Associations of IGF2 and DRD2 polymorphisms with laying traits in Muscovy duck
- Published
- Accepted
- Received
- Academic Editor
- Karl Broman
- Subject Areas
- Genetics, Zoology
- Keywords
- Insulin-like growth factor 2, Dopamine receptor 2, Polymorphisms, Association analysis, Muscovy duck, Egg production traits
- Copyright
- © 2017 Ye et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2017. Associations of IGF2 and DRD2 polymorphisms with laying traits in Muscovy duck. PeerJ 5:e4083 https://doi.org/10.7717/peerj.4083
Abstract
Insulin-like growth factor 2 (IGF2) and dopamine receptor 2 (DRD2) play important roles in ovarian follicular development. In this study, we analyzed tissue-specific expression of the Muscovy duck IGF2 and DRD2 genes and cloned those genes transcripts. Polymorphisms in these genes were tightly linked with egg production traits and both genes were highly expressed in the ovary. Moreover, we identified five single nucleotide polymorphisms (SNPs) for IGF1 and 28 for DRD2. Mutations A-1864G and C-1704G of IGF2 were positively correlated with increased egg laying at 59 weeks (E59W) (P < 0.05). The C+7T and C+364G mutations of DRD2 were highly and significantly associated with first-egg age (FEA) and egg numbers at 300 days (E300D) (P < 0.01). Moreover, C+3301G and C+3545G of DRD2 were highly significantly associated with FEA, E59W and E300D (P < 0.01). Other mutations were positively associated with FEA or E300D or E59W (P < 0.05). These data suggest specific roles for IGF1 and DRD2 polymorphisms in egg production in Muscovy ducks.
Introduction
Muscovy ducks are an excellent breed species because of their rapid growth, crude feed tolerance and high-quality meat. Although these ducks are raised on a large scale in China, low production performance affects the economic interests of farmers. Breeders have been looking for ways to improve Muscovy ducks egg production. In recent years, with the rapid development of genome sequencing technologies, molecular marker breeding and transgenic breeding technology have gradually become the mainstream of breeding. Traditional breeding mainly depends on breeding experience, which has a lot of unpredictability. Furthermore, molecular breeding can significantly improve breeding efficiency and shorten the breeding period, so using molecular marker breeding has a huge advantage in breeding. Nowadays, molecular markers are widely used in poultry breeding, such as green shell egg related molecular markers (Wang et al., 2013) and egg production related molecular markers (Han, An & Du, 2014). Due to the great prospects of molecular markers in breeding, using molecular markers to selecting high laying performance Muscovy ducks is a good decision.
Our research focuses on egg production related molecular markers that can be used to improve egg production for the Muscovy duck. Few researchers have paid attention to egg production traits in Muscovy ducks, which makes our research more meaningful. The first egg age (FEA), egg numbers at 300 days (E300D), and egg numbers at 59 weeks (E59W) are important traits in Muscovy ducks breeding. Muscovy ducks egg peak time is from 35 weeks to 53 weeks, and 59 weeks is the last stage of laying. Three hundred days is the peak time of laying, and 59 weeks is the end time of laying in Muscovy ducks, which covers most of the egg laying period. Therefore, we focus on FEA, E300D and E59W instead of egg production at other time points as important traits.
Insulin-like growth factor 2 (IGF2) plays a key roles in animal growth differentiation and proliferation (Kaneda et al., 2007), as well as reproduction and the regulation of ovarian follicle development. In mammals, IGF2 is highly expressed in the dominant follicle supporting key functions for follicular development (Mao et al., 2004). IGF2 may affect prolificacy in sows and cattle (Stinckens et al., 2010; Aad, Echternkamp & Spicer, 2013), and IGF2 may regulate ovarian development through follicle-stimulating hormone (FSH) (Baumgarten et al., 2015). But little research on the regulation of ovarian development by IGF2 has been conducted in birds. The present study is the first to report that IGF2 may be associated with ovarian development. Dopamine (DA) is an essential neurotransmitter and exists in the nerve center and its peripheral tissue. Dopamine receptor 2 (DRD2) may assist with the secretion of reproductive hormones through follicle-stimulating hormone (FSH) and luteinizing hormone (LH) in chicken (Youngren et al., 1996; Youngren, Chaiseha & El, 1998). Association studies between single nucleotide polymorphisms (SNPs) of IGF2 and DRD2 and reproduction traits have been carried out in poultry (Xu et al., 2011b; Xu et al., 2011a; Wang et al., 2014a; Wang et al., 2014b; Zhang et al., 2015; Zhu et al., 2015). However, until now very few studies have focused on the relevance of these genes to egg production in Muscovy ducks. Therefore, we aim to identify SNPs in these genes, and to reveal their associations with reproduction traits in Muscovy ducks. We hope these molecular markers may help to improve the production performance of Muscovy ducks in breeding.
Materials and Methods
Ethics statement
Ethical approval for all animal experiments was granted by the Animal Care Committee of South China Agricultural University (Guangzhou, People’s Republic of China) with approval number 20131019002.
Sample collection
Eight hundred white Muscovy ducks were offered by Wens Nanfang Poultry Breeding company (Yunfu, Guangdong, China) which were in the same run. All Muscovy ducks were reared under identical conditions of management and feeding. Ducks were maintained outside on the ground from four to 12 weeks of age, after which they were transferred to individual cages in a semiconfined house. Feed was provided by Wens (Xinxing, Guangdong, China). The first egg age (FEA), egg numbers at 300 days (E300D), and egg numbers at 59 weeks (E59W) were recorded for each female duck. Genomic DNA from each individual at 59 weeks was isolated from 0.5 ml blood stored with EDTA as an anticoagulant, using E.Z.N.A NRBC Blood DNA Kit (Omega, Norcross, GA, USA) according to the manufacturer’s instructions.
RNA isolation and cDNA synthesis
Muscovy duck tissues including pituitary, brain, lung, abdominal fat, liver, ovary, subcutaneous fat, spleen, kidney, leg muscle, hypothalamus, cerebellum, heart and breast muscle used for expression pattern analysis of the IGF2 and DRD2 genes, were sampled at first egg age. These ducks were raised under the same conditions, but in different batches from the eight hundred Muscovy ducks mentioned above. Total RNA was isolated from tissues using a TRIZOL Reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. RNA quality was evaluated by 2% agarose gel electrophoresis and then was reverse transcribed using Takara reverse transcription Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The cDNA was used as template to amplify the coding region of the IGF2 and DRD2 genes from Muscovy duck.
Cloning of Muscovy duck IGF2 and DRD2 genes
The Muscovy duck IGF2 and DRD2 genes were identified using Mallard duck gene sequences as a reference (Gene Bank accession No. XM_005019778 and XM_013109685). Primers were designed to amplify the coding regions of Muscovy duck IGF2 and DRD2 using Primer 5.0 (Primer IGF2-CDS and Primer DRD2-CDS; Table S1). PCR amplifications were conducted in a final volume of 50 µl with 2 µl cDNA, 25 µl 2 × Easy Taq SuperMix (TransGen, Beijing, China), and 0.5 µl each pair of primers, and 22 µl double distilled H2O. Optimum PCR amplification conditions were programmed as pre-denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s, and a final extension at 72 °C for 10 min. The PCR products were evaluated by electrophoresis using a 2% agarose gel and then gel purified using a HiPure Gel Pure DNA kit (TransGen, Beijing, China). The amplified fragments were cloned into pMD-18T vector (TaKaRa, Dalian, China), and sequenced by Majorbio, Shanghai, China. Sequence alignment and phylogenetic trees are constructed using MEGA5.
Expression pattern analysis of IGF2 and DRD2 mRNA
Total mRNA from 14 different tissues was extracted to investigate the mRNA expression profiles of Muscovy duck IGF2 and DRD2 genes using real-time qPCR. Muscovy duck β-actin gene was used as the internal reference gene. Primers for the IGF2, DRD2 and β-actin genes were designed using Primer 5.0 (Primer β-actin-duck, IGF2-Q and DRD2-Q; Table S1). The qPCR was performed using a standard SYBR Premix Ex Taq II (TaKaRa, Dalian, China) on a BioRad CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s protocol. The thermal cycling was 95 °C for 2 min, followed by 39 cycles of 95 °C for 15 s, 60 °C for 30 s, 72 °C for 30 s, and final cycle of 72 °C for 7 min. Relative expression of IGF2 and DRD2 genes was calculated relative to the expression of β-actin. Real-time PCR data were analyzed using the 2−ΔΔCt method.
SNPs detection by sequencing
We designed 7 primers to identify potential SNPs of IGF2 and DRD2 (Primer IGF2-P1, IGF2-P2, DRD2-P1, DRD2-P2, DRD2-P3, DRD2-P4 and DRD2-P5; Table S1). Twenty white Muscovy ducks were sampled and five individuals were selected as a mixed pool. PCR reactions were performed in a 50 µl final volume, containing 2 µl DNA, 25 µl 2 × Easy Taq SuperMix (TransGen Biotech, Beijing, China), 0.5 µl each pair of primers, and 22 µl double distilled H2O. PCR parameters were 3 min at 94 °C followed by 37 cycles of 94 °C for 30 s, annealing temperature for 60 s, 72 °C for 30 s and a final extension at 72 °C for 10 min. PCR products were evaluated by electrophoresis using 2% agarose gel and sequenced as described above. SNPs were identified by the Seqman program of DNASTAR 7.1.0 software (DNASTAR, Inc., Madison, WI, USA).
Genotyping and association analysis
The SNPs were genotyped in 800 female ducks with egg production records via sequencing. We designed 3 primers to genotyping SNPs of IGF2 and DRD2 (Primer IGF2-SNP, DRD2-SNP1 and DRD2-SNP2; Table S1). PCR reactions were identical to those used in SNP detection as described above. Genotypes were tested for Hardy-Weinberg equilibrium with the chi-square test. Linkage analysis was performed using Haploview software (Barrett et al., 2005). The associations between SNPs and egg production traits were calculated using the general linear model procedure of SAS v. 9.2 with the following model: where Yij is the observed value of different egg production traits, μ is the overall population mean, Gi is the effect of each genotype, and eij is the random error. For each egg production trait, the least-squares mean was estimated and differences between the genotypes were analyzed using a Bonferroni adjustment for multiple comparisons. Difference with P value ≤0.05 was considered to be significant in analyses.
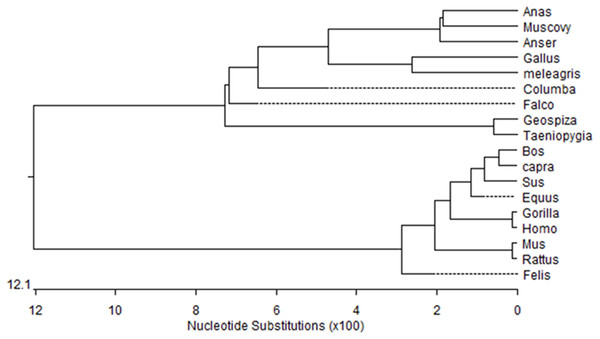
Figure 1: Phylogenetic tree of Muscovy duck DRD2 aligned amino acid sequences.
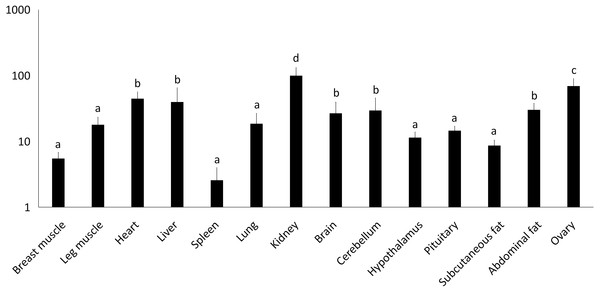
Orthologs were analyzed using ClustalW (Goujon et al., 2010).Figure 2: Total mRNA expression of the IGF2 gene in different tissues of the Muscovy duck.
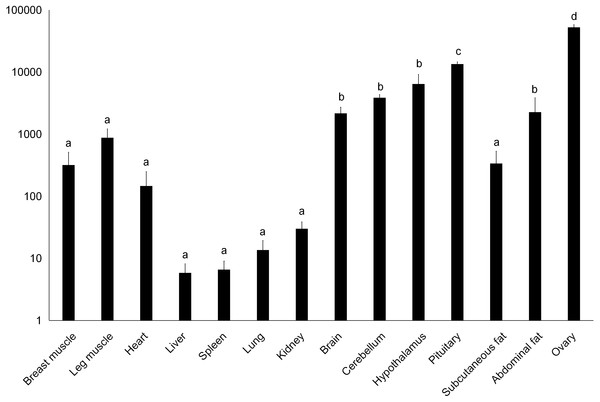
The value in the Y axis indicated 2−ΔΔCt value.Figure 3: Total mRNA expression of the DRD2 gene in different tissues of the Muscovy duck.
The value in the Y axis indicated 2−ΔΔCt value.Results
Characterization of Muscovy duck IGF2 and DRD2 coding region
We obtained a 311-bp partial cDNA of the IGF2 gene that was 98% and 95% identical to Anas platyrhynchos (XM_013191560.1) and Anser cygnoides domesticus (XM_005019778.2), respectively. We obtained the full-length cDNA of DRD2 including a 52-bp 5′-untranslated region (UTR), an 1,104-bp open reading frame (ORF) containing 368 codons and a 294-bp 3′-UTR. The Muscovy duck DRD2 cDNA sequence was 98% and 96% to Anas platyrhynchos (XM_013109686.1) and Anser cygnoides domesticus (XM_013187289.1), respectively. A phylogenetic tree constructed based on the DRD2 gene also revealed that the Muscovy duck was closely related with both animals above (Fig. 1).
Tissue expression of IGF2 and DRD2 genes
We examined tissue-specific expression of IGF2 and found that it was expressed in most tissues. The highest expression levels were found in the kidney and ovaries (Fig. 2). DRD2 expression was the highest in ovary, but it was also expressed in the cerebrum, cerebellum, hypothalamus and pituitary at lower levels. However, other tissues also had expression levels near detection limits including abdominal fat, sebum and breast and leg muscle. Expression in the spleen was negligible (Fig. 3).
Polymorphisms of IGF2 and DRD2 Genes
We identified 5 SNPs in the 5′ flanking region of IGF2, a level of one SNP per 449 bp on average. These SNPs were A-1864G, C-1704G, A-584G, A-227G and A-183G (Table 1). We found 28 SNPs in DRD2 giving rise to one SNP per 317 bp on average. Among them, the SNP C+7T in exon 1 was a missense mutation resulting in a P to S amino acid change (Table 1). We selected 2 SNPs of IGF2 and 11 SNPs of DRD2, based on mixed pool sequencing results which indicated that these were more likely to be associated with egg laying traits, for further association analysis.
Association of IGF2 and DRD2 with egg production traits
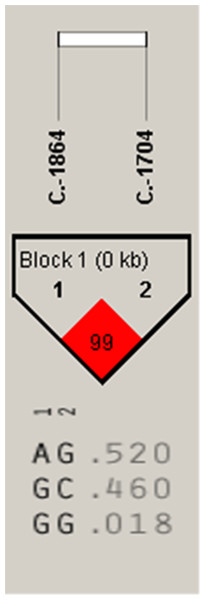
Association analysis indicated that the A-1864G and C-1704G SNPs of IGF2 gene were both significantly associated with E59W (P < 0.05) (Table 2), and linkage disequilibrium analysis indicated a high linkage block between A-1864G and C-1704G for IGF2 (Fig. 4). Multiple comparisons of different genotypes showed that the AG genotype individuals of A-1864G had 6–7 eggs more than GG genotype individuals for E59W (P < 0.01). The GG genotype individuals of C-1704G had 7–8 eggs more than individuals with the CC genotype for E59W (P < 0.05).
| No. | Gene | SNPsa | Locationb | Amino acid change |
|---|---|---|---|---|
| 1 | IGF2 | A-1864G | 5′regulatory region | No |
| 2 | IGF2 | C-1704G | 5′regulatory region | No |
| 3 | IGF2 | A-584G | 5′regulatory region | No |
| 4 | IGF2 | A-227G | 5′regulatory region | No |
| 5 | IGF2 | A-183G | 5′regulatory region | No |
| 6 | DRD2 | C-300G | 5′regulatory region | No |
| 7 | DRD2 | A-251T | 5′regulatory region | No |
| 8 | DRD2 | T-237G | 5′regulatory region | No |
| 9 | DRD2 | A-194G | 5′regulatory region | No |
| 10 | DRD2 | A-84G | 5′regulatory region | No |
| 11 | DRD2 | C+7T | Exon 1 | Yes (P-S) (ccc-tcc) |
| 12 | DRD2 | C+364G | Intron 1 | No |
| 13 | DRD2 | A+476T | Intron 1 | No |
| 14 | DRD2 | T+830G | Intron 1 | No |
| 15 | DRD2 | T+3024C | Intron 1 | No |
| 16 | DRD2 | A+3183C | Intron 2 | No |
| 17 | DRD2 | A+3262G | Intron 2 | No |
| 18 | DRD2 | C+3301G | Intron 2 | No |
| 19 | DRD2 | T+3423C | Intron 2 | No |
| 20 | DRD2 | T+3428C | Intron 2 | No |
| 21 | DRD2 | A+3484T | Intron 2 | No |
| 22 | DRD2 | A+3489G | Intron 2 | No |
| 23 | DRD2 | C+3545G | Intron 2 | No |
| 24 | DRD2 | T+6859G | Intron 5 | No |
| 25 | DRD2 | T+6986C | Intron 5 | No |
| 26 | DRD2 | T+7099C | Intron 5 | No |
| 27 | DRD2 | T+7295C | Intron 5 | No |
| 28 | DRD2 | T+7537C | Exon 6 | No |
| 29 | DRD2 | C+7654G | 3′regulatory region | No |
| 30 | DRD2 | T+8309G | 3′regulatory region | No |
| 31 | DRD2 | A+8442G | 3′regulatory region | No |
| 32 | DRD2 | T+8585C | 3′regulatory region | No |
| 33 | DRD2 | A+8770G | 3′regulatory region | No |
Notes:
Figure 4: The linkage status of 2 identified SNPs in IGF2 gene.
The color of block indicates the LD status of SNPs; deep red means high linkages between two SNPs.| SNPs1 | Traits2 | Least-squares mean ± SEM 3 | P-value | ||
|---|---|---|---|---|---|
| AA(n = 204) | AG(n = 308) | GG(n = 172) | |||
| A-1864G | FEA | 276.60 ± 1.41a | 275.67 ± 1.15a | 276.77 ± 1.54a | 0.8087 |
| E59W | 75.35 ± 1.92a | 76.01 ± 1.57a | 69.18 ± 2.10b | 0.0251 | |
| E300D | 21.43 ± 1.04a | 21.91 ± 0.85a | 20.84 ± 1.14a | 0.7533 | |
| CC(n = 158) | CG(n = 310) | GG(n = 216) | |||
| C-1704G | FEA | 276.50 ± 1.61a | 276.43 ± 1.15a | 275.72 ± 1.37a | 0.9067 |
| E59W | 68.92 ± 2.19b | 75.33 ± 1.56a | 76.11 ± 1.87a | 0.0254 | |
| E300D | 20.97 ± 1.19a | 21.18 ± 0.85a | 22.33 ± 1.01a | 0.6050 | |
Notes:
Data are summarized as means ± SEM.
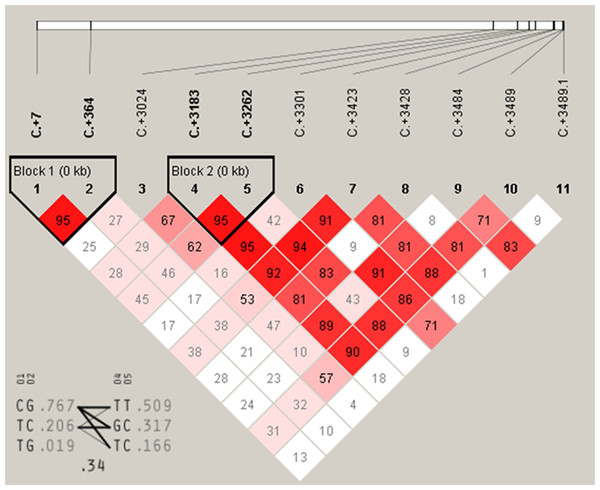
Association analysis for DRD2 gene further showed that C+7T and C+364G had highly significant associations with FEA and E300D (P < 0.01) and were significantly associated with E59W (P < 0.05) (Table 3). A+3489G, A+3484T and T+3428C were significantly associated with FEA and E300D (P < 0.05), and highly associated with E59W (P < 0.01). T+3423C and A+3262G were significantly associated with FEA (P < 0.05) and highly significantly associated with E59W (P < 0.01). A+3183C was significantly associated with E59W (P < 0.05), and T+3024C has no significant association with any of the three egg production traits. Moreover, it was notable that C+3301G and C+3545G were highly significantly associated with FEA, E59W and E300D (P < 0.01). Multiple comparisons among different genotypes showed that the GG genotypes of C+3301G and C+3545G were advantageous for earlier egg laying and egg production. There were two high linkage blocks (C+7T and C+364G, A+3183C and A+3262G) for DRD2 (Fig. 5).
| SNPs1 | Traits2 | Least-squares mean ± SEM3 | P-value | ||
|---|---|---|---|---|---|
| CC(n = 387) | CT(n = 237) | TT(n = 31) | |||
| C+7T | FEA | 272.95 ± 0.90c | 276.30 ± 1.15b | 295.94 ± 3.17a | <0.0001 |
| E59W | 74.62 ± 1.35a | 75.34 ± 1.73a | 61.13 ± 4.78b | 0.0187 | |
| E300D | 22.90 ± 0.71a | 21.81 ± 0.91a | 8.42 ± 2.51b | <0.0001 | |
| CC(n = 22) | CG(n = 239) | GG(n = 394) | |||
| C+364G | FEA | 297.00 ± 3.80a | 275.64 ± 1.15b | 273.79 ± 0.90b | <0.0001 |
| E59W | 62.32 ± 5.67b | 77.15 ± 1.72a | 73.14 ± 1.34ab | 0.0193 | |
| E300D | 9.86 ± 3.01b | 22.37 ± 0.91a | 22.16 ± 0.71a | 0.0003 | |
| TT(n = 130) | TC(n = 160) | CC(n = 410) | |||
| T+3024C | FEA | 276.27 ± 1.75a | 275.21 ± 1.58a | 277.19 ± 0.98a | 0.5547 |
| E59W | 79.02 ± 2.37a | 72.81 ± 2.13ab | 72.77 ± 1.33b | 0.0594 | |
| E300D | 21.94 ± 1.30a | 21.37 ± 1.17a | 20.80 ± 0.73a | 0.7271 | |
| CC(n = 143) | AC(n = 182) | AA(n = 375) | |||
| A+3183C | FEA | 276.84 ± 1.67a | 278.16 ± 1.48a | 275.67 ± 1.03a | 0.3816 |
| E59W | 69.38 ± 2.25b | 72.85 ± 2.00ab | 76.21 ± 1.39a | 0.0301 | |
| E300D | 21.02 ± 1.24a | 19.98 ± 1.09a | 21.75 ± 0.76a | 0.4124 | |
| GG(n = 205) | AG(n = 269) | AA(n = 226) | |||
| A+3262G | FEA | 278.86 ± 1.39a | 276.85 ± 1.21ab | 274.15 ± 1.32b | 0.0466 |
| E59W | 69.75 ± 1.88b | 73.88 ± 1.64ab | 77.80 ± 1.79a | 0.0084 | |
| E300D | 19.50 ± 1.03b | 21.06 ± 0.90ab | 22.72 ± 0.98a | 0.0766 | |
| GG(n = 132) | CG(n = 231) | CC(n = 337) | |||
| C+3301G | FEA | 272.05 ± 1.72b | 279.36 ± 1.30a | 276.42 ± 1.08a | 0.0033 |
| E59W | 80.70 ± 2.34b | 73.11 ± 1.77a | 71.86 ± 1.46a | 0.0052 | |
| E300D | 24.77 ± 1.28a | 19.26 ± 0.96b | 21.01 ± 0.80b | 0.0028 | |
| TT(n = 135) | TC(n = 245) | CC(n = 320) | |||
| T+3423C | FEA | 277.77 ± 1.71a | 278.80 ± 1.27a | 274.35 ± 1.11b | 0.0226 |
| E59W | 67.96 ± 2.31b | 73.13 ± 1.72ab | 77.08 ± 1.50a | 0.0038 | |
| E300D | 20.13 ± 1.27ab | 19.89 ± 0.94b | 22.53 ± 0.82a | 0.0730 | |
| TT(n = 47) | TC(n = 154) | CC(n = 499) | |||
| T+3428C | FEA | 270.64 ± 2.90b | 275.08 ± 1.60ab | 277.58 ± 0.89a | 0.0422 |
| E59W | 85.72 ± 3.92a | 76.16 ± 2.16b | 72.14 ± 1.20b | 0.0022 | |
| E300D | 25.87 ± 2.15a | 22.16 ± 1.19ab | 20.38 ± 0.66b | 0.0317 | |
| TT(n = 145) | AT(n = 141) | AA(n = 414) | |||
| A+3484T | FEA | 272.65 ± 1.65b | 278.96 ± 1.67a | 277.13 ± 0.97a | 0.0183 |
| E59W | 79.86 ± 2.23a | 74.19 ± 2.27a | 71.78 ± 1.32b | 0.0080 | |
| E300D | 24.17 ± 1.22a | 19.40 ± 1.24b | 20.67 ± 0.72b | 0.0141 | |
| GG(n = 140) | AG(n = 218) | AA(n = 342) | |||
| A+3489G | FEA | 278.66 ± 1.68a | 278.69 ± 1.34a | 274.35 ± 1.07b | 0.0159 |
| E59W | 67.56 ± 2.27b | 72.51 ± 1.82b | 77.46 ± 1.45a | 0.0008 | |
| E300D | 19.99 ± 1.24ab | 19.56 ± 1.00b | 22.61 ± 0.80a | 0.0343 | |
| GG(n = 198) | CG(n = 180) | CC(n = 322) | |||
| C+3545G | FEA | 272.70 ± 1.41b | 276.78 ± 1.47a | 278.81 ± 1.10a | 0.0029 |
| E59W | 79.74 ± 1.91a | 73.11 ± 2.00b | 70.84 ± 1.49b | 0.0011 | |
| E300D | 24.26 ± 1.04a | 21.02 ± 1.09b | 19.29 ± 0.82b | 0.0009 | |
Notes:
Data are summarized as means ± SEM.
Discussion
Muscovy duck is an excellent poultry, but its egg production is low, which has been plaguing farmers and breeders. In recent years, molecular marker breeding has gradually become the mainstream of breeding, and many breeders try to improve egg laying performances through breeding methods of molecular markers in poultry (Wang et al., 2014a; Wang et al., 2014b; Fulton et al., 2012; Uemoto et al., 2009). Using molecular marker to improve Muscovy ducks egg production is an effective method which will greatly improve the economic value of Muscovy ducks. Our study focused on egg production traits and related molecular markers, and we tried to find some molecular markers highly related to egg production in Muscovy ducks, with the hope that they can be used in Muscovy duck breeding. We believe that the relevant personnel of Muscovy ducks industry will have a strong interest in this study.
In the present study, we obtained the coding regions of IGF2 and DRD2 in Muscovy duck for the first time, which will be a great help in future research. IGF2 and DRD2 genes in humans, mice and chickens all have transcript variants (Kaalund et al., 2014; Wernersson et al., 2016; Johannessen et al., 2016). However, we only found one transcript in Muscovy duck. This may be caused by differences between different species.
High expression of IGF2 in the ovary may be related to follicular development in zebra fish (Irwin & Van Der Kraak, 2012). In our study, we found that IGF2 is widely expressed in different tissues with the highest expression in ovary. This suggests that IGF2 may be associated with ovarian development. The ovarian functions of birds are regulated by luteinizing hormone (LH) and follicle stimulating hormone (FSH). IGF2 can stimulate granule cell proliferation and related hormones synthesis and regulate follicle development with FSH in mammals (Lucy, 2011). Previous studies have found that IGF2 expression in the ovary directly affects the development of dominant follicles in rats (Wang, Asselin & Tsang, 2002). IGF1 can inhibit the apoptosis of granulosa cells, while IGF2 might regulate cell proliferation during follicular development in chicken (Johnson, Bridgham & Swenson, 2009). In addition, IGF2 expression in the follicles of highly productive chickens are significantly higher than that in lowly productive chickens. Therefore, a relationship exists between the expression of IGF2 in the ovary and egg production in chickens (Kim, Seo & Ko, 2004). It is also becoming clear from in vivo and in vitro studies carried out in birds that IGF2 plays an important role in ovarian follicular development (Wood, Schlueter & Duan, 2005). All these studies indicate that IGF2 is related to the development of ovary. Thus, we deduced that IGF2 might play a key role in ovarian follicular development of Muscovy ducks and regulate egg production.
In this study, we found that Muscovy DRD2 also had its highest expression in the ovary. The DRD2 gene belongs to the catecholamine neurotransmitter receptors that exist widely in central and peripheral nervous tissues. DRD2 is highly expressed in the ovary and this may be related to follicular and ovarian development in humans (Morton et al., 2006). Other studies identified high DRD2 expression in the regulation of reproductive functions in the grey mullet (Nocillado et al., 2007). DRD2 agonist can inhibit the production and secretion of vascular endothelial growth factor protein in human granulosa cells (Ferrero et al., 2014). Together these findings indicate that DRD2 may have a function in follicular and ovarian development. Therefore, we selected IGF2 and DRD2 as a candidate gene related to egg laying traits for further study.
Figure 5: The linkage status of 11 identified SNPs in DRD2 gene.
The color of block indicates the LD status of SNPs; deep red means high linkages between two SNPs.IGF2 is important in body growth and development. Most research on IGF2 has concentrated on growth studies and the association of IGF2 polymorphisms with growth related traits. Few studies have investigated the association between IGF2 and egg laying traits. But in the current study, we found the high linkage sites A-1864G and C-1704G of IGF2 were significantly associated with E59W. This indicated that IGF2 was positively related to egg laying traits. However, we have not studied how those two loci of IGF2 regulate egg laying performance. A future study should focus on the function of the two loci for egg laying performance. Recently, DRD2 polymorphisms have been related to poultry egg production. Our previous studies found the chicken DRD2 gene polymorphisms were correlated with the first egg age and the egg numbers at 300 days in chicken (Xu et al., 2011a; Xu et al., 2011b). SNPs of DRD2 were significantly associated with egg production at 38 weeks and egg weight at 300 days in chicken (Zhu et al., 2015). These studies suggest that the DRD2 is indeed associated with the laying performance of birds. In our study, we also found a link between DRD2 and the laying performance of birds. We found 10 SNPs of DRD2 gene (C+7T, C+364G, A+3183C, A+3262G, C+3301G, T+3423C, T+3428C, A+3484T, A+3489G and C+3545G) were significantly associated with egg production traits, and two high linkage blocks were found in haplotype analysis. According to our studies, IGF2 and DRD2 are indeed related to the laying performance of birds, but the specific functions of these SNPs remain to be studied.
In conclusion, we identified two SNPs of IGF2 and 11 for DRD2, which were highly correlated with egg laying performance in Muscovy ducks. These molecular markers highly associated with egg production traits can be used in Muscovy duck breeding. It is conducive to the development of the whole industry of Muscovy ducks. However, the functional mechanisms of these SNPs affecting egg production await further investigation.
Supplemental Information
Supplementary materials
Table S1. Fifteen pairs of primers used in this study.
Table S2. Raw data of association at IGF2 gene.
Table S3. Raw data of association at DRD2 gene.
Table S4. Raw data of association at DRD2 gene.