Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera
- Published
- Accepted
- Received
- Academic Editor
- Joseph Gillespie
- Subject Areas
- Bioinformatics, Genomics, Molecular Biology, Plant Science
- Keywords
- Lotus, bHLH gene family, Structural and evolutional analyses, Functional prediction, Stress
- Copyright
- © 2019 Mao et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera. PeerJ 7:e7153 https://doi.org/10.7717/peerj.7153
Abstract
Lotus (Nelumbo nucifera Gaertn.) is an economically important aquatic plant with multiple applications, but water salinity and cold stress seriously affect lotus yield and distribution. The basic helix-loop-helix (bHLH) transcription factors (TFs) play a vital role in plant growth and development, metabolic regulation processes and responses to environmental changes. However, systematic analyses of the bHLH TF family in lotus has not yet been reported. Here, we report the identification and description of bHLH genes in lotus (NnbHLHs) with a focus on functional prediction, particularly for those involved in stress resistance. In all, 115 NnbHLHs were identified in the lotus genome and classified into 19 subfamilies. The chromosomal distribution, physicochemical properties, bHLH domain, conserved motif compositions and evolution of these 115 NnbHLHs were further analyzed. To better understand the functions of the lotus bHLH family, gene ontology, cis-element, and phylogenetic analyses were conducted. NnbHLHs were predicted to be involved in plant development, metabolic regulation and responses to stress, in accordance with previous findings. Overall, 15 NnbHLHs were further investigated with functional prediction via quantitative real-time PCR analyses. Meanwhile, expression profiles of NnbHLHs in four tissues indicated that many NnbHLHs showed tissue preference in their expression. This study is supposed to provide a good foundation for further research into the functions and evolution of NnbHLHs, and identifies candidate genes for stress resistance in lotus.
Introduction
The basic helix-loop-helix (bHLH) family is the second largest gene family in plants, after the MYB family (Feller et al., 2011). With the greater availability of genome sequence data, genes in the bHLH family have been identified and characterized in various plant species (Li et al., 2006; Niu et al., 2017; Sun, Fan & Ling, 2015; Toledo-Ortiz, Huq & Quail, 2003). The bHLH gene family is named after its distinctive structure of a bHLH domain, consisting of two conserved regions, a basic region and a helix–loop–helix region (HLH region), of approximately 60 amino acids. The basic region is located at the N-terminus of the bHLH domain and consists of approximately 17 amino acids, six of which are basic amino acid residues. This domain is a DNA-binding region that enables bHLH transcription factors (TFs) to bind to E-box (CANNTG) (Carretero-Paulet et al., 2010; Niu et al., 2017). The HLH region, containing two amphipathic α-helices linked by a loop region with a variable sequences (Murre, Mccaw & Baltimore, 1989), is located at the C-terminus and participates in protein dimerization (Brownlie et al., 1997). The regions outside the bHLH domain are distinctly divergent. In animals, the bHLH genes are classified into six groups (from A to F), which contain 45 subgroups based on their various functions in the regulation of gene expression, target DNA elements, and phylogenetic analyses (Atchley, Terhalle & Dress, 1999). However, research on bHLH proteins in plants has been limited compared to that in animals, and therefore the exact organization of bHLH genes is unclear. Generally, the bHLH gene family in plants has been divided into 15–26 groups (Toledo-Ortiz, Huq & Quail, 2003), and sometimes up to 32 when atypical bHLH proteins are included (Carretero-Paulet et al., 2010). Based on sequence homology and phylogenetic relationships, 147 bHLH genes were identified in Arabidopsis and grouped into 21 subfamilies (Toledo-Ortiz, Huq & Quail, 2003), while 167 bHLH genes were detected in rice and formed 25 subfamilies (Li et al., 2006).
In plants, the bHLH gene family plays important roles in plant growth and development, metabolic regulation, and response to environmental changes. In Arabidopsis, the BR-Enhanced Expression and Phytochrome Interacting Factors (PIFs) had been reported to response to cold (Kim et al., 2010) and light (Paik et al., 2017). Paclobutrazol Resistance and Cryptochrome 2 Interacting BHLH are involved in flowering initiation and root initiation (Ikeda et al., 2012; Liu et al., 2013), with the MYCs acting as positive regulators of jasmonate biosynthesis (Fernández-Calvo et al., 2011; Song et al., 2014a). Furthermore, Glabra3, Enhancer of Glabra3 and Transparent Testa8 are involved in anthocyanin biosynthesis in Arabidopsis (Petridis et al., 2016; Wen et al., 2018). Recently, numerous bHLH TFs in plants have been suggested to respond to diverse abiotic stresses and improve plant stress tolerance, including to cold, salt and drought (Sun, Wang & Sui, 2018). Under salt stress, bHLH39 increased the expression levels of stress-response genes in wheat, and thus improved the salt tolerance of bHLH39-overexpressing wheat plants (Zhai et al., 2016). A total of 18 bHLH genes in poplar have been found to respond to salt stress (Zhao et al., 2018a). In addition, CgbHLH001 in Chenopodium glaucum can interact with CgCDPK in a signal transduction pathway under salt stress (Wang et al., 2017b). A study on grape validated the activity of VabHLH1 and VvbHLH1 as positive regulators under cold stress (Xu et al., 2014). NtbHLH123 in Nicotiana tabacum is a transcriptional activator that can bind to the G-box/E-box motif in the NtCBF gene promoter, thus regulating the expression of stress-responsive genes and increasing the cold tolerance of Nicotiana tabacum (Zhao et al., 2018c). FtbHLH2 in Fagopyrum tataricum is significantly induced under cold treatment and overexpression of FtbHLH2 results in better cold/oxidative tolerance in transgenic Arabidopsis (Yao et al., 2018). Furthermore, StbHLH45 in Solanum tuberosum (Wang et al., 2018) acts as a positive regulatory factor under cold stress. The functions of bHLH gene family in plant under biotic and abiotic stresses are gaining increasing attention, and will be an important direction of future research (Sun, Wang & Sui, 2018).
Lotus is an economically important aquatic plant that has been widely used for food, medicinal, and ornamental purposes. As a basal eudicot plant with numerous monocot characteristics, lotus has been an important subject of evolutionary and taxonomic studies (The Angiosperm Phylogeny Group, 2009; Ming et al., 2013; Zhao et al., 2018b). Meanwhile, lotus plays a vital role in cultural and religious activities and is extensively distributed throughout Asia and Northern Australia. Soil and water salinity are some of the most serious environmental stresses affecting crop yields worldwide, so it is essential to improve the tolerance of plants to salinization of soil and water (Munns & Tester, 2008). In addition, low temperatures can severely impact the growth, yield and distribution area of plants (Sanghera et al., 2011). The dry matter content of lotus is minimal during colder seasons at high latitudes (Bullard & Crawford, 2010; Halling, Topp & Doyle, 2010), causing substantial economic losses. Hence, identification of salt and cold tolerance and response genes is expected to improve salt and cold resistance of lotus, which may increase the yield and expand the distribution of lotus to some extent. As noted above, the bHLH gene family responds to environmental stimuli, and thus structural and functional analyses of the NnbHLH gene family in lotus are necessary to improve stress tolerance in lotus. However, the bHLH gene family has not yet been comprehensively studied. Although the identification and structural properties of some bHLH family members were reported in 2014, systematic structural and functional analyses of the bHLH gene family in lotus have remained scarce (Hudson & Hudson, 2014). In this study, the bHLH gene family of lotus was systematically identified using bioinformatics methods. Then, thorough analyses of the gene sequences, gene structures and conserved motifs, phylogenetic relationships, gene ontology (GO) annotations, function prediction, and expression patterns were conducted. The results offer an effective framework for further functional characterization of the lotus bHLH gene family, and in particular their role under stress.
Materials and Methods
Plant materials and treatments of the stress experiments
Seedlings (25 days old) of Nelumbo nucifera were grown in artificial climate chambers (Model:RXZ-500C, NingboJiangnan) with 16 h light and 8 h dark at 24 °C, following the conditions for lotus cultivation described in Diao et al. (2014). For cold treatment, seedlings were placed in an Intellus Ultra Controller (Model:LT-36VLC8, Percival Scientific, Inc., Perry, IA, USA) with the temperature at 4 °C, a common condition for cold treatment, and the same photoperiod as described above. Leaves were collected before cold exposure (control), after 2, 4, 8, and 12 h of cold exposure for RNA extraction before expression analyses. For salt stress treatment, roots of the lotus seedlings were exposed to 50 mM NaCl (Diao et al., 2014), and the leaves were collected before treatment (control) and after 2, 4, 8, and 12 h of salt exposure. All of the samples were collected with three biological replications and were immediately frozen in liquid nitrogen. The samples were stored at −80 °C for RNA isolation.
Identification of the NnbHLH gene family
The lotus genome sequence and the gene annotation data were obtained from the lotus genome database (Lotus-DB: http://lotus-db.wbgcas.cn) (Wang et al., 2015). First, a Hidden Markov Model search (HMMsearch) was performed using HMMER software with the seed profile of the bHLH domain (PF00010) (Cheng et al., 2018; Sun, Fan & Ling, 2015), which was downloaded from the PFAM database (http://pfam.xfam.org/) (Finn et al., 2014). HMMER software was used to search for bHLH protein in the entire protein dataset with the E-value cut-off set to 10−5. Then, a Basic Local Alignment Search Tool protein (BLASTp) alignment against all lotus protein sequences was subjected to perform an extensive search for candidate bHLH genes using bHLH protein sequences from Arabidopsis and rice as queries. The protein sequences of Arabidopsis and rice were downloaded from PlantTFDB (http://planttfdb.cbi.pku.edu.cn) (Jin et al., 2017). The Simple Modular Architecture Research Tool (Letunic, Doerks & Bork, 2015) and Conserved Domains Search (Marchler-Bauer et al., 2017) were employed to detect the bHLH domain in candidate protein sequences.
Chromosomal distribution, gene duplication analyses and calculation of synonymous (Ks) and non-synonymous (Ka) substitution rates
The distribution of each NnbHLH on megascaffolds was obtained based on the GFF3 file (Wang et al., 2015) and analyzed using Map Gene 2 Chromosome V2 (http://mg2c.iask.in/mg2c_v2.0/). Gene duplication analyses for lotus was conducted using the Multiple Collinearity Scan Toolkit (MCScanX) (Wang et al., 2012). To identify candidate homologous gene pairs (E < 1e−5), BLASTp alignment was carried out across the whole lotus genome. The potential homologous gene pairs were identified, and then loaded into the program MCScanX with the default parameters to identify syntenic chains. MCScanX was used to further distinguish among whole-genome duplication (WGD)/segmental, dispersed proximal, and tandem duplication events in the NnbHLH gene family. Candidate homologous gene pairs identified in the same synteny block were applied to calculation of Ka and Ks values using DNAsp5 (Librado & Rozas, 2009).
Sequence alignment and phylogenetic, gene structure and conserved motif analyses
Multiple domain alignments were performed using MEGA (vision 6.0) (Tamure et al., 2013) and loaded into Geneious to visualize. The phylogenetic tree was constructed using MEGA 6.0 with the neighbor joining (NJ) method and the following parameters: pairwise deletion and 1,000 bp replications. The phylogenetic tree was visualized by plotting it using the EvolView tool (http://www.evolgenius.info).
The intron-exon organization of NnbHLHs was visualized using the Gene Structure Display Server 2.0 (GSDS: http://gsds.cbi.pku.edu.cn/) (Hu et al., 2014). Conserved motifs in NnbHLHs were identified using the MEME (http://meme-suite.org/index.html) (Bailey et al., 2009) server with the maximum number of motifs set to 20.
GO annotation and analyses of cis-regulatory elements
Gene ontology analyses of NnbHLHs was performed using the Blast2GO program (Conesa et al., 2005), and the NCBI database was selected as the reference database. The sequences 1,500 bp upstream of the translation initiation codon ATG for each NnbHLH were selected for analyses of the promoters using a C script. Cis-regulatory elements for each promoter sequence were predicted through searching the PlantCARE database (Lescot et al., 2002) with the false discovery rate <0.1%.
Expression profiles and qRT-PCR analyses of NnbHLHs
RNA-seq data of four lotus tissues (leaf, petiole, rhizome, root) were downloaded from Lotus-DB. Total RNA was isolated using an RNA extraction kit (Aidlab, Beijing, China) according to the manufacturer’s instructions. 5X All-In-One RT MasterMix (abm) was used for reverse transcription. Specific primers for quantitative real-time PCR (qRT-PCR) were designed using Primer Premier 5 software (Lalitha, 2000). qRT-PCR was conducted using the LightCycler®/LightCycler®96 System Real Time PCR (Roche, Basel, Switzerland) with SYBR Premix Ex Taq II (TaKaRa, Kusatsu, Japan). The actin gene (GeneBank: XM_010252745) was used as an internal control for normalization of the expression levels of candidate NnbHLHs among different samples, following previous studies (Cheng et al., 2013; Diao et al., 2014; Jin et al., 2016), and expression levels were calculated using the delta–delta CT method (Livak & Schmittgen, 2001).
Results
Identification, chromosomal distribution and physicochemical properties of NnbHLHs
Members of the bHLH family in lotus were identified in the lotus genome using two strategies, that is, HMM search and BLASTp search, as described in the Materials and Methods section. Then, to verify the sequences, all candidates were checked for the presence of a complete bHLH domain via CDD and SMART. In total, 115 sequences were confirmed as lotus bHLHs and named NnbHLH1 to NnbHLH115. The encoded putative NnbHLH proteins were predicted to be 89 (NnbHLH110) to 779 (NnbHLH61) amino acids in length, with molecular weights ranging from 10.07 kDa (NnbHLH110) to 82.64 kDa (NnbHLH61) (Table S1). The grand average of hydropathicity values (GRAVY) of all candidate NnbHLH proteins were negative, ranging from −0.887 to −0.059, representing a hydrophilic characteristic. The predicted isoelectric points of NnbHLH proteins were 4.72 (NnbHLH72) to 10.62 (NnbHLH110). The predicted numbers of negatively charged residues (Asp and Glu) were 7 (NnbHLH110) to 104 (NnbHLH1), while the predicted number of positively charged residues (Arg and Lys) ranged from 18 (NnbHLH110) to 83 (NnbHLH13) (Table S1). Detailed information about these characteristics is listed in Table S1. The ratio of NnbHLHs in the Nelumbo nucifera genome was about 0.425%, similar to that in rice (0.44%) (Li et al., 2006) and poplar (0.40%) (Carretero-Paulet et al., 2010) but lower than the ratios in Arabidopsis (0.59%) (Toledo-Ortiz, Huq & Quail, 2003) and Brachypodium distachyon (0.55%) (Niu et al., 2017). Based on the results of lotus genome annotation (Wang et al., 2015), the 115 predicted NnbHLHs were distributed unevenly, localized on 20 megascaffolds of lotus, with 168 megascaffolds in the lotus genome (Table S1). Among them, megascaffolds 1, 2, 3, 4, 5, 6, 7, 8, 10, 12, and 13 contained most NnbHLHs, while megascaffolds 9, 11, 15, 17, 24, 33, 43, 59, and 114 and scaffolds 554 and 634 only possessed 1–2 NnbHLHs each (Table S1).
Multiple sequence alignment, prediction of DNA-binding and protein dimerization activity of NnbHLHs
To further clarify the structural characteristics of NnbHLHs, multiple sequence alignment analyses of the bHLH domain were carried out. As shown in Fig. 1, one basic region, one loop region and two helix regions were detected in all 115 NnbHLH proteins. And conserved amino acids in bHLH domains, with sequence identity more than 50%, were shaded in grey or black color (Fig. 1A). A sequence logo of the conserved amino acids in the NnbHLH domain was presented in Fig. 1B. The bHLH gene family in Arabidopsis, rice and Brachypodium distachyon all contain 17 conserved amino acids in the bHLH domain, which were also detected in the NnbHLH proteins (Fig. 1C; Table 1). The consensus ratios of the 17 conserved amino acids among the three species were also calculated (Table 1). Arg-16, Arg-17, Leu-27, and Leu-61 showed extremely high sequence identity (98%, 93%, 100%, and 94%, respectively) among the 115 NnbHLH proteins (Table 1). The HLH region may be essential for dimerization, particularly Leu-27 in helix 1 and Leu-61 in helix 2 (Carretero-Paulet et al., 2010). Thus, we speculated that NnbHLH proteins may also have the capacity to form protein complexes.
Figure 1: Conserved amino acids and multiple sequence alignment schematic diagrams of the NnbHLHs bHLH domains.
(A) Conserved amino acids across NnbHLH domain. The amino acid with sequence identity more than 50% was labeled with gray or black shading (B) sequence logo of NnbHLH domains. The overall height of each stack represents the conservation of the sequence at that position. (C) Multiple sequence alignment of the NnbHLH doamins. Shading represents the degree of amino acids identity at each position, with the black shading indicating 100% sequence identity.| Position in the alignment | Consensus amino acids and their ratios within the lotus bHLH domain | Position in the alignment | Consensus amino acids and their ratios within the Arabidopsis bHLH domain Toledo-Ortiz, Huq & Quail (2003) | Position in the alignment | Consensus amino acids and their ratios within the rice bHLH domain Li et al. (2006) | Position in the alignment | Consensus amino acids and their ratios within the Brachypodium distachyon bHLH domain Niu et al. (2017) |
|---|---|---|---|---|---|---|---|
| 13 | E (82%) | 13 | E (76%) | 13 | E (68%) | 14 | E (74%) |
| 14 | R (85%) | 14 | R (74%) | 14 | R (67%) | 15 | R (73%) |
| 16 | R (98%) | 16 | R (91%) | 16 | R (90%) | 18 | R (88%) |
| 17 | R (93%) | 17 | R (86%) | 17 | R (84%) | 19 | R (88%) |
| 27 | L (100%) | 27 | L (100%) | 27 | L (99%) | 29 | L (99%) |
| 30 | L (81%) | 30 | L (65%) | 30 | L (68%) | 32 | L (69%) |
| 32 | P (92%) | 32 | P (88%) | 32 | P (82%) | 34 | P (56%) |
| 36 | K (70%) | 39 | K (76%) | 38 | K (64%) | ||
| 46 | D (66%) | 41 | D (64%) | 50 | D (71%) | 59 | D (68%) |
| 48 | A (77%) | 43 | A (73%) | 52 | A (74%) | 61 | A (69%) |
| 49 | S (63%) | 44 | S (53%) | 53 | S (57%) | 62 | S (51%) |
| 51 | L (75%) | 46 | L (76%) | 55 | L (86%) | 64 | L (84%) |
| 54 | A (51%) | 49 | A (60%) | 58 | A (57%) | 67 | A (58%) |
| 55 | I (72%) | 50 | I (63%) | 59 | I (64%) | 68 | I (55%) |
| 57 | Y (77%) | 52 | Y (78%) | 61 | Y (83%) | 70 | Y (77%) |
| 59 | K (61%) | 54 | K (55%) | 63 | K (62%) | 72 | K (53%) |
| 61 | L (94%) | 56 | L (93%) | 65 | L (96%) | 77 | L (99%) |
Based on the criteria developed by Toledo-Ortiz, Huq & Quail (2003), bHLH proteins with more than five basic amino acid residues in the basic region were identified as DNA-binding proteins. NnbHLH proteins were divided into two major groups accordingly, including 101 DNA-binding proteins and 14 non-DNA-binding proteins. Then we subdivided the 101 DNA-binding NnbHLH proteins into two groups: 89 E-box-binding proteins (based on the presence of Glu-6 and Arg-9) and 12 non-E-box-binding proteins (without the simultaneous presence of Glu-6 and Arg-9), according to previous research (Ellenberger et al., 1994). The E-box-binding NnbHLH proteins were further subdivided into two groups, including 75 G-box-binding proteins and 14 non-G-box-binding proteins, based on the presence or absence of His/Lys-9, Glu-13, and Arg-17, which may be critical for binding to the G-box (CACGTG) (Niu et al., 2017; Sun, Fan & Ling, 2015).
Gene structure and conserved motif analyses of NnbHLHs
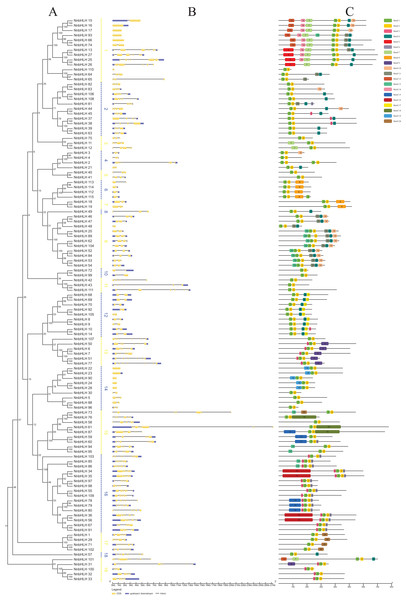
A neighbor-joining phylogenetic tree was constructed using the alignment results of the NnbHLH proteins (Fig. 2A). Based on the bootstrap values, the NnbHLH proteins were divided into 19 subfamilies (Fig. 2A). Among those subfamilies, subfamily 16 was the largest, containing 16 proteins, while subfamilies 8 and 18 had only had 1 protein each.
Figure 2: Gene structures, phylogenetic relationships and conserved motifs analyses of the NnbHLHs.
(A) Neighbor-joining phylogenetic tree of NnbHLHs. (B) Gene structure of NnbHLHs. Orange box represent exon, blue box represent UTR and the black line represent intron. The sizes of exons can be estimated by the scale at bottom. (C) Conserved motifs in the NnbHLH proteins. A total of 20 predicted motifs are represented by different colored boxes and motif sizes can be estimated by the scale at the bottom.Next, the exon/intron structures of NnbHLHs were analyzed using the GSDS 2.0 online tool. NnbHLHs in the same subfamily appeared to share similar numbers of exons and introns (Fig. 2B). Members of subfamilies 6, 7, 10, and 12 all contained the same number of exons and introns, while the majority of subfamilies 2, 9, 11, and 13 shared the same number of exons and introns. Meanwhile, the numbers of exons and introns were variable among subfamilies 1, 3, 15, and 19 (Fig. 2B).
To further reveal the specific regions of NnbHLH proteins, 20 conserved motifs that varied from 16 to 100 residues in length were detected using the MEME tool. All predicted motifs were identified only once in each NnbHLH protein sequence. NnbHLH proteins contained different numbers of conserved motifs, ranging from two to eight, and all NnbHLH proteins possessed motifs 1 and 2, representing the location of the bHLH domain (Fig. 2C). Meanwhile, NnbHLH proteins in the same subfamily always shared similar motif composition. For example, members of subfamily 9 all contained motifs 1, 2, 4, 10, and 11, while motifs 5, 17, and 19 were found exclusively in subfamilies 1, 12, and 14, respectively (Fig. 2C).
Gene duplication analyses of NnbHLHs
Duplication events usually contribute to the expansion of gene families and genome evolution, and in particular, WGD/segmental and tandem duplication events have been important in the expansion of multigene families (Kong et al., 2010). In this study, almost all 115 detected NnbHLHs had experienced duplication events, except NnbHLH21. Because duplication usually contributes to the expansion of gene families, we further investigated the duplication patterns of each NnbHLH. In total, 80% (92/115) of the NnbHLHs were retained from WGD/segmental duplication events (Table S2), with only 3.47% (4) from tandem, 13.9% (16) from dispersed, and 1.73% (2) from proximal duplication events.
The substitution rate ratio, Ka/Ks, is an effective criterion for the selective pressure during gene duplication. Ka/Ks values less than one indicate negative selection, those equal to one indicate neutral selection, and values greater than one indicate positive selection (Yang, 2007). Among the 118 pairs of WGD, the Ka/Ks ratios of 27 pairs were greater than one, with those of NnbHLH1 and NnbHLH59, NnbHLH6 and NnbHLH97, NnbHLH7 and NnbHLH76, NnbHLH26 and NnbHLH93 being 1.808, 1.640, 1.510, and 1.679, respectively, indicating that these duplication pairs have been strongly positively selected during their evolutionary history (Cheng et al., 2018). The remaining NnbHLH genes, with Ka/Ks < 1, may have undergone negative selection, suggesting that most bHLH genes have evolved slowly (Table S2). For the two tandem duplication pairs, the Ka/Ks of the gene pair NnbHLH112 and NnbHLH115 was 1.334, while that of NnbHLH117 and NnbHLH120 was 0.453.
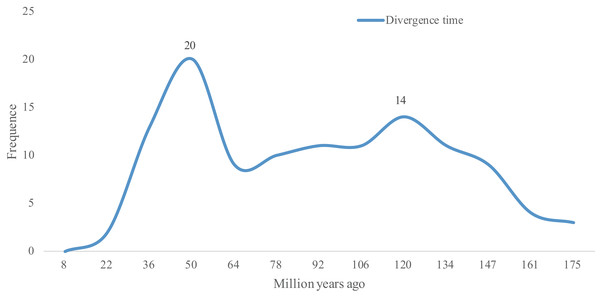
As Ks values can be further be applied to trace the divergence time of duplication events (Lynch & Conery, 2000), the divergence times of 118 WGD gene pairs were calculated. As shown in Table S2, the divergence times of these 118 WGD pairs ranged from 8.39 to 167.35 Mya. The two peaks observed in Fig. 3 show that the lotus bHLH gene family has undergone two larger duplication events, an ancient one that occurred ∼120 Mya, and a recent one that occurred ∼50 Mya. The earliest duplication event in the NnbHLH family appeared at 176 Mya. Since 147 Mya, bHLH family genes began to duplicate frequently, and the expansion of the gene family accelerated.
Figure 3: Distribution of divergence time of WGD pairs of the NnbHLHs.
The x-axis represents divergence time; y-axis represents the density of the distribution.GO annotation and cis-element analyses of NnbHLH proteins
To understand the specific functions of bHLH proteins, GO annotation of NnbHLH proteins was performed. As shown in Table S7, most NnbHLH proteins were annotated as being associated with protein dimerization activity, the development process, and response to stimulus. Within the cellular component, most genes were assigned to the nucleus (95/115). Very few genes had predicted distributions in organelles, such as the chloroplast envelope (8), mitochondrion (8), cytosol (4), and endosome (1). In addition, 19 NnbHLH proteins were predicted to be located on the plasmodesma (Table S7). In terms of molecular functions, almost all NnbHLH proteins were predicted to be involved in protein dimerization activity (113/115), with 83 proteins related to DNA binding TFs and 79 proteins associated with DNA-binding activity. These results are consistent with the DNA-binding analyses described in “Multiple sequence alignment, prediction of DNA-binding and protein dimerization activity of NnbHLHs.” Within the biological process category, 102 NnbHLHs were predicted to be involved in development processes, including the development of the roots (52/115), carpels (43/115), floral organs (34/115), and petals (25/115). Moreover, 104 NnbHLH proteins were predicted to respond to stimuli, with 65 supposedly reacting to abiotic stresses. A total of 28 NnbHLH proteins could respond to low temperature, 35 were associated with responses to radiation (blue light, red light, or far-red light) and 13 were predicted to respond to salt stress. The number of NnbHLH proteins predicted to respond to abiotic stresses was high, suggesting that the NnbHLH gene family may play a vital role in lotus tolerance to abiotic stresses (Table S7).
Cis-element analyses were also conducted applying the 1,500 bp upstream sequence of NnbHLHs in the PlantCARE database. In the NnbHLH promoter region, G-box and Sp1 (response to light), MBS (response to drought), Skn-1_motif (required for endosperm expression), and ARE (essential for anaerobic induction) were the most common elements (Table S4). All NnbHLH family members except NnbHLH 58, 86, 90, 100, and 101 possessed at least one cis-element involved in stress responses (Table S4). The cis-regulatory elements present within NnbHLH promoters could be divided into three main categories. The first category was a ubiquitous class of plant light-responsive elements (such as G-box and Sp1), and the second category included plant growth- and development-responsive elements (such as Skn-1_motif and ARE) (Table S4). The last category included elements that respond to diverse stresses (such as TC-rich), including elements responding to biotic stresses (CGTCA-motif and ABRE) and abiotic stresses (HSE, MBS, and LTR). Elements in this category were widely distributed throughout the NnbHLH gene family (Table S4).
Function prediction of NnbHLHs based on phylogenetic analyses
To date, the biological functions of most NnbHLHs have remained unclear. Meanwhile, in Arabidopsis and rice, the functions of many bHLH proteins have been characterized and verified. In this study, phylogenetic analyses allowed us to identify putative orthologous and paralogous bHLH genes in lotus, Arabidopsis and rice. In general, homologous genes share similar structures and are clustered in the same clades, and these genes possess similar functions. To predict the gene functions of each NnbHLH, another NJ phylogenetic tree was constructed based on the protein of 115 NnbHLHs, 132 OsbHLHs of rice, and 160 AtbHLHs of Arabidopsis (Li et al., 2006; Toledo-Ortiz, Huq & Quail, 2003) (Fig. S1). In total, 24 subgroups were clustered, and the functions of NnbHLHs were predicted based on their homologs with verified functions in the same cluster (Table S5).
In summary, the majority of the members of subgroups 1a, 3, 4, 6, 7, 15a, and 19 may be able to enhance stress tolerance (Kim et al., 2005) and response to diverse abiotic and biotic stresses (Sasaki-Sekimoto et al., 2014; Song et al., 2013), including cold (Kim et al., 2010; Deng et al., 2017), salt (Ahmad et al., 2015; Sakai et al., 2013; Jiang, Yang & Deyholos, 2009), and drought (Le Hir et al., 2017). In subgroups 8 and 18, proteins may be related to Fe regulation, modulating the homeostasis of Fe content (Kurt & Filiz, 2018; Wang et al., 2017a). Members of subgroups 1a, 2, and 15a were predicted to regulate flower development of flower (Sharma et al., 2016; Choi et al., 2018; Ito et al., 2012), while those in subgroups 1b, 3, 4, and 17 may be involved in the development of various plant organs (Karas et al., 2009; Le Hir et al., 2017; An et al., 2014; Lee, Jung & Park, 2017; Ohashi-Ito & Bergmann, 2006; Cui et al., 2016; Feng et al., 2017). Subgroup seven members may regulate the flavonoid (Rai et al., 2016) and anthocyanin biosynthesis (Wang et al., 2017a). In subgroup 10, many proteins were predicted to be PIFs, which are related with photo-induced signal transduction and may optimize plant growth and development (Paik et al., 2017). Other members of subgroup 10 and subgroup 13 have also showed regulatory abilities (Leivar et al., 2008; Zumajo-Cardona, Ambrose & Pabón-Mora, 2017; Cifuentes-Esquivel et al., 2014). Members of subgroups 15b and 16 have been predicted to be involved in diverse processes (Mai et al., 2015; Ferguson et al., 2017; Gaillochet et al., 2018; Zhu et al., 2016). NnbHLHs in subgroup 21 should negatively control thermospermine biosynthesis and xylem differentiation (Cai et al., 2016; Yamamoto & Takahashi, 2017), while members of subgroup 24 may play an essential role in establishing vascular cells (Ohashi-Ito, Matsukawa & Fukuda, 2013). The detailed function prediction of NnbHLHs can be found in Table S5.
Expression profiles of NnbHLHs
Transcriptional data of four different lotus tissues were employed to analyze the expression patterns of NnbHLHs (Table S6). In all tissues, NnbHLHs with FPKM > 1 were used for further analyses (Zhuo et al., 2018). Among 115 NnbHLH genes, 91 NnbHLHs were expressed in at least one of the tissues, while the expression levels of NnbHLH 2, 12, 19, 21, 23, 30, 36, 40, 44, 45, 47, 48, 49, 61, 64, 65, 84, 88, 89, 90, 92, 96, 104, and 110 were low, with FPKM < 1 in all four tissues (Table S6). Some NnbHLH genes exhibited tissue preferences, with expression levels in the preferred tissue that were more than two times higher than those in other tissues (Sun, Fan & Ling, 2015). Overall, NnbHLH 20, 52, 84, 106, 108, and 115 in the leaves; NnbHLH 9, 24, 55, 97, 98, and 109 in petioles, 14 genes (NnbHLH 1, 4, 16, 29, 38, 50, 57, 59, 72, 74, 81, 93, 99, and 102) in rhizomes; and 22 genes (NnbHLH 5, 11, 14, 26, 34, 39, 41, 46, 53, 54, 56, 60, 63, 67, 68, 69, 80, 82, 83, 91, 113, and 114) in roots were suggested to have tissue expression preferences (Table S6). In addition, most members of subfamilies 4, 5, 6, 7, 8, 14, and 18 had no expression or low expression in the four tissues. The majority of the members of subfamilies 10, 11, and 19 exhibited high expression levels in all four tissues (Table S6).
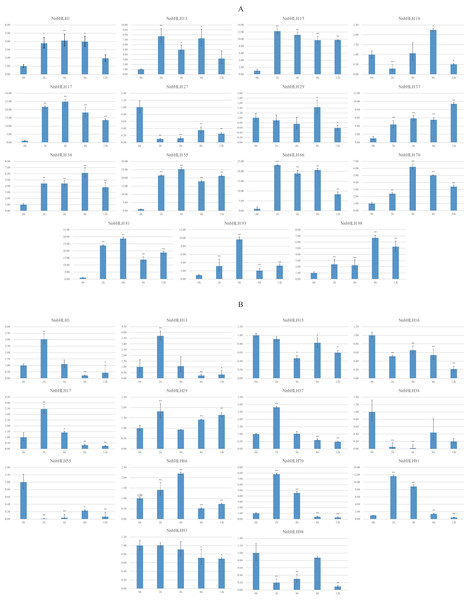
Because stress-resistance is very important in lotus, many resistance-related NnbHLHs were predicted based on the structural and predictive analysis. In all, 15 NnbHLHs were selected for analyses of their expression under low temperatures and salt stress, and the specific primers used are listed in Table S3. The expression levels of the 12 NnbHLHs increased initially under 4 °C treatment (Fig. 4). Most were upregulated and reached their maximum values after 4 h of treatment, decreasing thereafter. The expression levels of NnbHLH13 and NnbHLH66 reached a maximum after just 2 h of treatment, while the expression levels of NnbHLH16, NnbHLH38, and NnbHLH98 were highest after 8 h. We also found that NnbHLH 15, 17, 37, 38, 55, 66, 70, 81, 93, and 98 were significantly (P < 0.01) upregulated at all time points. Among these genes, the expression levels of NnbHLH 15, 17, 55, 66, and 81 were significantly increased by 10-fold or more (Fig. 4); in particular, NnbHLH17 and NnbHLH81 were strongly induced under 4 °C treatment, with expression levels that increased by nearly 25-fold and 30-fold, respectively. Under NaCl treatment, eight NnbHLHs (NnbHLH1, NnbHLH13, NnbHLH17, NnbHLH29, NnbHLH37, NnbHLH66, NnbHLH70, and NnbHLH81) were upregulated initially and then decreased. Among these genes, only the expression level of NnbHLH81 increased more than 10-fold compared to its untreated level. The other six NnbHLHs were downregulated. Although some NnbHLHs were significantly upregulated (P < 0.01) at one or two time points, none of them were upregulated at all time points (Fig. 4).
Figure 4: Expression patterns of the NnbHLH candidate genes in lotus under cold and salt treatment by qRT-PCR.
(A) Expression patterns of NnbHLH candidate genes under cold stress treatment. (B) Expression patterns of NnbHLH candidate genes under salt stress treatment. y-axis: relative expression levels; x-axis: the time course of stress treatments; Error bar represent standard errors from three biological replicates. The asterisks indicate significant differences (P < 0.05) between treatment group and control group (0h) with two asterisks indicate extremely significant differences (P < 0.01).Discussion
Comprehensive genome-wide detection of NnbHLHs in lotus
Numerous studies of the bHLH family in various species, such as Arabidopsis (Toledo-Ortiz, Huq & Quail, 2003), rice (Li et al., 2006), peanut (Gao et al., 2017), chinses cabbage (Song et al., 2014b), tomato (Sun, Fan & Ling, 2015), apple (Yang et al., 2017), and bamboo (Cheng et al., 2018), have been reported in recent years. In our study, 115 bHLH genes of lotus were identified, which is two fewer than in a previous study (Hudson & Hudson, 2014). This difference may be the result of a more restrictive selection in NnbHLH in our study, in which genes without complete bHLH domain were removed. These 115 genes were further classified into 19 subfamilies based on phylogenetic analyses (Fig. 2A). Multiple sequence alignment of the full-length NnbHLH protein sequences showed that all putative NnbHLHs contained the classic bHLH domain, and the number and ratio of the conserved amino acid were consistent with the Arabidopsis, rice, and Brachypodium distachyon (Table 1). Further structural analyses indicated that most of the NnbHLHs were associated with DNA-binding and homodimer formation activities. In addition, the conserved motif analyses showed that motifs representing the conserved domain were apparent in almost all the NnbHLH family members (Fig. 2C). Together, these results indicate that the 115 NnbHLHs all show characteristics of the bHLH family, which confirms the accuracy of the detection of bHLH gene family in lotus. Meanwhile, the expression profiles of NnbHLHs in four different tissues were analyzed, revealing that expression of many NnbHLH showed tissue preferences.
NnbHLHs may play a special role in lotus evolution
The duplication pattern of a gene usually reveals how the gene was generated, how its function has evolved, and what roles it may play in plant growth and development (Xia et al., 2017). In our analyses, 80% (92/115) of NnbHLHs were duplicated through WGD (Table S2), similar to previous studies that have reported 48% in rice and 58.2% in Brachypodium distachyon (Li et al., 2006; Niu et al., 2017). Because the major duplication type in lotus was WGD, the bHLH gene family in lotus is presumed to have existed in ancient times (Xia et al., 2017). New genes produced through WGD usually increase the ability of plants to adapt to various growth conditions, suggesting that NnbHLHs have been indispensable to the growth and development of lotus (Flagel & Wendel, 2009; Van De Peer, Maere & Meyer, 2009). Meanwhile, most Ka/Ks values of WGD NnbHLHs gene pairs were less than 1 (Table S2), showing that the lotus bHLH gene family has evolved slowly (Wang et al., 2013).
Based on Fig. 3, the NnbHLH gene family underwent a large WGD duplication event in modern times (about 50 Mya). Interestingly, two extant species of Nelumbonaceae, the Nelumbo nucifera and Nelumbo lutea, reportedly diverged in modern times (Xue et al., 2012). Thus, the WGD duplication event observed in our study may be related to the divergence event of Nelumbo nucifera and Nelumbo lutea. Meanwhile, another peak appears around 120 Mya in Fig. 3, which suggests that the NnbHLH family experienced relatively frequent duplication events. Coincidentally, the Nelumbonaceae are closely related to Platanaceae phylogenetically, and their divergence time has been estimated to be around 75.2–122.8 Mya (Xue et al., 2012). A duplication event appeared around 120 Mya in our study, which may be related to the divergence of the Nelumbonaceae and Platanaceae. WGD events are related to the divergence of plant taxa and often appear to be accompanied by marked and sudden increases in species richness (Van De Peer, Maere & Meyer, 2009), and therefore the high frequency of WGD events in the NnbHLH gene family are likely to have been important to lotus evolution.
Functional prediction of NnbHLHs in lotus
Research on the functional and structural genomics of Arabidopsis and rice has shown that bHLH TFs are involved in stress responses and plant development, including cold stress, heat stress, abscisic acid, jasmonic acid, and light response signaling pathways (Ikeda et al., 2012; Kim et al., 2010; Liu et al., 2013; Paik et al., 2017). Meanwhile, bHLH genes are involved in plant metabolite biosynthesis and trait development, including the formation of root hairs, anther development, and axillary meristem generation (Fernández-Calvo et al., 2011; Song et al., 2014a; Wen et al., 2018). However, little is known about the functions of the bHLH gene family in lotus. To better understand the second-largest gene family in plants, preliminary analyses of three aspects were conducted to reveal the functions of NnbHLH family genes in lotus for the first time.
Cis-element analyses showed that elements that can respond to diverse stresses (such as LTR, ABRE, TC-rich and HSE) were widely distributed in the NnbHLH gene family (Table S4). In addition, GO annotation revealed that most NnbHLH proteins were probably involved in the plant development process and responses to stimuli (Table S7). Furthermore, the functions of 69 NnbHLHs were predicted based on their known and verified homologs in Arabidopsis and rice (Table S5), which were mainly associated with development processes (root hair development, seed dormancy, fruit dehiscence, and flowering initiation) and stress responses (responses to low-temperature and salt stress and enhancement of stress tolerance) (Table S5). These analyses suggest that the bHLH gene family is also related to plant development, metabolic regulation, and the response to stress in lotus, in accordance with previous studies (Li et al., 2006; Niu et al., 2017; Toledo-Ortiz, Huq & Quail, 2003; Zhang et al., 2015). Next, we analyzed the candidate stress-response NnbHLHs, as improving stress tolerance in lotus is vital. Based on cis-element analyses, TC-rich cis-elements that may be involved in defense and stress responses were detected in the promoter regions of 58 NnbHLH genes (Table S4). Meanwhile, 38 NnbHLHs contained LTR box, which responds to cold stress and 44 members contained HSE box, which responds to heat stress (Table S4). The results of GO annotation suggested that 109 member genes can respond to stimuli (Table S7), with 13 and 28 NnbHLHs predicted to play roles in salt and cold stress, respectively, (Table S7). Phylogenetic analyses further suggested that 25 NnbHLHs may respond to stresses, including cold, salt, and drought, based on their homologs in Arabidopsis and rice (Table S5). Comprehensive analyses of these three datasets suggested that NnbHLH 1, 15, 37, 38, 55, 70, 81, 93, and 98 and NnbHLH13, 15, 16, 17, 27, 29, 66, 70, and 93 were likely to respond to cold and salt stress, respectively.
To validate the functional prediction of NnbHLHs, qRT-PCR analyses were conducted for 15 NnbHLH genes under cold (4 °C) and salt (50 mM NaCl) treatments. NnbHLH 1, 15, 37, 38, 55, 70, 81, 93, and 98 were all significantly (P < 0.01) upregulated at all treatment time points, and were thus candidates of NnbHLHs that respond to cold stress. NnbHLH 13, 17, 29, 66, and 70 were also upregulated under salt stress, as expected. We found that NnbHLH 1, 13, 17, 37, 66, 70, and 81 were upregulated under both cold and salt stresses, in accordance with the results of GO annotations and cis-element and homolog analyses. Thus, NnbHLH 1, 13, 17, 37, 66, 70, and 81 are the strongest candidates for lotus resistance genes due to their responsiveness to various stressors. The results of qRT-PCR analyses suggested that functional prediction of the NnbHLH gene family could provide valuable reference data for further functional research of this gene family.
Conclusions
In summary, we conducted a genome-wide evaluation of the bHLH gene family in lotus. The structural characteristics of this gene family were thoroughly investigated. Functional prediction of the NnbHLH family was systematically conducted for the first time using three methods. We also analyzed the expression patterns of 15 candidate genes under cold and salt treatments at several time points based on functional prediction. Taken together, the results and findings described in this study provide a strong basis for further investigation of the function and evolution of NnbHLHs. In addition, candidate genes for stress resistance in lotus were identified.
Supplemental Information
Phylogeny analysis of the bHLHs from lotus, Arabidopsis and rice.
The Ka/Ks ratios and estimated divergence time for WGD/segmentally duplicated NnbHLHs.
Predicted functions of the 69 NnbHLHs and their functional verified homologs in Arabidopsis and rice by phylogenetic analysis.
FPKM values of the NnbHLHs in four tissues.
FPKM: fragments per kilobase of exon per million fragments mapped.