (UFID 49342) from the type locality as well as five individuals from Hoai Bay (UF 49343) have been deposited.
Type locality: Kalihiwai Beach, Kaua‘i (D1; Lat.: 22°13′05.30″N; Long.: 159°25′31.15″W)
Type: The syntype deposited by Dana appears to be a female in extremely poor condition (i.e., less than half the specimen remains) and with no locality information beyond “Hawaiian Archipelago” available for it. Thus, a neotype has been deposited with the expressed purpose of providing clarity to this particular taxon. The neotype deposited belongs to the lineage as that of Dana’s original syntype and was collected from one of the two islands sampled by said author. It is 17.7 mm long and 6.8 mm wide (body length to width ratio of ∼2.6) has been designated as a neotype. Eyes appear to be moderate in size (eye length is ∼0.5 greatest width of cephalon) and spacing (inter-eye distance ∼0.7 times eye length). Posterolateral processes of the pereionite 7 extend ∼0.6 length of the pleonite 3. Antennae are long, extending almost the entire length of the body. The UFID for the neoype is 49339.
Diagnostic molecular characters:
| COI: 108-G; 165-T; 177-C; 279-G; 426-A; 459-G; 492-C; 522-G; 591-C. |
| 16S: 6(6)-T; 19(19)-A; 174(186)-C; 224(236)-C; 230(260)-C; 272(302)-A; 306(349)-C; 391(434)-T; 414(457)-T |
| Cyt-b: 115-C; 125-T; 175-A; 250-C. |
| 12S: 32(35)-G; 297(297)-G; 318(321)-A; 376(379)-A; 475(492)-C. |
Partially diagnostic molecular characters:
| COI: 87-G; 90-A; 126-T; 201-C; 300-G; 306-T; 327-G; 393-C; 405-C; 411-T; 417-G; 586-C; 606-G; 706-C. |
| 16S: 110(110)-C; 270(300)-A; 273(303)-T; 360(403)-G; 439(482)-C. |
| Cyt-b: 37-G; 109-C; 154-C; 155-C; 187-A; 196-C; 290-G; 295-C; 349-T; 352-C. |
| 12S: 33(36)-T; 83(86)-T; 301(304)-G; 319(322)-C. |
Distribution: this species appears to be geographically limited to the island of Kaua‘i, where it appears to be the only endemic coastal Ligia species. It is widely distributed across the island.
Etymology: The name was originally proposed by Dana to reflect the Hawaiian distribution of this species.
| Ligia dante nov. sp. |
| LSID: urn:lsid:zoobank.org:act:29042EF5-3FB4-4B34-AE4D-39E9B3462CD4. |
| BOLD BINs : ADO0227; ACQ3367. |
Materials examined: 11 individuals from two localities in the island of Hawai‘i (A2, A5). Both males and females were included. The holotype (UFID 49315), a paratype (UFID 49316), and a lot of five individuals (UFID 49317) from the type locality have been deposited.
Type locality: Miloli’i Beach Park, Hawai‘i, USA (A5; Lat.: 19°10′58.10″N; Long.: 155°54′25.10″W).
Type: small male individual (11.27 mm long) that is 4.40 mm wide at the widest point of the pereionite 4 (body length to width ratio of ∼2.5). Eyes appear to be smaller (eye length is ∼0.4 greatest width of cephalon) and more widely spaced (inter-eye distance ∼1.1 times eye length) than in other Ligia in the area. Posterolateral processes of the pereionite 7 extend length of the pleonite 3. Antennae does not extend past pleonites being ∼0.9 of body length. The holotype is deposited in the FLMNH under UFID 49315. GenBank Accession numbers for sequences obtained from the holotype are as follows: MK034481 (COI); MK032557 (16S rDNA); MK032624 (12S rDNA); MK034569 (Cytb); MK034643 (NaK).
Diagnostic molecular characters:
| COI: 126-G. |
| 16S: 224(236)-G; 291(321)-A. |
| Cyt-b: N/A |
| 12S: 9(9)-G. |
Partially diagnostic molecular characters:
| COI: 123-G; 237-C; 241-C; 259-C; 606-T. |
| 16S: 146(158)-A; 269(299)-C. |
| Cyt-b: 55-G; 82-G. |
| 12S: 85(88)-C; 105(108)-G. |
Distribution: Rocky intertidal habitats in the southwestern region of the island of Hawai‘i: Napo’opo’o Park (A2) and Miloli’i Beach Park (A5).
Etymology: This species is named after Dante Santamaria, a dear friend to the author who recently passed away.
| Ligia eleluensis nov. sp. |
| LSID: urn:lsid:zoobank.org:act:D9C872EB-8BCD-4B16-8FA7-08B1B805E529. |
| BOLD BINs : ADO6183. |
Materials examined: 12 individuals, both males and females, from three localities in the island of Maui (A1, A6, and A7). The holotype (UFID 49328), a paratype (UFID 49329), and a lot of five individuals (UFID 49330) from the type locality have been deposited.
Type locality: Koki Beach Park, Maui (A7; Lat.: 20°43′41.62″N; Long.: 155°59′06.71″W).
Type: male individual that is 14.98 mm long and 5.41 mm wide at pereionite 4 (body length to width ratio of ∼2.8). Eyes appear to be moderate in size (eye length is ∼0.5 greatest width of cephalon) but somewhat more distant than for most other Ligia in the area (inter-eye distance ∼0.8 times eye length). Posterolateral processes of the pereionite 7 extend about length of the pleonite 3. Antennae does not extend past pleonites and is ∼0.7 of body length. The holotype has been deposited under UFID 49328, with sequences for the holotype found under GenBank Accession numbers: MK034485 (COI); MK032499 (16S rDNA); MK032598 (12S rDNA); MK034608 (NaK); MK034656 (H3A).
Diagnostic molecular characters:
| COI: 81-G; 225-T; 324-A; 336-C; 360-G; 456-C; 513-C; 549-C; 696-A. |
| 16S: 13(13)-G; 45(45)-G; 168(180)-T; 189(201)-C; 289(319)-C. |
| Cyt-b: 4-C; 22-C; 289-T. |
| 12S: 1(1)-C; 19(19)-C; 30(33)-A; 166(169)-A. |
Partially diagnostic molecular characters:
| COI: 88-C; 129-A; 333-T; 411-G; 504-C; 558-G; 573-T; 663-C. |
| 16S: 146(158)-T; 280(310)-G; 291(321)-C. |
| Cyt-b: 52-G; 184-G; 187-G; 257-A; 287-C; 304-A; 307-G; 355-C. |
| 12S: 82(85)-G; 85(88)-T; 86(89)-C; 87(90)-T; 382(399)-T. |
Distribution: this species has only been identified in three localities in the eastern coastline of Maui: Wai‘Ōpae (A1), Waianapanapa State Park (A6), and Koki Beach Park (A7).
Etymology: Ligia isopods are often referred to as “wharf roaches” in common parlance. The proposed species name honors this by incorporating the Hawaiian name for a cockroach (‘elelū) into the species epithet.
| Ligia honu nov. sp. |
| LSID: urn:lsid:zoobank.org:act:2C6958E3-C573-4F5A-AB97-7AFC19384603. |
| BOLD BINs : ACQ3366. |
Materials examined: 11 Ligia individuals, both male and female, collected in two localities in the southern coast of the island of Hawai‘i were examined (A3, A4). The holotype (UFID 49318), a paratype (UFID 49319), and five individuals (UFID 49320) from the type locality have been deposited.
Type locality: Punalu’u Black Sand Beach Park, Hawai‘i, U.S.A. (A3; Lat.: 19° 08′00.60″N; Long.: 155°30′18.30″W).
Type: a 4.97 mm long male individual that is 3.76 mm wide at its widest point (pereionite 4; body length:width ratio of ∼2.6). Eyes appear to be moderate in size (eye length is ∼0.5 greatest width of cephalon) and separation (inter-eye distance is ∼0.7 eye length) when compared to other Ligia from the area. Posterolateral processes of the pereionite 7 extend about length of the pleonite 3. Antennae does not extend past pleonites and is ∼0.7 of body length. The UFID for the holotype is 49318, while GenBank Accession numbers for sequences obtained from this individual are as follows: MK034514 (COI); MK032566 (16S rDNA); MK032627 (12S rDNA); MK034582 (Cytb); MK034677 (H3A).
Diagnostic molecular characters:
| COI: 123-A; 126-A; 189-C; 222-C; 234-C; 429-C; 433-C; 597-C; 675-T. |
| 16S: N/A |
| Cyt-b: N/A |
| 12S: 43(46)-C; 140(143)-G. |
Partially diagnostic molecular characters:
| COI: 120-T; 321-T; 402-G; 648-C; 678-G. |
| 16S: 105(105)-A. |
| Cyt-b: 112-G; 238-G; 281-A; 304-G; 341-T |
| 12S: 82(85)-T; 312(315)-C. |
Distribution: This species has only been identified in two localities in the southern coastline of Hawai‘i: Punalu’u Black Sand Beach Park (A3) and Isaac Hale Beach Park (A4).
Etymology: The epitet “honu” is derived from the Hawaiian word for turtle and is in reference to the green sea turtles often found resting in the shores of the type locality.
| Ligia kamehameha nov. sp. |
| LSID: urn:lsid:zoobank.org:act:2EBFFFA4-3BA0-497C-BF97-A09B472893EF. |
| BOLD BINs : ADN0096; ADN6487; ACQ8239; ACQ8240; ACQ8241. |
Materials examined: Forty-three individuals, both males and females, from eight localities in the island of Hawai‘i were examined (F4–11). A holotype (UFID 49321), a paratype (UFID 49322), and a lot of five individuals (UFID 49323) from the type locality as well as five individuals from Onekahakaha Beach Park (UFID 49324) have been deposited.
Type locality: Spencer Beach Park, Hawai‘i (F11; Lat.: 20°01′22.41″N; Long.: 155°49′21.50″W)
Type: a female individual that is ∼2.5 longer than wide at pereionite 4. Eyes are relatively small (∼0.4 times greatest width of cephalon) yet moderately spaced (inter-eye distance ∼0.7 eye length). Posterolateral processes of the pereionite 7 extend to the middle of pleonite 3. Antennae does not extend past pleonites and is ∼0.7 times the body length. The holotype has been deposited under UFID 49321 with sequences available under GenBank Accession numbers: MK034535 (COI); MK032585 (16S rDNA); MK032637 (12S rDNA); MK034592 (Cytb); MK034649 (NaK); MK034684 (H3A).
Diagnostic molecular characters:
| COI: 207-A; 243-G. |
| 16S: N/A. |
| Cyt-b: N/A. |
| 12S: 224(227)-G; 236(239)-C. |
Partially diagnostic molecular characters:
| COI: 633-G. |
| 16S: N/A. |
| Cyt-b: N/A. |
| 12S: 352(355)-C. |
Distribution: The distributional range of this species appears to be limited to the island of Hawai‘i where it is widespread, particularly across its north and west coasts.
Etymology: The species epithet honors Kamehameha I, founder and first ruler of the Kingdom of Hawaii, who was born in the Kohala region of the island of Hawai‘i where the type location for this species is located.
| Ligia mauinuiensis nov. sp. |
| LSID: urn:lsid:zoobank.org:act:B8D9EC07-5127-45A4-94D4-8B14C392A2C0. |
| BOLD BINs : AAD0844. |
Materials examined: 40 individuals from ten localities across the islands of Maui (E5–9), Moloka‘i (E2, E4), Lana‘i (E3), and O‘ahu (E10). A holotype (UFID 49344) from the type locality as well as a paratype (UFID 49331) and a lot of five individuals (UFID 49332) from Hanakao’o Park have been deposited.
Type locality: DT Fleming Beach Park, Maui (E8; Lat.: 21°00′20.82″N; Long.: 156°38′58.43″W)
Type: male individual that is ∼2.8 times longer than wide with average sized and spaced eyes (eye length is ∼0.5 greatest width of cephalon, inter-eye distance ∼0.7 eye length). Posterolateral processes of the pereionite 7 extend about the length of the pleonite 3. Antennae does not extend past pleonites and is ∼0.70 of body length. Body is finely granular. The holotype has been deposited under UFID 49344, with sequences obtained from this individual available under GenBank Accession numbers: MK034550 (COI); MK032503 (16S rDNA); MK032602 (12S rDNA); MK034599 (Cytb); MK034611 (NaK); MK034659 (H3A).
Diagnostic molecular characters:
| COI: 99-G; 303-G; 450-C; 535-T; 564-T; 567-C; 609-G. |
| 16S: 93(93)-T; 143(143)-A; 258(288)-C; 335(378)-T. |
| Cyt-b: 283-G; 340-A; 346-G. |
| 12S: 139(142)-G; 246(249)-A; 262(265)-G; 306(309)-G; 349(352)-A. |
Partially diagnostic molecular characters:
| COI: 120-G; 318-T; 429-T; 525-A; 631-T. |
| 16S: 235(265)-G; 294(324)-C; 440(483)-T. |
| Cyt-b: 181-T; 217-T; 223-T; 275-A; 292-C; 301-C; 304-C. |
| 12S: 148(151)-A; 199(202)-T. |
Distribution: This species appears to be widespread across the islands of the Maui-Nui group as well as the eastern coastlines of O‘ahu.
Etymology: The species name proposed reflects the distribution of this species primarily across the islands of the Maui-Nui group.
| Ligia pele nov. sp. |
| LSID: urn:lsid:zoobank.org:act:9E02F410-21B0-4D86-92D3-1CC4E3FBAF05. |
| BOLD BINs : ADN2023. |
Materials examined: eight individuals from two localities in north Maui were examined (A6, A7). The holotype (UFID 49325), a paratype (UFID 49326) and a lot of five individuals (UFID 49327) from the type locality have been deposited.
Type locality: Baby Beach, Spreckelsville, Maui (F12; Lat.: 20°54′45.09″N; Long.: 156°24′16.01″W).
Type: male specimen that is 14.95 mm long and 5.69 mm wide at its widest point (pereionite 4; body length to width ratio of ∼2.6). Eyes appear smaller than other Ligia from the area (eye length is ∼0.4 greatest width of cephalon) and wide set (inter-eye distance is equal to eye length). Posterolateral processes of the pereionite 7 extend more than length of the pleonite 3. Antennae does not extend past pleonites and is ∼0.8 body length. The holotype is deposited under UFID 49325. Sequences produced from this individual are available under GenBank Accession numbers: MK034562 (COI); MK032482 (16S rDNA).
Diagnostic molecular characters:
| COI: 228-C; 234-A; 369-T; 462-G; 474-C; 564-G. |
| 16S: 127(127)-G; 340(383)-T; 355(398)-G. |
| Cyt-b: N/A |
| 12S: 13(13)-G; 207(210)-G. |
Partially diagnostic molecular characters:
| COI: 43-T; 81-A; 141-A; 318-C; 519-C; 681-G. |
| 16S: 11(11)-G; 102(102)-G; |
| Cyt-b: N/A |
| 12S: 223(226)-G; 305(308)-G; 474(491)-C. |
Distribution: This species has been recorded in two localities in northern Maui: Honomanu Bay (F3) and Baby Beach in Spreckelsville (F12).
Etymology: The name of this species honors the Hawaiian deity Pele.
| Ligia rolliensis nov. sp |
| LSID: urn:lsid:zoobank.org:act:760DB916-8442-42B8-A6E5-09E32773001A |
| BOLD BINs : AAD0843. |
Materials examined: 37 individuals, both males and females, from six localities across O‘ahu were examined (F1, F2, F13–16). A holotype (UFID 49334), paratype (UFID 49336) and a lot of five individuals (UFID 49337) from the type locality as well as an additional five individuals from Kaena Point (North; UFID 49338) have been deposited.
Type locality: Pupukea, O‘ahu (F1; Lat.: 21°38′59.70″N; Long.: 158°03′45.48″W).
Type: This specimen is a male that is 21.50 mm long and 8.18 mm wide at its widest point (pereionite 4) resulting in a body length to body width ratio of ∼2.6. Eyes appear to be of moderate size when compared to other Ligia from the area (eye length is ∼0.4 greatest width of cephalon) with a distance between the eyes that also appears comparable to most other Ligia in the region (inter-eye distance is ∼0.7 of eye length). Posterolateral processes of the pereionite 7 extend about halfway of the length of the pleonite 3. Antennae does not extend past pleonites and is ∼0.7 body length. The neotype is deposited under UFID 49334. GenBank Accession numbers for the neotype specimen are as follows: MK034494 (COI); MK032521 (16S rDNA); MK032611 (12S rDNA); MK034622 (NaK); MK034665 (H3A).
Molecular diagnostic characters:
| COI: 520-T. |
| 16S: N/A. |
| Cyt-b: 302-A. |
| 12S: N/A. |
Partially diagnostic molecular characters:
| COI: 579-T; 672-C. |
| 16S: N/A. |
| Cyt-b: 40-C; 325-C. |
| 12S: 225(228)-G; 351(354)-G. |
Distribution: Widespread across the island of O‘ahu, excluding the eastern tip of the island.
Etymology: This species reflects both a term commonly used for terrestrial isopods in the U.S.A (i.e., rollie pollies) and that of a beloved pet who recently passed away: Rollie.
Discussion
Phylogeographic work on Ligia from the Hawaiian archipelago has uncovered several genetically divergent yet morphologically cryptic lineages in L. hawaiensis, suggesting this coastal species endemic to the region represents a cryptic species complex (Taiti et al., 2003; Santamaria et al., 2013). Results of MSDAs implemented on a molecular dataset comprised of mitochondrial and nuclear markers lend further support to this idea, as 5–57 putative species were identified from L. hawaiensis individuals, further underscoring the need for a taxonomic revision of the Ligia from Hawaii. Ideally, such taxonomic revisions would entail an integrative approach incorporating several lines of evidence (e.g., morphology, mDNA, nDNA; Schlick-Steiner et al., 2010); however, morphological variation appears to be severely limited across genetically divergent L. hawaiensis lineages (Taiti et al., 2003; Santamaria et al., 2013). Similarly, variation in nuclear markers appears to be limited perhaps as a result of the young age of the Ligia lineages in the region (Santamaria et al., 2013). Thus, species descriptions in this study relied primarily on mitochondrial data, a suitable approach for delineating cryptic species and/or organisms with a young evolutionary history (Schlick-Steiner et al., 2010; Jörger & Schrödl, 2013; Grabowski, Wysocka & Mamos, 2017).
In the case of coastal Hawaiian Ligia, results from MSDAs, phylogenetic patterns, distributional data, and molecular diagnostic characters analyses led to the re-description of L. hawaiensis and the description of seven novel species in the area. Ligia hawaiensis is herein re-described to represent the sole coastal Ligia lineage found in the island of Kaua‘i, as both COI and 16S rDNA sequences obtained from the syntype of L. hawaiensis was identified as a member of Clade D (greens in all figures). This finding cements the status of L. kauiensis as a junior synonym of L. hawaiensis, as Clade D also includes Ligia individuals from the type locality of L. kauaiensis (Kalihiwai Bay, Kaua‘i). It also underlines the potential of non-destructive approaches in producing molecular data from historical specimens, as the DNA obtained for the >150 year old syntype was extracted from its fixative using a variant of the protocol proposed by Shokralla, Singer & Hajibabaei (2010).
The seven novel species described mainly appear to be allopatric species that are primarily found in the younger Hawaiian islands. Three novel species are found solely on the island of Hawai‘i: L. dante from the South Kona district in the south-west, L. honu in the Ka‘u and Puna districts along the south and southeastern coast, and L. kamehameha in the coastlines of the Kohala, Hamakua, and Hilo districts. Another two newly described species exhibit geographic ranges limited to the island of Maui: L. pele from the north of the island, and L. eleluensis from its eastern coastline. Lastly, L. mauinuiensis is described from localities in the islands of Maui, Lana‘i, Moloka‘i, and the eastern coastline of O‘ahu, while L. rollie is found solely in O‘ahu. These general patterns suggest that most of the Ligia species in the Hawaiian archipelago may be relatively young species (most species found in islands <2 myo; (Carson & Clague, 1995)) largely confined to a single island where they exhibit disjunct geographical distributions, and may thus be informative on the processes driving diversification for coastal organisms across local scales in Hawai‘i.
Although additional work remains needed to fully delineate the distributional limits for these new species, particularly in islands where more than one species is found (e.g., O‘ahu, Hawai‘i), distributional patterns observed to date suggest local-scale allopatric events (e.g., volcanic or hydrogeographic processes) may have driven the diversification of Ligia populations at local scales. For instance, Ligia species in the island of Hawai‘i exhibit distributional breaks that partially match volcanic rift zones: L. dante appears to be distributed solely within the western coast line of the Mauna Loa rift zone, L. honu within the Kilauea rift zone, and L. kamehameha primarily found in the Mauna Kea and Kohala rift zones (see Fiske, Jackson & Sutton, 1972). Similar phylogeographic patterns have been observed for Halocaridina rubra (Craft et al., 2008) and vermetid snails (Faucci, 2007) in the island of Hawai‘i, suggesting that allopatric processes associated with volcanic rift zones may have played a role in the diversification of other poorly dispersing coastal organisms in the Hawaiian island, even across small distances. Additional sampling and the application of genomic approaches may not only further elucidate the role of these allopatric events but also whether any introgression amongst species is occurring.
Highly divergent genetic lineages representing possible cryptic species have been reported from several Ligia species (Jung et al., 2008; Hurtado, Mateos & Santamaria, 2010; Eberl et al., 2013; Raupach et al., 2014; Santamaria, Mateos & Hurtado, 2014; Santamaria et al., 2017; Greenan, Griffiths & Santamaria, 2018; Hurtado et al., 2018); however, this represents the first attempt at formally describing such lineages as species. Findings of this study underscore the value molecular taxonomic approaches hold for completing taxonomic revisions in this isopod genus. Thus, their application in taxonomic evaluations of other Ligia species known to harbor highly divergent lineages is recommended. Doing so may help clarify the taxonomy of these isopods while updating it to match the levels of genetic diversity that have been reported so far in these species and may aid in the conservation of coastal biodiversity across the globe.
Conclusion
Phylogenetic reconstructions combined with molecular species delimitation analyses using both mitochondrial and nuclear gene fragments led to the re-description of L. hawaiensis and the description of seven novel coastal species in the area. These species are largely allopatric, exhibiting non-overlapping geographic ranges in the Hawaiian Archipelago. As such, they appear to represent another case of local diversification in a Hawaiian marine organism. Further studies of these organisms may thus be informative on the processes responsible for diversification in Hawaiian coastlines. Lastly, the successful application of MSDAs in this study suggest these approaches may be suitable to formally describe cryptic species in this genus in other regions of the world where high levels of genetic divergence have been reported from.