Genome-wide identification, characterization and expression analysis of the HD-Zip gene family in the stem development of the woody plant Prunus mume
- Published
- Accepted
- Received
- Academic Editor
- Ramamoorthy Siva
- Subject Areas
- Agricultural Science, Bioinformatics, Genomics, Plant Science, Forestry
- Keywords
- Prunus mume, Genetic evolution, Expression pattern, Stem development, HD-Zip TFs
- Copyright
- © 2019 Li et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Genome-wide identification, characterization and expression analysis of the HD-Zip gene family in the stem development of the woody plant Prunus mume. PeerJ 7:e7499 https://doi.org/10.7717/peerj.7499
Abstract
The homeodomain-leucine zipper (HD-Zip) gene family, a group of plant-specific transcriptional factors (TFs), participates in regulating growth, development, and environmental responses. However, the characteristics and biological functions of HD-Zip genes in Prunus mume, which blooms in late winter or early spring, have not been reported. In this study, 32 HD-Zip genes, named PmHB1–PmHB32 based on their chromosomal positions, were identified in the genome of P. mume. These genes are distributed among seven chromosomes and are phylogenetically clustered into four major groups. Gene structure and motif composition were mostly conserved in each group. The Ka/Ks ratios showed that purifying selection has played a leading role in the long-term evolution of the genes, which maintained the function of this family. MicroRNA target site prediction indicated that the genes of the HD-Zip III subfamily may be regulated by miR165/166. Expression pattern analysis showed that the 32 genes were differentially expressed across five different tissues (leaf, flower bud, stem, fruit, and root) and at different stages of stem and leaf-bud development, suggesting that 10 of the genes may play important roles in stem development. Protein–protein interaction predictions showed that the subfamily III genes may regulate vascular development and shoot apical meristem (SAM) maintenance. Promoter analysis showed that the HD-Zip III genes might be involved in responses to light, hormones, and abiotic stressors and stem development. Taken together, our results provide an overview of the HD-Zip family in P. mume and lay the foundation for the molecular breeding of woody ornamental plants.
Introduction
During growth and development, plants are controlled by many transcriptional factors. The plant-specific homeodomain-leucine (HD-Zip) proteins transcriptional factors affect various biological processes including development, cell division, and responses to abiotic stresses in plants (Elhiti & Stasolla, 2014). HD-Zip proteins are characterized by a C-terminal homeodomain (HD) domain, which is responsible for sequence-specific DNA-binding, and an adjacent leucine zipper (LZ), which facilitates protein–protein interactions. Based on sequence similarity and additional conserved motifs, HD-Zip proteins can be classified into four subfamilies: HD-Zip I, II, III, and IV (Ariel et al., 2007). Compared with HD-Zip I proteins, HD-Zip II proteins contain conserved CPSCE motifs and N-terminal consensus sequences. Both HD-Zip III and HD-Zip IV proteins have a steroidogenic acute regulatory protein (StAR)-related lipid transfer domain (START), which has an unknown function in plants. Compared with HD-Zip III proteins, HD-Zip IV proteins lack the C-terminal MEKHLA motif, but contain the PAS domain, which may be involved in the circadian clock and signal transduction (Mukherjee & Bürglin, 2006; Hefti et al., 2004). To date, many HD-Zip genes have been identified, and many attempts have been made to elucidate the functions of various HD-Zip genes in several species, such as Arabidopsis thaliana (Henriksson, 2005; Nakamura et al., 2006; Ciarbelli et al., 2008), rice (Oryza sativus) (Agalou et al., 2008), maize (Zea mays) (Javelle et al., 2011), tomato (Solanum lycopersicum) (Zhang et al., 2014a; Zhang et al., 2014b), Brassica rapa (Khan et al., 2018), cotton (Gossypium hirsutum) (He et al., 2018), soybean (Glycine max) (Chen et al., 2014), foxtail millet (Setaria italica) (Chai et al., 2018), sunflower (Helianthus annuus) (Manavella et al., 2010; Dezar et al., 2011), poplar (Populus trichocarpa) (Hu et al., 2012), grape (Vitis vinifera) (Jiang et al., 2014), pear (Pyrus betulifolia) (Wang et al., 2015) and peach (Prunus persica) (Zhang et al., 2014a; Zhang et al., 2014b).
Genes within the same subfamily tend to exhibit functional conservation due to structural similarities. Several members of the HD-Zip I subfamily are involved in responses to abiotic stresses and phytohormones, embryogenesis, and de-etiolation and response to blue light. These genes include AtHB6, AtHB7, and AtHB12 from Arabidopsis (Söderman, Mattsson & Engström, 1996; Söderman et al., 1999; Lee et al., 2001), which are induced by drought and abscisic acid (ABA), and HaHB4 from sunflower, which responds to methyl jasmonate (MeJA), ethylene, and drought stress (Manavella et al., 2010). Furthermore, overexpression of AtHB13 in Arabidopsis and HaHB1 in sunflower can significantly improve the cold resistance of transgenic plants (Cabello, Arce & Chan, 2012); the maize HD-Zip gene Zmhdz10 enhances the drought resistance of transgenic rice and Arabidopsis (Zhao et al., 2014). In contrast, MtHB1 reduces the drought resistance of alfalfa by inhibiting lateral root formation (Tina et al., 2017). Moreover, ATHB52 can respond to light and regulate photomorphogenesis and de-etiolation (Henriksson, 2005), and AtHB1 responds to short-day photoperiods and promotes hypocotyl and root elongation (Capella et al., 2015). In addition to the above functions, HD-Zip I genes are associated with the development of plant organs. For example, Arabidopsis hb21/hb40/hb53 triple mutants produced more lateral buds than wild-type plants under short-day conditions, which showed that AtHB21/AtHB40/AtHB53 negatively regulate bud formation (Gonzalez-Grandio et al., 2017).
HD-Zip II genes have key roles in plant responses to light and auxin signalling. For instance, ATHB4 and HAT3 from Arabidopsis, two auxin-induced genes, are involved in shade-induced growth (Bou-Torrent et al., 2012). The sunflower HD-Zip II gene HaHB10 responded to dark and light conditions and was associated with the induction of flowering (Dezar et al., 2011). In addition, there is evidence showing that many HD-Zip II genes are relevant to the regulation of organ growth and development. The AtHAT3, AtHB2, and AtHB4 genes co-regulate shoot apical meristem (SAM) and cotyledon development in Arabidopsis seedlings (Turchi et al., 2013). The Arabidopsis loss-of-function mutant athb4/hat3 results in severely abaxialized leaves (Bou-Torrent et al., 2012).
HD-Zip class III proteins act as regulators of apical meristem development, embryogenesis, lateral organ initiation, and vascular bundle development. The functions of some HD-Zip III genes have been investigated both in herbaceous and woody plants. There are five (REV, PHB, PHV, CAN/ATHB-15, and ATHB-8) and eight (PtrHB1–8) HD-Zip III genes in Arabidopsis and Populus, respectively (Prigge et al., 2005; Hu et al., 2012). REV, PHB and PHV, along with KANADI, are believed to control the abaxial-adaxial patterning of lateral organs, such as leaves and embryos (Emery et al., 2003a). All five members of the HD-Zip III subfamily are thought to be directly involved in vascular development in Arabidopsis by up- or down-regulation. For example, REV, PHB, and PHV promote, while CAN and ATHB-8 inhibit, the formation of interfascicular cambium at the base of the inflorescence stem (Baima, 2001; Prigge et al., 2005). PopREVOLUTA (PRE, PtrHB2), a REV- homologous gene in poplar, plays a key role in regulating the initiation of cambia and the pattern of secondary vascular tissues (Robischon et al., 2011). In contrast, PtrHB5 (PCN), a CAN-homologous gene, inhibits the development of secondary vascular tissue (Du et al., 2011). A PtrHB4 mutation caused a defect in interfascicular cambium in transgenic poplar (Zhu et al., 2018). Because of changes in vascular patterns, some transgenic plants exhibit various morphological characteristics, such as tortuous stems and leaves, dwarfism, and shortened internodes (Du et al., 2011; Zhu et al., 2013; Zhu et al., 2018).
HD-Zip class IV proteins control epidermal cell differentiation and cuticle and trichome formation. Most HD-Zip IV genes are specifically expressed in epidermal or subepidermal layers: these genes include Arabidopsis GL2 (GLABRA2), ATHB-10, ANL2 (ANTHOCYANINLESS 2), ATML1 (ARABIDOPSIS THALIANA MERISTEM LAYER 1), and PDF2 (PROTODERMALFACTOR 2) (Abe, 2003) and OCL1 (OUTER CELL LAYER 1) and OCL4 (OUTER CELL LAYER 4) from maize (Vernoud et al., 2009; Depègefargeix et al., 2011). The atml1/pdf2 double mutant exhibits a few defects in shoot epidermal cell differentiation in Arabidopsis. GL2 and ATHB-10 suppress hair formation to determine trichome and root-hair patterning in A. thaliana by coordinating with single-repeat R3-MYBs (Ohashi et al., 2010). The latest findings suggest that AaHD8 not only promotes the initiation of glandular trichomes but also facilitates leaf cuticle development in Artemisia annua (Yan et al., 2018).
Prunus mume is an economically important fruit and ornamental plant that has been cultivated in China for thousands of years; it is widely distributed and used in East Asia. P. mume has acquired favourable ornamental characteristics, including colourful corollas, a pleasant fragrance, and various flower shapes. In addition, it has different types of plant architecture including the upright, weeping, and tortuous types (Chen, 1996). It is important to improve plant architectural traits that are associated with the development of tissues and organs. HD-Zip transcriptional factors can not only enhance resistance to abiotic stress but also improve morphological features to control the growth and development of multiple tissues and organs, such as the apical meristem, vascular tissue, epidermal tissue, and lateral organs. Compared to abundant annotations of Arabidopsis HD-Zip genes, no P. mume HD-Zip has been analysed in detail. In this study, we identified 32 HD-Zips genes from P. mume and performed detailed structural, evolutionary, miRNA165/166 target gene, and expression pattern analyses in different organs and at different stem development stages. Our study provides an overview of the HD-Zip family in P. mume and information to aid understanding of HD-Zip functions in woody plants.
Materials & Methods
HD-Zip gene identification in P. mume and other species
Candidate genes of the HD-Zip family were extracted from the A. thaliana, P. mume (Zhang et al., 2012), Prunus avium (Shirasawa et al., 2017), and Prunus persica (International Peach Genome et al., 2013) genome databases using the software HMMER (Johnson, Eddy & Portugaly, 2010) based on a Hidden Markov Model (HMM) of the HD-domain profile (PF00046). The genome database of P. mume was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome/13911) (Zhang et al., 2012). The genomes of P. avium and P. persica were downloaded from the GDR database (https://www.rosaceae.org/), and the genome of A. thaliana was downloaded from NCBI databases (https://www.ncbi.nlm.nih.gov/). All obtained protein sequences were further analysed by SMART (http://smart.embl-heidelberg.de/) and CD-search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) softwares to confirm the presence of the leucine-zipper (LZ) domains. Sequences lacking the LZ domains were discarded.
Gene classification and phylogenetic analysis
Multiple sequence alignments of the full-length HD-Zip protein sequences from P. mume, A. thaliana, P. avium, and P. persica were performed by the NAFFT 7 program (Katoh, Rozewicki & Yamada, 2017). The maximum-likelihood (ML) phylogenetic tree was constructed using MEGA 6.0 under the Jones–Taylor–Thornton (JTT) amino acid substitution model with 1,000 bootstrap replicates. The full-length sequences of all HD-Zip proteins used in this study is provided in Data S1.
Gene characteristics, structure, and conserved motif analyses
The genomic sequences, ID numbers, and chromosomal locations of the P. mume HD-Zip genes were obtained from the P. mume genome. The physical parameters of putative proteins, including the lengths of the amino-acid sequences (aa), molecular weights (MWs), theoretical isoelectric points (pIs), and grand averages of hydropathicity (GRAVYs), were calculated using the online ExPASy tool (http://www.expasy.org/tools/protparam.html). The gene structure display server (GSDS) program (http://gsds.cbi.pku.edu.cn/) was applied to illustrate the exon–intron structures by alignment of the CDS with the DNA sequences of individual HD-Zip genes. The orthologues of P. mume HD-Zip in Arabidopsis mapping and GO annotation were conducted using the software Blast2GO (Conesa et al., 2005). The detailed information of all orthologues is provided in Data S2.
The online MEME program was used to display the motif structures of HD-Zip proteins (http://meme-suite.org/tools/meme) using the default parameters, except that the optimum motif widths were set from 6 to 200 residues. SMART (http://smart.embl-heidelberg.de) and CD-search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) web servers were used for structural motif annotation.
Chromosomal location and gene duplication analyses
The chromosomal location information of HD-Zip genes was obtained from the P. mume genome database, and the chromosomal location map was produced by TBtools (Chen et al., 2018). Tandem duplicated genes were identified, and paralogues were genes separated by five or fewer genes in a 100-kb region, as proposed by Tu et al. (2010). Identification of segmentally duplicated genes was conducted using the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/index/downloads). Searches for transposons and retrotransposons in DNA sequences 10-kb upstream and downstream of each HD-Zip gene were conducted with RepeatMasker software (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker/).
Estimation of Ka/Ks ratios
The non-synonymous substitution (Ka) and synonymous substitution (Ks) values of duplicated pairs were calculated on PAL2NAL (http://www.bork.embl.de/pal2nal) (Suyama, Torrents & Bork, 2006; Yang, 2007) after the alignment of protein sequences. The duplication time of duplicated genes was calculated by the following formula: T = Ks∕2λ × 106 Mya (λ = 1.5 × 10−8 for dicots) (Blanc & Wolfe, 2004).
Cis-elements in promoters and miR165/166 target site prediction
The online database PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot, 2002) was employed to investigate putative cis-regulatory elements in the sequences approximately 1,500-bp upstream of the translational start site (ATG).
Full-length HD-Zip nucleic acid sequences (including introns and UTRs) were obtained, and psRNATarget software (http://plantgrn.noble.org/psRNATarget/?function) was used to predict miR165 and miR166 target sites (Wang et al., 2014b).
Expression profile analysis based on transcriptome data
To gain insight into the tissue-specific expression patterns of PmHB genes, our previous RNA-Seq data (GEO No. GSE40162) were used to generate a heat map based on the reads per kilobase per million (RPKM) values of individual gene in all tissue samples. HD-Zip gene expression levels were analysed in five organs of P. mume: roots, stems, leaves, flower buds, and fruits (Zhang et al., 2012). HD-Zip gene expression quantities in flower buds at the endodormancy stage (ED) and natural flush stage (NF) were obtained by RNA sequencing during bud dormancy (Zhang et al., 2018). The heat maps were illustrated using the HemI 1.0 software with the default settings (http://hemi.biocucukoo.org/faq.php).
Preparation of plant materials
P. mume cv. ‘Fei Lve’ was cultivated in the greenhouse of Beijing Forestry University. To investigate the expression of PmHBs, leaf buds and young stems at different developmental stages were sampled from one-year-old branches (Fig. S1). All samples were collected and frozen immediately in liquid nitrogen and then stored at −80 °C until needed for RNA extraction.
RNA extraction and qRT-PCR
Total RNA was extracted from leaf buds and stem tissues using the RNA extraction kit (Tiangen, Beijing, China). After testing for concentration and quality, 500 ng total RNA was used to synthesize first-strand cDNA using the PrimeScript RT Reagent kit (TaKaRa, Dalian, China). The SYBR Premix Ex Taq II kit (TaKaRa, Dalian, China) was used for qRT-PCR according to the manufacturer’s instructions. PP2A and actin were selected as reference genes based on previous reports (Wang et al., 2014a; Xu et al., 2015). Three biological replicates were carried out, and expression levels were calculated by the 2−ΔΔCt method (Schmittgen & Livak, 2008). Gene-specific primers used in the qRT-PCR analyses were designed by IDT (https://sg.idtdna.com/scitools/Applications/RealTimePCR/) and are listed in Table S1.
| Gene name | Gene ID | Locus | Start | End | pI | MW | Strand | Subcellular localization | Gene length (bp) | ORF (bp) | Protein length (aa) | Exons | Introns | Homolog in Arabidopsis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PmHB1 | Pm001037 | Pm1 | 6524197 | 6530678 | 6.02 | 92.54 | – | Nucleus | 6482 | 2527 | 842 | 18 | 17 | REV |
| PmHB2 | Pm002097 | Pm1 | 16624414 | 16625975 | 5.42 | 38.08 | – | Nucleus | 1562 | 1012 | 337 | 3 | 2 | ATHB-6 |
| PmHB3 | Pm002327 | Pm1 | 18696993 | 18698133 | 8.62 | 31.60 | – | Nucleus | 1141 | 856 | 285 | 3 | 2 | HAT22 |
| PmHB4 | Pm004476 | Pm2 | 5264456 | 5265317 | 5.46 | 28.82 | – | Nucleus | 862 | 763 | 254 | 2 | 1 | ATHB-6 |
| PmHB5 | Pm005163 | Pm2 | 9571242 | 9576314 | 5.83 | 92.89 | – | Nucleus | 5073 | 2521 | 840 | 18 | 17 | ATHB-8 |
| PmHB6 | Pm005455 | Pm2 | 11136937 | 11138132 | 6.24 | 35.78 | – | Nucleus | 1196 | 949 | 316 | 3 | 2 | HAT22 |
| PmHB7 | Pm006530 | Pm2 | 17359670 | 17361143 | 5.72 | 32.89 | – | Nucleus | 1474 | 871 | 290 | 3 | 2 | ATHB-13 |
| PmHB8 | Pm006578 | Pm2 | 17692588 | 17694392 | 4.69 | 36.36 | – | Nucleus | 1805 | 964 | 321 | 3 | 2 | HAT5 |
| PmHB9 | Pm008838 | Pm2 | 36540330 | 36541338 | 9.31 | 27.57 | – | Nucleus | 1009 | 724 | 241 | 3 | 2 | HAT22 |
| PmHB10 | Pm009758 | Pm3 | 782406 | 785901 | 5.96 | 82.14 | – | Nucleus | 3496 | 2251 | 750 | 10 | 9 | ATML1 |
| PmHB11 | Pm010515 | Pm3 | 4909848 | 4916241 | 5.76 | 86.72 | – | Nucleus | 6394 | 2374 | 791 | 18 | 17 | ATHB-14 |
| PmHB12 | Pm011328 | Pm3 | 10393933 | 10394448 | 7.1 | 19.97 | – | Nucleus | 516 | 514 | 171 | 1 | 0 | ATHB-52 |
| PmHB13 | Pm013001 | Pm4 | 1041360 | 1045089 | 6.3 | 72.62 | – | Nucleus | 3730 | 1957 | 652 | 10 | 9 | HDG11 |
| PmHB14 | Pm013438 | Pm4 | 4125685 | 4131654 | 6.06 | 92.11 | – | Nucleus | 5970 | 2512 | 837 | 18 | 17 | ATHB-15 |
| PmHB15 | Pm013549 | Pm4 | 4819146 | 4823243 | 5.99 | 84.58 | – | Nucleus | 4098 | 2272 | 757 | 11 | 10 | GLABRA 2 |
| PmHB16 | Pm015192 | Pm4 | 17990442 | 17995263 | 5.78 | 78.05 | – | Nucleus | 4822 | 2137 | 712 | 12 | 11 | HDG2 |
| PmHB17 | Pm015729 | Pm4 | 21131194 | 21133120 | 4.95 | 30.69 | – | Nucleus | 1927 | 820 | 273 | 3 | 2 | HAT5 |
| PmHB18 | Pm015861 | Pm4 | 21777276 | 21778882 | 6.57 | 35.05 | – | Nucleus | 1607 | 925 | 308 | 3 | 2 | HAT7 |
| PmHB19 | Pm017678 | Pm5 | 13200909 | 13202636 | 6.96 | 33.31 | – | Nucleus | 1728 | 898 | 299 | 4 | 3 | HAT3 |
| PmHB20 | Pm017987 | Pm5 | 15486990 | 15492069 | 6 | 90.37 | – | Nucleus | 5080 | 2488 | 829 | 9 | 8 | ANL2 |
| PmHB21 | Pm018314 | Pm5 | 17547953 | 17548796 | 4.93 | 27.76 | – | Nucleus | 844 | 724 | 241 | 2 | 1 | ATHB-7 |
| PmHB22 | Pm019365 | Pm5 | 23522721 | 23527271 | 6.03 | 88.19 | – | Nucleus | 4551 | 2425 | 808 | 9 | 8 | ANL2 |
| PmHB23 | Pm019693 | Pm5 | 25211414 | 25212172 | 7.82 | 25.37 | – | Nucleus | 759 | 658 | 219 | 2 | 1 | ATHB-12 |
| PmHB24 | Pm023161 | Pm7 | 2755202 | 2759847 | 5.42 | 92.33 | – | Nucleus | 4646 | 2494 | 831 | 11 | 10 | HDG5 |
| PmHB25 | Pm023405 | Pm7 | 4736073 | 4737543 | 7.62 | 34.88 | – | Nucleus | 1471 | 934 | 311 | 4 | 3 | HAT4 |
| PmHB26 | Pm024816 | Pm7 | 14444420 | 14446077 | 4.64 | 36.77 | – | Nucleus | 1658 | 988 | 329 | 3 | 2 | HAT5 |
| PmHB27 | Pm025254 | Pm7 | 16763051 | 16766231 | 6.3 | 78.27 | – | Nucleus | 3181 | 2128 | 709 | 10 | 9 | HDG11 |
| PmHB28 | Pm025933 | Pm8 | 6532622 | 6534440 | 8.86 | 38.76 | – | Nucleus | 1819 | 1054 | 351 | 4 | 3 | HOX11 |
| PmHB29 | Pm026270 | Pm8 | 8869226 | 8870539 | 8.31 | 26.16 | – | Nucleus | 1314 | 676 | 225 | 3 | 2 | ATHB-51 |
| PmHB30 | Pm026578 | Pm8 | 10635982 | 10637531 | 6.47 | 24.76 | – | Nucleus | 1550 | 643 | 214 | 3 | 2 | ATHB-40 |
| PmHB31 | Pm029201 | scaffold205 | 1964174 | 1967025 | 9.13 | 25.62 | – | Nucleus | 2852 | 685 | 228 | 3 | 2 | HOX3 |
| PmHB32 | Pm030054 | scaffold427 | 2076 | 3624 | 6.32 | 40.29 | – | Nucleus | 1549 | 1102 | 367 | 4 | 3 | HOX11 |
Results
Identification, characteristics, and phylogenetic analysis in P. mume and other species
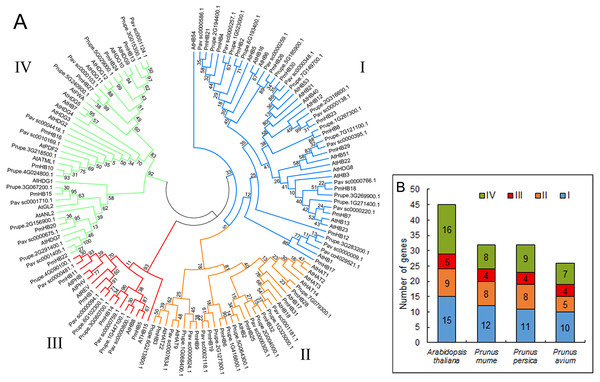
In total, 32, 32, 26, and 45 putative proteins containing the HD and LZ domains were identified in P. mume, P. persica, P. avium, and A. thaliana, respectively. The putative proteins of P. mume were named PmHB1–PmHB32 based on their chromosomal positions in the order of successive chromosomes (Table 1). Among the putative PmHBs, physicochemical characteristics were variable. Their encoded proteins had diverse lengths, from 171 aa to 842 aa, with an average length of 463 aa. The molecular weights ranged between 11.98 kDa and 92.32 kDa, and the isoelectric points ranged from 4.64 to 9.31. The number of HD-Zip genes in P. mume was 0.7-fold less than that in A. thaliana (45), whereas it was similar to that in P. persica (32) and less than that in P. avium (26). To investigate the classification of HD-Zip proteins in P. mume and their evolutionary relationships in P. mume, P. persica, P. avium, and A. thaliana, full-length HD-Zip proteins were used to construct a maximum-likelihood phylogenetic tree using MEGA software (Fig. 1A). The HD-Zip proteins of the four species were divided into four subfamilies (HD-Zip I to IV), and the proportions of each subfamily were similar in all four species (Fig. 1B). The HD-Zip I gene family was the largest subfamily in P. mume (37.5%), P. persica (34.4%), and P. avium (38.5%), while HD-Zip IV genes composed the largest proportion in A. thaliana (35.6%). HD-Zip III genes had the fewest representatives in all four species: there were 4 (12.5%), 4 (12.5%), 4 (15.4%), and 5 (11.1%) members in P. mume, P. persica, P. avium, and A. thaliana, respectively.
Chromosomal location and gene duplication of PmHBs
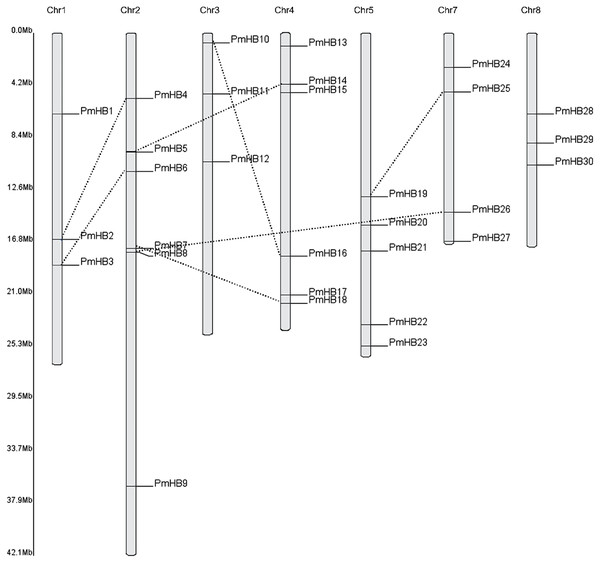
Thirty of the P. mume HD-Zip genes were distributed among all the chromosomes, except for Chr6 (Fig. 2), and only two genes (PmHB31 and PmHB32) remained on unmapped scaffolds. Chr2 and Chr4 contained the most HD-Zip genes, accounting for 18.75% (6) of the entire HD-Zip family; there were five genes on Chr5 and four on Chr7. In addition, Chr1, Chr3, and Chr8 each contained one gene (9.3%).
Figure 1: Phylogenetic tree analysis of HD-Zip sequences from P. mume and other plant species.
(A) Phylogenetic tree of HD-Zip proteins in P. mume; the Group I, Group II, Group III, and Group IV subfamilies are indicated by blue, orange, red, and green branch lines, respectively. At, A. thaliana; Pm, P. mume; Pav, P. avium; Prupe, P. persica; The phylogenetic tree was produced with MEGA 6.0 by the maximum likelihood (ML) method under the Jones-Taylor-Thornton (JTT) amino acid substitution model with 1,000 bootstrap replications. (B) The gene numbers of subfamily in A. thaliana, P. mume, P. avium, and P. persica.Because genome duplication is absent in P. mume, segmental duplication, tandem duplication, and transposons are the three mechanisms that resulted in gene expansion in P. mume (Clark & Donoghue, 2018; Del Pozo & Ramirez-Parra, 2015; Qiao et al., 2019). To identify potential segmental duplications in the HD-Zip family, pairs of P. mume and P. persica genes were obtained from the Plant Genome Duplication Database. The distribution of PmHBs relative to their corresponding duplicate genes is illustrated in Fig. 2 and Table 2. Among the 30 PmHBs mapped, seven PmHB gene pairs (43.8%) were found in segmental repeats (PmHB2–PmHB4, PmHB8–PmHB26, PmHB7–PmHB18, PmHB3–PmHB6, PmHB5–PmHB14, PmHB10–PmHB16, and PmHB19–PmHB25), representing all the HD-Zip subfamilies. No genes had been tandemly duplicated, suggesting that tandem duplications were not involved in the expansion of the HD-Zip gene family in P. mume.
Figure 2: Chromosome localization of HD-Zip genes in P. mume.
Thirty PmHB genes were located in seven chromosomes. The chromosome 6 has no PmHB genes. The dotted line indicates the duplicated gene pair.| LOCUS 1 | LOCUS 2 | Ka | Ks | Ka/Ks | Select pressure | MYA |
|---|---|---|---|---|---|---|
| PmHB2 | PmHB4 | 0.3466 | 4.2421 | 0.081705 | Purifying selection | 141.40 |
| PmHB8 | PmHB26 | 0.3217 | 1.1706 | 0.274816 | purifying selection | 39.02 |
| PmHB7 | PmHB18 | 0.2416 | 1.2732 | 0.189758 | purifying selection | 42.44 |
| PmHB3 | PmHB6 | 0.3759 | 2.5532 | 0.147227 | purifying selection | 85.11 |
| PmHB5 | PmHB14 | 0.0941 | 1.1559 | 0.081408 | purifying selection | 38.53 |
| PmHB10 | PmHB16 | 0.1989 | 1.7398 | 0.114323 | purifying selection | 57.99 |
| PmHB19 | PmHB25 | 0.2835 | 1.2658 | 0.223969 | purifying selection | 42.19 |
| PmHB2 | ppa008318m | 0.0089 | 0.0528 | 0.168561 | purifying selection | 1.76 |
| PmHB3 | ppa008984m | 0.3583 | 2.0541 | 0.174432 | purifying selection | 68.47 |
| PmHB4 | ppa009419m | 0.0083 | 0.0192 | 0.432292 | purifying selection | 0.64 |
| PmHB6 | ppa009614m | 0.3838 | 2.4082 | 0.159372 | purifying selection | 80.27 |
| PmHB7 | ppa020465m | 0.2535 | 1.2274 | 0.206534 | purifying selection | 40.91 |
| PmHB8 | ppa018002m | 0.008 | 0.0405 | 0.197531 | purifying selection | 1.35 |
| PmHB9 | ppa016510m | 0.0895 | 0.067 | 1.335821 | positive selection | 2.23 |
| PmHB12 | ppa017711m | 0.0126 | 0.036 | 0.35 | purifying selection | 1.2 |
| PmHB17 | ppa009747m | 0.1 | 0.1506 | 0.664011 | purifying selection | 5.02 |
| PmHB18 | ppa009498m | 0.2416 | 1.37 | 0.17635 | purifying selection | 45.67 |
| PmHB19 | ppa024161m | 0.0073 | 0.0497 | 0.146881 | purifying selection | 1.66 |
| PmHB21 | ppa010647m | 0.007 | 0.0346 | 0.202312 | purifying selection | 1.15 |
| PmHB23 | ppa011221m | 0.0236 | 0.0513 | 0.460039 | purifying selection | 1.71 |
| PmHB25 | ppa008774m | 0.0099 | 0.0319 | 0.310345 | purifying selection | 1.06 |
| PmHB26 | ppa008495m | 0.0039 | 0.0321 | 0.121495 | purifying selection | 1.07 |
| PmHB28 | ppa008075m | 0.0038 | 0.0548 | 0.069343 | purifying selection | 1.83 |
| PmHB29 | ppa015952m | 0.014 | 0.0503 | 0.27833 | purifying selection | 1.68 |
| PmHB30 | ppa011343m | 0.01 | 0.0366 | 0.273224 | purifying selection | 1.22 |
| PmHB31 | ppa011006m | 0.0873 | 0.1827 | 0.477833 | purifying selection | 6.09 |
Interspersed duplication, mainly attributable to transposons, was another mechanism that contributed to the expansion of the HD-Zip gene family. A total of 22 transposons or retrotransposons are found upstream and downstream of 15 PmHB genes. Details about the transposons and retrotransposons are shown in Table S2, implying that transposons may have played an important role in the expansion of HD-Zip genes in P. mume.
The ratio of nonsynonymous to synonymous mutation rates (Ka/Ks) was used to estimate the selection pressure that promoted the evolution of the gene family. The Ka, Ks, and Ka/Ks of 19 orthologous gene pairs between P. mume and P. persica and seven paralogous gene pairs in P. mume were calculated (Table 2). All the Ka/Ks of the paralogous gene pairs were <1, which indicates that the HD-Zip genes of P. mume have mainly experienced purifying selection pressure with limited functional divergence. In addition, most of the Ka/Ks ratios of the orthologous genes were <1, while the Ka/Ks value of PmHB9/ppa016510 m was >1, suggesting that HD-Zip genes evolved primarily under purifying selection that occurred after the divergence of P. mume and P. persica; adaptive evolution may have occurred in PmHB9.
Conserved domains and gene structure analysis of PmHBs
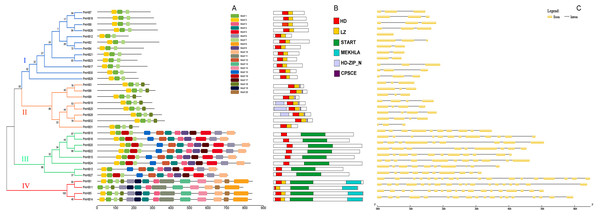
The conserved motifs of the HD-Zip protein sequences were analysed by MEME (Fig. 3A). Motif 1 and motif 2 were found to encode the homeobox domain. Motif 1 was found in all 32 genes. In addition, motif 7 was found to encode the LZ domain. Motifs 3, 4, 9, and 12 encode START domains and are present in subfamilies III and IV. Motif 8 was only found in subfamily III and encodes the MEKHLA domain. In addition, it was suggested that some unannotated subfamily-specific motifs might contribute to the diversification of the subfamily. A schematic representation of the HD-Zip family based on the HD domain, LZ domain, START domain and MEKHLA domain regions is shown in Fig. 3B.
Figure 3: Conserved domains and gene structure analyses of HD-Zip genes in P. mume.
(A) Conserved motif analysis of HD-Zip genes in P. mume; (B) conserved domains analysis of HD-Zip genes in P. mume; (C). intron/exon construction of HD-Zip genes in P. mume.The number of introns is similar in the different subfamilies (Fig. 3C). Most of the HD-Zip I genes have two introns, except PmHB12. In the HD-Zip II subfamily, PmHB3, 6, 9, and 31 have two introns, while PmHB19, 25, 28, and 32 have three introns. HD-Zip IV subfamily genes have 8–11 introns, whereas all the HD-Zip III genes have 17 introns, which was the maximum number among the HD-Zip family in P. mume. The results above divided the HD-Zip genes into four subfamilies, which were consistent with the result of the phylogenetic analysis.
MicroRNA165/166 target-site prediction and cis-acting elements of the PmHBs promoters
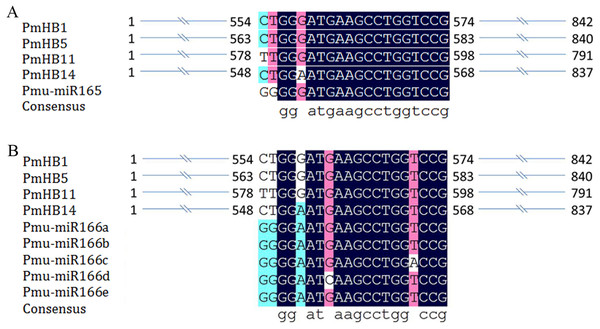
Previous studies revealed that the activity of HD-Zip III proteins was regulated by miR165/166 (Kidner & Martienssen, 2004; Mallory et al., 2014). Here, miR165 and miR166a–e target sites were found in four P. mume HD-Zip III subfamily genes (PmHB1, PmHB5, PmHB11, and PmHB14) (Fig. 4), suggesting that P. mume HD-Zip III subfamily genes may be regulated by miR165 and miR166.
Figure 4: MiRNA target site in HD-ZIP III genes in P. mume.
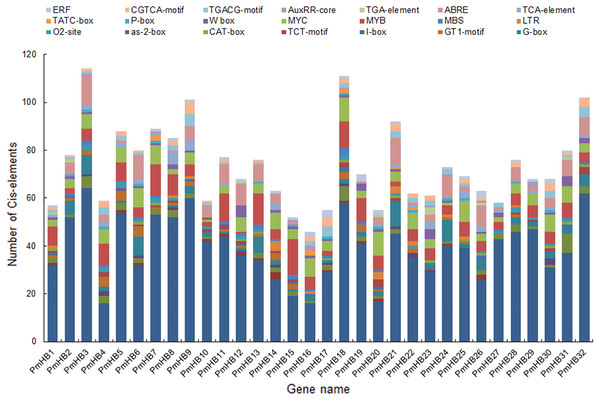
(A) miR165 target site in HD-ZIP III subfamily member; (B) miR166 target site in HD-ZIP III subfamily member and the secondary structures of miR166 a-e.The cis-acting elements of promoters are essential for determining tissue-specific expression and are involved in the regulation of gene expression. A region of 1500 bp upstream of the start codon (ATG) of HD-Zip genes was used to analyse cis-acting regulatory elements. Many of the cis-acting elements identified in the PmHB gene promoters were related to the light response, including the AE-box, Box 4, the I-box, the GATA-motif, the G-box, the GT1-motif, and the TCT-motif (Fig. 5), suggesting that PmHBs may respond to light and participate in plant light morphogenesis. Because many cis-elements associated with hormone responses (the P-box, TATC-box, TCA-element, ABRE, TGA-element, AuxRR-core, TGACG-motif, CGTCA-motif, and ERF) were distributed in the promoter regions, the expression of PmHB genes may be induced by several plant hormones, such as salicylic acid, MeJA, abscisic acid, gibberellins, and auxin. In addition, the presence of shoot- and meristem-specific expression elements (as-2-box, CAT-box, and O2-site) in the promoters of several PmHB genes indicated that those genes might participate in regulating the development of the meristem and stem. HD-Zips are deemed to be crucial TFs involved in meristem and stem development.
Expression analysis of PmHBs in different tissues of P. mume
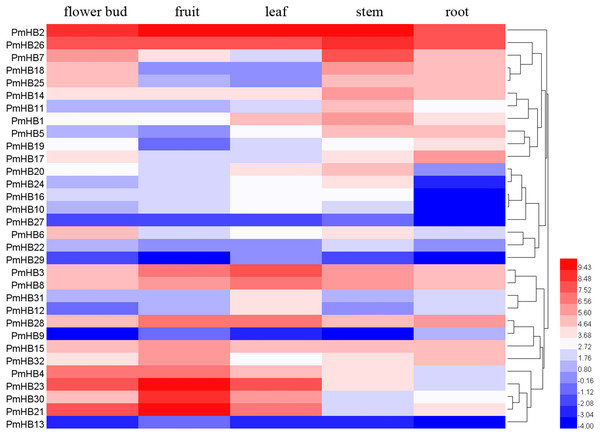
To explore the expression patterns of PmHBs in different organs and their biological functions during plant growth and development, the expression levels of PmHBs in five tissues (leaf, flower bud, fruit, root, and stem) were analysed by RNA-seq. Heatmaps showed that HD-Zip gene expression levels are diverse in the five tissues (Fig. 6). PmHB29 transcripts were not present in fruits and roots, and PmHB9 was not present in buds and stems; however, other HD-Zip genes could be detected in all five tissues. These results indicated that some HD-Zip genes have multiple functions in the development of different organs. PmHB2 and PmHB26 were highly expressed in all tissues, whereas PmHB9, 13, and 27 were expressed at low levels in all tissues. Twelve genes (PmHB1, 5, 7, 11, 14, 16, 18, 20, 22, 25, 26, and 27) were most highly expressed in the stem, which showed that these genes might have functions in regulating stem development. It is notable that PmHB10, 16, 20, and 24 were expressed in all aboveground organs but were expressed either at low levels or not at all in roots. In addition, some genes of the same subfamilies exhibited similar expression patterns. For instance, all HD-Zip III subfamily genes (PmHB1, 5, 11, and 14) showed high RPKM values in the stem, and five HD-Zip IV subfamily genes (PmHB16, 17, 20, 22, and 24) showed high RPKM values in both leaf and stem tissues.
Figure 5: Types and number of cis-elements analysis involved in the stress response, hormone effects, and stem development.
The x-axis represents 1.5 kb upstream promoter region of PmHB genes. The y-axis represents number of cis-promoters.Figure 6: Hierachical clustering of expression profiles of PmHBs in leaf, flower bud, fruit, stem, and root.
Color scale represents log2 rate expression values.HD-Zip genes may also regulate flower bud dormancy. Screening for differentially expressed genes (DEGs) by RNA sequencing in flower buds at the endodormancy (ED) and natural flush (NF) stages was carried out in our previous study (Zhang et al., 2018). Eleven genes (PmHB6, 7, 8, 11, 19, 26, 27, 29, 30, and 31) were more highly expressed at the ED stage, and four genes (PmHB3, 17, 18, and 23) were upregulated at the NF stage (Table S3).
Expression pattern analysis of PmHBs in stem development
To verify the functions of PmHBs in the development of leaf buds and stems, RNA-seq on leaf buds and stem tips was conducted in P. mume. HD-Zip family genes that were differentially expressed the between leaf bud and stem tip are shown in Table S4. Seven genes (PmHB10, 16, 20, 22, 24, 27, and 29) were upregulated in leaf buds, while another six genes (PmHB3, 13, 17, 18, 21, and 30) were upregulated in stem tips. The results above suggest that HD-Zip genes play putative roles in the development of leaf buds and stems.
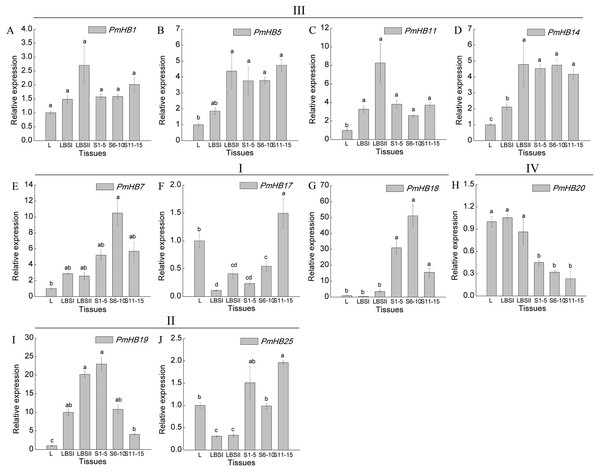
Expression levels of 10 selected genes in different organs (leaf, leaf bud, and differentiating stem) were determined by qRT-PCR (Fig. 7). The results showed that genes in the HD-Zip III subfamily had similar expression patterns, which were highly expressed in both LBSII and stems, and these patterns were not significantly different at different stages of stem development; PmHB7 and PmHB18, two HD-Zip I genes, were predominantly expressed in S6–10 in stem, while another HD-Zip I gene, PmHB17, exhibited high levels of expression in leaf and in S11–15. PmHB20 of the HD-Zip IV subfamily had higher expression levels in leaves, leaf buds, and large leaf buds and lower expression levels in different stages of stem development. PmHB19 is a HD-Zip II gene with high expression levels in both large leaf buds and stem tips (S1–5). Another HD-Zip II gene, PmHB25, was expressed maximally in S11–15, while it exhibited its lowest expression levels in LBSI and LBSII (Fig. 7).
Figure 7: Expression pattern analysis of HD-Zip genes in P. mume at different development stages.
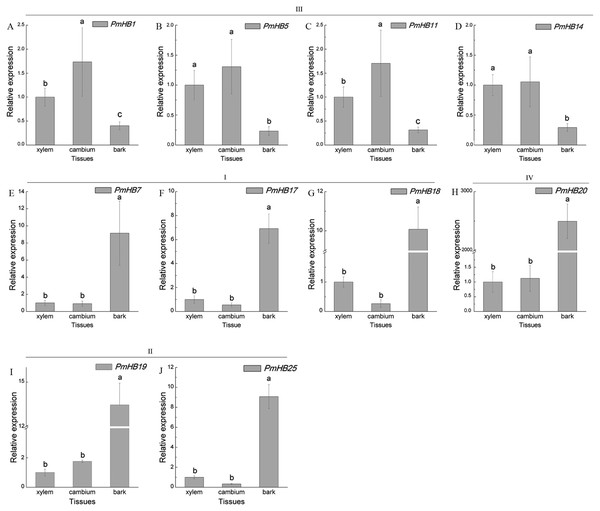
Ten HD-Zip genes were selected to analysis the expression patterns in leaf, leaf buds, and stems at different development stages (A-J). L, leaf; LBSI (leaf buds in stage I), leaf bud in sprouted phase; LBSII (Leaf bud in stage II), leaf buds before leaf unfolding; S1-5, stems from 1 to 5 internodes; S6-10, stems from 6 to 10 internodes; S11-15, stems from 11 to 15 internodes. PmPP2A and Pmactin were used as reference genes. Vertical lines represent S.E. (n = 3). Different lower case letters indicate statistically significant differences at the P < 0.01 level.Expression in different tissues of the stem (xylem, cambium, and bark) were determined by qRT-PCR (Fig. 8). HD-Zip III subfamily genes (PmHB1, 5, 11, and 14) may participate in xylem differentiation because they are expressed predominantly in the xylem. However, PmHB7, 17, 18, 19, 20, and 25 were mainly expressed in bark, suggesting that they might be involved in the development of phloem, endodermis, or epidermis (Fig. 8).
Figure 8: Expression pattern analysis of HD-Zip genes in P. mume in different tissues of young stems.
Ten HD-Zip genes were selected to analysis the expression patterns in different tissues of young stems (A-J). PmPP2A and Pmactin were used as reference genes. Vertical lines represent S.E. (n = 3). Different lower case letters indicate statistically significant differences at the P < 0.01 level.Interaction network between ten P. mume and A. thaliana HD-Zip proteins
The GO annotation of HD-Zip genes in P. mume were conducted by Blast2GO software basing on the orthologues in Arabidopsis (Fig. S2). PmHBs were divided into three GO categories: biological processes, cell components and molecular functions. In the biological process category, biological regulation was the most highly represented groups. The cell, cell part, and organelle classes were abundant in cellular component category. Within the molecular function category, genes that corresponded to binding were the most abundant.
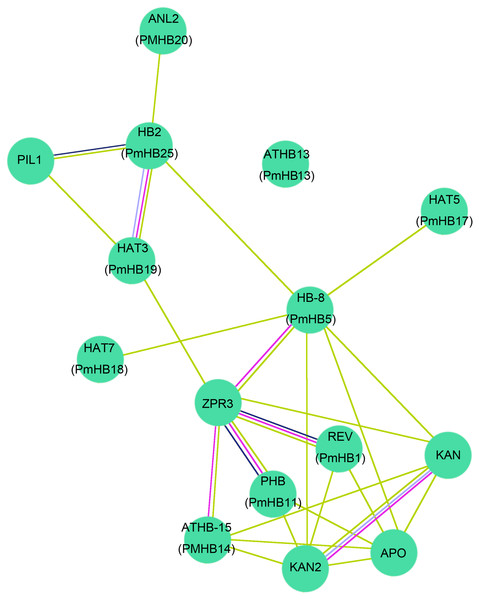
To further predict P. mume HD-Zip proteins functions during plant stem development, the protein–protein interactions between ten HD-Zip proteins of P. mume and A. thaliana were analysed using STRING software (http://string-db.org) (Fig. 9). The homologous genes were matched with the highest bit score, the results were corresponding to those of Blast2GO and the lines of different colours represent different types of evidence for the proteins interaction. Ten HD-Zip proteins were found to interact in a network, suggesting that they might have complex connections in the co-regulation of stem development. Previous studies showed that HD-Zip III activity can promote the expression of ZPR proteins in A. thaliana (Husbands et al., 2016; Wenkel et al., 2007). Specifically, REV and PHB can interact with ZPR3 to participate in the regulation of leaf polarity and vascular development (Kim et al., 2008; Wenkel et al., 2007). Moreover, interactions between the HD-Zip III and KANDAI proteins to regulate vascular development have been reported in A. thaliana (Emery et al., 2003a; Emery et al., 2003b). The HD-Zip III subfamily genes PmHB1, 5, 11, and 14 had highly similar amino sequences to those of REV, HB-8, PHB, and ATHB-15, respectively, and might regulate leaf and vascular development. In addition, HB-2 was induced by the PIFs and participated in the shade avoidance response (Kunihiro et al., 2011). PmHB25, which is closely related to HB-2, may play an important role in regulating light-mediated stem elongation.
Figure 9: Interaction networks of ten HD-Zip proteins in P. mume according to the orthologs in Arabidopsis.
The nodes with different colors represent different proteins and the lines with different color represent different types of evidence for the interaction. Light blue lines show known interactions from curated databases, rose red lines show known interactions experimentally determined, blue lines show predicted interactions of gene co-occurrence, black lines show interactions of co-expression, and yellow lines show interactions of text mining.Discussion
HD-Zip, containing a homeodomain (HD) domain and an adjacent leucine zipper (LZ), is a major group of transcriptional factors present in higher plants that has a significant impact on the light reduction, abiotic stress, and organic development processes of plants. Most studies on HD-Zip proteins have focused on herbaceous plants, while the functions of only a few HD-Zip proteins have been verified in woody plants, such as poplar and peach (Hu et al., 2012; Zhang et al., 2014a; Zhang et al., 2014b). However, a great deal is unknown about these genes in P. mume. A total of 32 genes were identified, which is 0.7-fold more than the number found in Arabidopsis (Emery et al., 2003b; Henriksson, 2005; Nakamura et al., 2006; Ciarbelli et al., 2008), 0.5-fold more compared to Populus (Hu et al., 2012), and similar to the number found in P. persica (Zhang et al., 2014a; Zhang et al., 2014b). The number of HD-Zip genes varies widely among these species. Nevertheless, the P. mume genome (280 Mb) is approximately two-fold larger than that of Arabidopsis (125 Mb) (Zhang et al., 2012; Initiative, 2000). To explore the phylogenetic relationship between the HD-Zip proteins of P. mume and Arabidopsis, PmHB orthologues in Arabidopsis with the highest levels of homology were extracted (Table 1), showing that a single HD-Zip gene in Arabidopsis had multiple orthologues in P. mume. For instance, ATHB-6, HDG11, ANL2, and HOX11 had two orthologues each in P. mume, whereas HAT22 and HAT3 had three orthologues each in P. mume. These results suggested that, such as peach, some HD-Zip genes may have been lost in the P. mume lineage during evolution (Zhang et al., 2014a; Zhang et al., 2014b). The number of HD-Zip genes in P. mume was similar to that in peach, which might be because of their closer genetic relationship.
The 32 putative genes were classified into four subfamilies according to the results of phylogenetic analysis, and the length and intron number of the genes in different subfamilies were conserved (Figs. 1 and 3). For example, the lengths of HD-Zip I and II genes were shorter than those of HD-Zip III and IV. HD-Zip III genes have 17 introns. In addition, proteins in the same subfamily showed similar structures. In general, most of the motifs and domains in the same subfamily were similar, which may be consistent with the phylogenetic analysis. However, some motifs were specific to subfamilies. For example, motif 18 was only detected in the HD-Zip II subfamily; motifs 3 and 4 were found in the HD-Zip III and IV subfamilies, and motifs 5, 6, 9, 10, 12, 15, and 17 were only found in the HD-Zip IV subfamily, which might be associated with the functional diversity of the HD-Zip family. These findings clearly support the results of phylogenetic analysis. Tandem duplication, segmental duplication, and transposons have resulted in the genome evolution. It was revealed that segmental duplication and transposons may have contributed to the expansion of the HD-Zip gene family in P. mume, accounting for 92% of duplications (Fig. 2 and Table 2).
Proteins can interact with other proteins to co-regulate biological processes. A network of ten P. mume and Arabidopsis HD-Zip TF pairs was established. In response to shade, plants can react by altering elongation during growth and development. It has been found that HD-Zip genes positively regulate the plant response to shade. In Arabidopsis, the HD-Zip II gene HB-2 can be activated by PIF4 and PIF5, two phytochrome-interacting transcriptional factors that participate in the regulation of hypocotyl elongation under short-day conditions (Kunihiro et al., 2011). As the orthologues of HB-2 and HAT3, PmHB25 and PmHB19 may also play roles in shade avoidance. Moreover, Arabidopsis hat3/athb4/athb2 loss-of-function triple mutants showed developmental defects in white light, such as adaxialized leaves and reduced shoot apical meristem activity (Turchi et al., 2013). Previous research showed that the expression of several HD-Zip II transcriptional factors, including HAT3 and HB-2/HAT4, was regulated by the HD-Zip III gene REVOLUTA (REV) (Bou-Torrent et al., 2012). Mutation of REV, PHABULOSA (PHB), and PHAVOLUTA (PHV) enhanced the bilateral symmetry and SAM defects in hat3/athb4 mutants. These results indicated that HD-Zip II and HD-Zip III genes cooperate in establishing bilateral symmetry and patterning in the embryo, as well as in controlling SAM activity in plants. Six genes in both the HD-Zip II and III subfamilies (PmHB1, 5, 11, 14, 19, and 25) had higher expression levels in leaf buds before leaf unfolding (LBSII) or in stem tips (S1–5), suggesting that PmHB1, 5, 11, 14, 19, and 25 might participate in the formation and maintenance of SAM polarity in P. mume.
HD-Zip III transcriptional factors also play roles in controlling primary and secondary vascular cell differentiation and SAM (shoot apical meristem) maintenance (Baima, 2001; Kim et al., 2005), which is essential for the formation of plant architecture. Overexpression of ATHB-8 promotes vascular cell differentiation and xylem tissue production in inflorescence stems of Arabidopsis (Baima, 2001). HD-Zip III TFs are key factors that regulate the secondary growth of vascular bundles in Populus. PtrHB7, an ATHB-8 homologue, regulates the balance of vascular patterning by inhibiting the differentiation of secondary xylem while promoting the differentiation of secondary phloem (Zhu et al., 2013). REV, which has overlapping functions with PHV and PHB, regulates meristem initiation in lateral organs in opposition to KANADI (Ilegems et al., 2010). Overexpression of PRE, the Populus homologue of the Arabidopsis REV gene, in Populus causes the patterning of secondary vascular tissues to be altered (Robischon et al., 2011). In addition, genes of the four HD-Zip III subfamilies of P. mume were expressed preferentially in xylem and cambium; thus, we speculated that four genes from the HD-Zip III subfamily were closely associated with stem development in P. mume. The expression of HD-Zip III genes was controlled by multiple molecular mechanisms. First, the small ZIP protein ZPR3 reduced HD-Zip III protein activity by direct interaction to form nonfunctional heterodimers in the SAM of Arabidopsis (Kim et al., 2008). Moreover, overexpression of KANADI negatively affected the expression of HD-Zip III genes during embryogenesis, whereas HD-Zip III genes and KANADI co-regulated auxin flow by controlling the distribution of PIN-FORMED1 (PIN1) (Izhaki & Bowman, 2007). In addition, protein-interaction networks showed that REV, HB-8, PHB, and ATHB-15, the orthologues of PmHB1, PmHB5, PmHB11, and PmHB14, respectively, interacted with ZPR3, and ATHB-15 interacted with KANADI (Fig. 9). Finally, MiR165/166 negatively regulated the activity of HD-Zip III genes in both Arabidopsis (Kim et al., 2005; Mallory et al., 2014) and Populus (Ko, Prassinos & Han, 2006). Because the target sites were present in the sequences of HD-Zip III genes, we speculated that HD-Zip III genes might also be regulated by MiR165/166. However, the molecular mechanisms of the HD-Zip III gene regulation of vascular development and interactions between proteins need further investigation.
HD-Zip IV proteins have been reported to regulate the differentiation and maintenance of outer cell layers (Nakamura et al., 2006; Yan et al., 2018). PDF2 and ATML1 regulate shoot epidermal cell differentiation and embryo development in Arabidopsis (Ogawa et al., 2015). In P. mume, the expression level of PmHB20 in bark was more than 2,000 times that in cambium and xylem (Fig. 8), which is similar to Arabidopsis HD-Zip IV genes that are preferentially or specifically expressed in plant epidermal or sub-epidermal cells (Rombolácaldentey et al., 2014).
Conclusions
Thirty-two HD-Zip genes were identified in P. mume. Phylogenetic analysis showed that there were four subfamilies in the HD-Zip family: subfamilies I, II, III, and IV. Gene structure and motif analysis suggested that genes in the same subfamily were conserved. Furthermore, expression profile analysis showed that HD-Zip genes may be involved in stem development and flower bud dormancy. Otherwise, all HD-Zip III genes were highly expressed in cambium and xylem, suggesting that HD-Zip III genes may be indispensable in vascular differentiation. The prediction of microRNA target sites indicated that HD-Zip III subfamily genes may be regulated by miR165/166. Promoter analysis showed that HD-Zip III genes may be involved in responses to light, hormones, and abiotic stressors, as well as plant development. Our study identified the members of the HD-Zip family in P. mume and laid the foundation for molecular breeding programmes for woody ornamental plants.
Supplemental Information
Sequence of forward and reverse primer pairs for real-time quantification PCR
Transposons and retrotransposons in genomic sequences of 10-kb upstream and downstream of each HD-ZIP gene
Differentially expressed genes (DEGs) in flower buds with endodormancy stage (ED) and natural flush stage (NF) in P. mume
Differentially expressed genes (DEGs) in leaf bud and stem tip of P. mume
Samples of leaf buds and young stems in different stages
L, leaf; LBSI (leaf buds in stage I), leaf bud in sprouted phase; LBSII (Leaf bud in stage II), leaf buds before leaf unfolding; S1-5, stems from 1 to 5 internodes; S6-10, stems from 6 to 10 internodes; S11-15, stems from 11 to 15 internodes