Temporal regulation of proteome profile in the fruit fly, Drosophila melanogaster
- Published
- Accepted
- Received
- Academic Editor
- Juan Riesgo-Escovar
- Subject Areas
- Biochemistry, Bioinformatics, Entomology, Molecular Biology
- Keywords
- Proteome, Drosophila melanogaster, Circadian, Metabolism, Mass spectrometry
- Copyright
- © 2016 Subramanian et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2016. Temporal regulation of proteome profile in the fruit fly, Drosophila melanogaster. PeerJ 4:e2080 https://doi.org/10.7717/peerj.2080
Abstract
Background. Diurnal rhythms of protein synthesis controlled by the biological clock underlie the rhythmic physiology in the fruit fly, Drosophila melanogaster. In this study, we conducted a proteome-wide investigation of rhythmic protein accumulation in D. melanogaster.
Materials and Methods. Total protein collected from fly samples harvested at 4 h intervals over the 24 h period were subjected to two-dimensional gel electrophoresis, trypsin digestion and MS/MS analysis. Protein spots/clusters were identified with MASCOT search engine and Swiss-Prot database. Expression of proteins was documented as percentage of volume contribution using the Image Master 2D Platinum software.
Results. A total of 124 protein spots/clusters were identified using MS/MS analysis. Significant variation in the expression of 88 proteins over the 24-h period was observed. A relatively higher number of proteins was upregulated during the night compared to the daytime. The complexity of temporal regulation of the D. melanogaster proteome was further reflected from functional annotations of the differently expressed proteins, with those that were upregulated at night being restricted to the heat shock proteins and proteins involved in metabolism, muscle activity, protein synthesis/folding/degradation and apoptosis, whilst those that were overexpressed in the daytime were apparently involved in metabolism, muscle activity, ion-channel/cellular transport, protein synthesis/folding/degradation, redox homeostasis, development and transcription.
Conclusion. Our data suggests that a wide range of proteins synthesized by the fruit fly, D. melanogaster, is under the regulation of the biological clock.
Introduction
A broad spectrum of physiological, cellular, biochemical, endocrinological and molecular functions in living systems display temporal 24 h rhythms. As a consequence, the extent of biological processes regulated by biological clock vary from sleep-wake patterns, body temperature, activities of numerous enzymes, hormones, synthesis of nucleic acids and cell division (Moore-Ede, Sulzman & Fuller, 1982). As various aspects of cell metabolism and cell division cycle are regulated by biological clock (Asher & Schibler, 2011; Bass & Takahashi, 2010) it could be easily hypothesized that proteome profile of a living organism could be circadian in nature.
Previous studies showed that 30% of mRNA transcripts exhibit circadian variation (Lück et al., 2014). The circadian regulation of posttranslational processes has also been revealed by proteomic studies of the circadian rhythm (Robles, Cox & Mann, 2014) and studies on the variation of the cerebrospinal fluid over the light-dark cycle have been reported (Teixeira-Gomes et al., 2015). Earlier, the global level of circadian proteome of whole mouse liver (inclusive of various functional parts—left and right triangular ligaments, fissure for ligamentum teres, fissure for ligamentum venosum, hepatic veins, etc.,) has been investigated by Reddy et al. (2006). This report revealed a contrasting variation of protein profile between day and night. Whilst many genes have been demonstrated to coordinate rhythms in RNA synthesis, splicing and translation, numerous others also exhibited significant temporal disconnections between these functions (reviewed in Beckwith & Yanovsky, 2014). Hence, circadian oscillations represent a perfect system to comprehend how manifold transcriptional and post-translational processes are integrated rhythmically to maximize the fine-tuning of functions of organisms to the environment cycle.
Although rigorous research has been carried out on molecular genetics and developmental studies in the fruit fly, Drosophila melanogaster, little is known about the proteome of the fly. Proteomics is a central aspect in systems biology of the fruit fly that appends a distinctive dimension in investigating gene function and regulatory mechanisms. Hence, the temporal pattern of proteome profile would add useful information for further research and analysis. Search of PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) references illustrated that by the end of October 2015 articles on Drosophila proteomics constitute only 0.95% of all published papers on proteomics.
The circadian clock is conventionally thought to exist in the lateral neurons of the fly brain (Kaneko, Helfrich-Forster & Hall, 1997). Intriguingly, brain-independent circadian oscillations have been perceived in almost all peripheral tissues of D. melanogaster. For instance, the same circadian rhythm of β-galactosidase expression has been demonstrated in the Malpighian tubules of decapitated and non-decapitated flies bearing the per-lacZ reporter transgene (Hege et al., 1997). In addition, Plautz et al. (1997a) has demonstrated the appearance, disappearance and reappearance of PERIOD protein in a rhythmic (24 h) pattern in the head, abdomen, thorax, legs and wings of the fly, indicating that numerous cellular processes all over the body of the fly are regulated by the temporally oscillating circadian clock gene, period. It is in this context, the present study has been carried out.
Currently, very little has been documented about the rhythmic build-up of proteins in D. melanogaster (Mauvoisin et al., 2014). To investigate the overall circadian transcriptional regulation in D. melanogaster, Rodriguez et al. (2013) separated nascent RNA from fly heads at six time points over a 24 h period (00:00, 04:00, 08:00, 12:00, 16:00 and 20:00 in 12:12 h light-dark cycle) and their data specified a key role of posttranscriptional control to fly’s circadian mRNA oscillation. Following this temporal schedule, we have investigated the overall pattern of temporal proteome (i) to complement the available data in fly literature, and (ii) to reveal the integrated pattern/regulation of circadian proteome in the whole body of the fly.
Materials and Methods
Drosophila culture and sample collection
D. melanogaster (wild type-Canton S) flies were maintained on medium comprising maize powder, sucrose, yeast and nepagin (anti-fungal agent) at 21 ± 2 °C under 12 h:12 h (light:dark) phases. We have used the whole fly for the proteomic study as performed in typical proteotypic peptide (PTP) studies reported earlier (Brunner et al., 2007) and followed the same protocol with trypsin digestion. The adult male flies (seven days old) were collected at 4 h intervals (Rodriguez et al., 2013) over a 24 h period (at 00:00, 04:00, 08:00, 12:00, 16:00 and 20:00). The flies collected at each time point (n = 15) were suspended in sample solubilization solution (100 µL) containing equal volume of SDS (1%) and β-mercaptoethanol (5%) and were swiftly frozen in liquid nitrogen. The flies were homogenized and the homogenate was kept at −80 °C until analysis. The proteins were solubilized at 95 °C in sample solubilization solution and vortexed. The miniature cuticle residues were sedimented by centrifugation at 8,200 g (5 min). The solubilized proteins were precipitated in TCA (20%) in cold acetone (90%) along with dithiothreitol (DTT, 20 mM) on ice (Jessie, Hashim & Rahim, 2008).
Assay for protein estimation
The total protein content of fly homogenate was estimated (Bradford, 1976) after pre-treatment and re-solubilization of protein pellet with sodium hydroxide (0.2 M) and rehydration buffer (urea (7 M), thiourea (2 M), CHAPS (4%, 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate) and bromophenol blue (0.002%)), respectively, as previously described (Jessie, Hashim & Rahim, 2008).
2-D gel electrophoresis (2DE) and silver staining
2DE was performed with 50 µg of precipitated (TCA/acetone) fly proteins. The proteins were incubated in rehydration buffer (urea (7 M), thiourea (2 M), CHAPS (4%), IPG buffer (pH 3–10 NL), DTT (65 mM) and bromophenol blue (0.002%)) at 25 °C for about 12 h and loaded onto 13 cm rehydrated precast immobilized dry strips (pH 3–10 non-linear, GE Healthcare BioSciences, Uppsala, Sweden). The strips were subjected to isoelectric focusing with EttanIPGhor 3 Isoelectric Focusing Unit (GE Healthcare, Uppsala, Sweden) for a total time of 20 kV/h. Focused strips were equilibrated in Tris–HCl (1.5 M, pH 8.8 with urea (6 M), SDS (2%), glycerol (30%) and DTT (0.06 M)) for 20 min and subsequently incubated in a similar equilibration solution but containing iodoacetamide (4.5%) in lieu of DTT for 20 min. The equilibrated strips were then overlaid onto homogenous polyacrylamide gel (12.5%) and electrophoresis was carried using the SE 600 Ruby Electrophoresis System and Power Supply-EPS601 (GE Healthcare), following a protocol as reported earlier (Jessie, Hashim & Rahim, 2008). The 2DE gels were then developed by silver staining (Heukeshoven & Dernick, 1988). All samples (at each time point) were examined independently in triplicate. A modified silver staining protocol was performed for visualization of proteins well-suited for MALDI-ToF/ToF mass spectrometry (MS) investigation (Yan et al., 2000).
Image analysis
2DE gels (silver stained) were scanned using Imaging Densitometer GS690 (Bio-Rad Laboratories, Hercules, CA, USA). Expression level of proteins was calculated in terms of percentage of volume contribution using the Image Master 2D Platinum software, version 7.0 (GE Healthcare Biosciences, Uppsala, Sweden) by selecting the particular spot of the 2DE, matching it with the same spot of another replicate (of the same time point) for which a matchset has been already created using the software. Cut-off parameters for this analysis were: Smooth—2; Saliency—1; Min area—5 (Jayapalan et al., 2012; Jayapalan et al., 2013).
Trypsin digestion and mass spectrometry
Identification of protein spots of interest was performed as described previously (Jayapalan et al., 2012; Jayapalan et al., 2013). Further, spots were carefully excised from 2DE gels and destained with potassium ferricyanide (15 mM) and sodium thiosulfate (50 mM) for 20 min at about 20 °C. The proteins were reduced with 10 mM DTT (10 mM) and ammonium bicarbonate (100 mM) for 25 min, and alkylated with 55 mM iodoacetamide (55 mM) for 15 min, at 60 °C and in the dark, respectively. This was followed by subsequent washings with 50 and 100% acetonitrile (ACN, 50 and 100%) in 100 mM ammonium bicarbonate (100 mM), and dehydration of the gel plugs using vacuum centrifugation. The spots were digested with trypsin (6 ng/µl in ammonium bicarbonate solution (50 mM)) at 37 °C, for about 12 h. Peptides were extracted from the gels using ACN (50 and 100 %) subsequently. Extracted peptides were lyophilized, treated with formic acid (0.1%) and desalted using ZipTip columns containing C18 reversed phase media (Millipore, Madison, USA). The sample peptide was mixed with α-cyano-4-hydroxycinamic acid (5 mg/ml) at a ratio of 1:1, and 0.7 µl of the mixture was immediately spotted onto an OptiToF 384-well insert and analyzed using 5,800 MALDI ToF/ToF analyser (ABSciex, Toronto, Canada).
Identification of proteins
The proteins in the spots/clusters were identified using the MASCOT search engine (Jayapalan et al., 2012; Jayapalan et al., 2013). The MS data acquired was searched against Drosophila melanogaster in the Swiss-Prot database (last update: 21 April 2015, 3,067 sequences) in accordance with the following selection parameters: enzyme-trypsin, missed cleavage—1, variable modification—2; (i) carbamidomethylation of cysteine and (ii) oxidation of methionine, MS precursor ion mass tolerance—100 ppm, MS/MS fragment ion tolerance −0.2 Da and inclusion of monoisotopic masses only.
Statistical analysis
Percentage of volume contribution was expressed as mean ± SD. The Statistical Package for Social Sciences (SPSS) version 22.0 (IBM Corporation, New York, NY, USA) was used to analyze the data. The test of homogeneity was employed to evaluate the sample distribution of the dataset. In the study, maximum numbers of protein spots were recorded at 04:00 and 20:00. Hence, the percentages of volume contribution of spots/clusters of these time points were used for comparison with other time points. The Student’s t-test was subsequently used to compare means of percentage of volume contribution of the spots (04:00 and 20:00) with other time points (individually) of all datasets that showed normal distribution. A p value of <0.01 was deemed significant.
Results
Figures 1A–1F demonstrate the representative 2DE profiles of D. melanogaster at 00:00, 04:00, 08:00, 12:00, 16:00 and 20:00. Marked variations in intensity of the protein spots/ clusters were apparent over the 24 h period. A total of 124 protein spots/clusters, which were classified into 11 groups based on known or predictive functions, were identified by MS/MS analysis and database query (Table 1). In this analysis, matched peptides for 2 or more reflect better confidence of the results. A few protein spots, which were not resolved at all time points, were excluded from the analysis. Multiple hits for single protein spots (e.g., spot/cluster Nos. 57 and 200, 104 and 135, 86 and 175 and 120 and 204) were also observed.
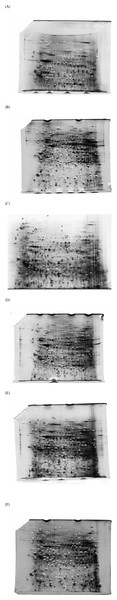
Figure 1: Representative 2DE photographs of proteome profile of Drosophila melanogaster at 4-h intervals over a period of 24-h.
The protein spots/cluters are labeled in black (or) white for easy visualization. (A) 00:00, (B) 04:00, (C) 08:00, (D) 12:00, (E) 16:00 and (F) 20:00. At each time point, 2DE was performed in triplicate. Expression levels of the spots/clusters are designated as percentage of volume contribution using the Image Master 2D Platinum software, version 7.0.When image analysis of the 2DE gels was performed, expression of 88 proteins was found to demonstrate significant variation over the 24 h period (Table 2). Whilst 45 appeared upregulated during the nighttime, 43 were overexpressed in the daytime (Table 2 and Fig. 2). The complexity of temporal regulation of D. melanogaster proteome was further reflected from functional annotations of the differently expressed proteins, with those that were upregulated during the nighttime being restricted to the heat shock proteins and proteins involved in metabolism, muscle activity, protein synthesis/folding/degradation and apoptosis, whilst those that were overexpressed in the daytime were apparently involved in metabolism, muscle activity, ion-channel/cellular transport, protein synthesis/folding/degradation, redox homeostasis, development and transcription. In addition, proteins which showed significant variation in percentage of volume contribution in at least 2 time points were apparently from six different functional groups (i.e., metabolism, muscle activity, ion-channel/cellular transport, heat shock proteins, protein synthesis/folding/degradation and miscellaneous (redox homeostasis, apoptosis, development and transcription); (Table 2 and Figs. 3A–3I). The percentage of volume contribution of all 124 protein spots/clusters are shown in the Table S1.
| S. No. | Spot/ Cluster ID/No. | Protein identification | Primary accession number | Theoretical mass (Da) | Calculated pI | Peptide score | No. of peptides matched | Sequence coverage (%) |
|---|---|---|---|---|---|---|---|---|
| Metabolism | ||||||||
| 1 | 12 | 6-phosphofructokinase | P52034 | 86593 | 6.41 | 43 | 3 | 5 |
| 2 | 25 | Succinate dehydrogenase | Q94523 | 72297 | 6.65 | 149 | 10 | 21 |
| 3 | 32 | N-glycanase | Q28YQ7 | 73443 | 8.15 | 26 | 1 | 3 |
| 4 | 41 | Maltase H | P07190 | 66344 | 4.75 | 82 | 5 | 10 |
| 5 | 44 | Vacuolar ATP synthase catalytic subunit A | Q27331 | 68259 | 5.23 | 148 | 13 | 22 |
| 6 | 51 | Vacuolar ATP synthase subunit B | P31409 | 54515 | 5.25 | 131 | 10 | 27 |
| 7 | 61 | Enolase | P15007 | 54276 | 8.68 | 415 | 11 | 32 |
| 8 | 63 | Phosphoglycerate kinase | Q01604 | 43834 | 7.01 | 223 | 6 | 18 |
| 9 | 64 | ATP synthase subunit α | ACP35381 | 59384 | 9.09 | 332 | 5 | 15 |
| 10 | 68 | Arginine kinase | P48610 | 39841 | 6.04 | 363 | 7 | 26 |
| 11 | 69 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 525 | 10 | 34 |
| 12 | 71 | Arginine kinase | P48610 | 39841 | 6.04 | 291 | 10 | 38 |
| 13 | 72 | Glyceraldehyde-3-phosphate dehydrogenase | P07486 | 35328 | 8.26 | 46 | 2 | 8 |
| 14 | 74 | Glyceraldehyde-3-phosphate dehydrogenase | P07486 | 35328 | 8.26 | 332 | 5 | 25 |
| 15 | 75 | Glyceraldehyde-3-phosphate dehydrogenase 1 | P07486 | 35328 | 6.44 | 306 | 3 | 15 |
| 16 | 79 | Alcohol dehydrogenase | P00334 | 27744 | 7.64 | 358 | 6 | 43 |
| 17 | 80 | Stellate protein CG33247 | Q7KV23 | 19396 | 7.64 | 28 | 1 | 5 |
| 18 | 81 | Stellate protein CG33247 | Q7KV23 | 19396 | 7.64 | 28 | 1 | 5 |
| 19 | 83 | Triosephosphate isomerase | Q7JNS1 | 26609 | 6 | 178 | 7 | 43 |
| 20 | 90 | ATP synthase subunit β | Q05285 | 54074 | 5.14 | 67 | 5 | 8 |
| 21 | 97 | ATP synthase D chain | Q24251 | 20188 | 6.1 | 81 | 5 | 33 |
| 22 | 98 | Tyrosine kinase 2 | Q9V3D5 | 79559 | 9.13 | 23 | 1 | 1 |
| 23 | 105 | ATP synthase subunit β | Q05825 | 54074 | 5.14 | 178 | 7 | 16 |
| 24 | 111 | Protein l(2)37Cc | P18432 | 23700 | 4.67 | 56 | 1 | 6 |
| 25 | 117/118 | ATP synthase subunit β | Q05825 | 54074 | 5.14 | 573 | 11 | 28 |
| 26 | 123 | ATP synthase subunit β | Q05825 | 54074 | 5.14 | 61 | 6 | 17 |
| 27 | 124 | Inorganic pyrophosphatase | O77460 | 37915 | 6.52 | 99 | 3 | 12 |
| 28 | 126 | ATP synthase subunit β | Q05825 | 54074 | 5.14 | 48 | 2 | 3 |
| 29 | 131 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 92 | 5 | 21 |
| 30 | 132 | Alcohol dehydrogenase | P00334 | 27744 | 7.74 | 198 | 5 | 28 |
| 31 | 133 | Pyruvate kinase | O62619 | 57404 | 7.13 | 59 | 3 | 7 |
| 32 | 140 | Phosphoglycerate kinase | Q01604 | 43834 | 7.01 | 195 | 7 | 15 |
| 33 | 152 | Phosphoglycerate kinase | Q01604 | 43834 | 7.01 | 217 | 5 | 15 |
| 34 | 155 | Glycerol-3-phosphate dehydrogenase | P13706 | 39659 | 6.17 | 96 | 7 | 26 |
| 35 | 156 | Isocitrate dehydrogenase | Q9VWH4 | 40818 | 6.96 | 63 | 7 | 21 |
| 36 | 157 | Glyceraldehyde-3-phosphate dehydrogenase | P07486 | 35328 | 8.26 | 106 | 2 | 8 |
| 37 | 159 | ATP synthase subunit α | P35381 | 59384 | 9.09 | 380 | 10 | 20 |
| 38 | 161 | Thioredoxin reductase I | P91938 | 64282 | 8.11 | 79 | 7 | 12 |
| 39 | 162 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 55 | 3 | 16 |
| 40 | 163 | Alcohol dehydrogenase | P00334 | 27744 | 7.74 | 105 | 3 | 18 |
| 41 | 165 | ATP synthase subunit α | P35381 | 59384 | 9.09 | 196 | 5 | 14 |
| 42 | 166 | Maltase | P07190 | 66344 | 4.75 | 92 | 7 | 15 |
| 43 | 170 | Pyruvate kinase | O62619 | 57404 | 7.13 | 35 | 2 | 4 |
| 44 | 171 | Glycerol-3-phosphate dehydrogenase | Q27556 | 38298 | 6.33 | 27 | 6 | 20 |
| 45 | 173 | ATP synthase subunit β | Q05825 | 54074 | 5.14 | 263 | 7 | 14 |
| 46 | 177 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 22 | 7 | 22 |
| 47 | 178 | ATP synthase subunit α | P35381 | 59384 | 9.09 | 77 | 3 | 6 |
| 48 | 183 | Alcohol dehydrogenase | P00334 | 27744 | 7.74 | 22 | 1 | 7 |
| 49 | 185 | ATP synthase subunit β | Q05825 | 54074 | 5.14 | 32 | 1 | 2 |
| 50 | 186 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 176 | 6 | 16 |
| 51 | 187 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 107 | 4 | 13 |
| 52 | 192 | Glyceraldehyde-3-phosphate dehydrogenase 1 | P07486 | 35328 | 8.26 | 40 | 3 | 12 |
| 53 | 193 | Cytochrome P450 | Q9VGB5 | 55583 | 9.39 | 27 | 2 | 2 |
| 54 | 195 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 62 | 4 | 15 |
| 55 | 196 | Fructose bisphosphate aldolase | P07764 | 39023 | 6.97 | 58 | 4 | 17 |
| 56 | 199 | Inorganic pyrophosphatase | O77460 | 37915 | 6.52 | 93 | 5 | 19 |
| 57 | 209 | Arginine kinase | P48610 | 39841 | 6.04 | 50 | 1 | 2 |
| Muscle activity | ||||||||
| 58 | 57 | Actin-87E | P10981 | 41775 | 5.3 | 132 | 6 | 20 |
| 59 | 59 | Actin-88F | P83967 | 41673 | 5.29 | 421 | 7 | 26 |
| 60 | 94 | Myosin light chain alkali | P06742 | 17513 | 4.29 | 89 | 2 | 16 |
| 61 | 102 | Actin-57B | P53501 | 41808 | 5.23 | 22 | 5 | 17 |
| 62 | 104 | Actin-88F | P83967 | 41673 | 5.29 | 413 | 5 | 22 |
| 63 | 106 | Actin-5C | P10987 | 41795 | 5.3 | 27 | 2 | 6 |
| 64 | 114 | Myosin regulatory light chain-2 | P10987 | 41795 | 5.3 | 215 | 6 | 20 |
| 65 | 115 | Actin-5C | P02574 | 41760 | 5.3 | 194 | 4 | 14 |
| 66 | 116 | Actin-79B | Q540X7 | 41760 | 5.3 | 194 | 4 | 14 |
| 67 | 125 | Actin-5C | P10987 | 41795 | 5.3 | 30 | 2 | 6 |
| 68 | 135 | Actin-88F | P83967 | 41673 | 5.29 | 316 | 6 | 20 |
| 69 | 136 | Actin-5C | P10987 | 41795 | 5.3 | 25 | 2 | 5 |
| 70 | 149 | Actin-5C | P10987 | 41795 | 5.3 | 182 | 3 | 11 |
| 71 | 151 | Synapse associated protein | Q960T2 | 56946 | 4.45 | 46 | 1 | 1 |
| 72 | 153 | Actin-5C | P10987 | 41795 | 5.3 | 27 | 1 | 2 |
| 73 | 154 | Tubulin α-1 chain | P06603 | 49876 | 5 | 173 | 5 | 14 |
| 74 | 158 | ADP-ribosylation factor-8 | Q9VHV5 | 21240 | 6.74 | 28 | 1 | 4 |
| 75 | 168 | Actin-79B | P02574 | 41760 | 5.3 | 190 | 8 | 23 |
| 76 | 169 | Actin-88F | P83967 | 41673 | 5.29 | 43 | 2 | 9 |
| 77 | 172 | Tubulin β-1 chain | Q24560 | 50115 | 4.76 | 30 | 2 | 3 |
| 78 | 174 | Actin-57B | P53501 | 41808 | 5.23 | 117 | 3 | 10 |
| 79 | 182 | Actin-87E | P10981 | 41775 | 5.3 | 46 | 2 | 7 |
| 80 | 184 | Actin-57B | P53501 | 41808 | 5.23 | 136 | 5 | 15 |
| 81 | 188 | Actin-57B | P53501 | 41808 | 5.23 | 103 | 5 | 17 |
| 82 | 189 | Actin-5C | P10987 | 41795 | 5.3 | 108 | 4 | 14 |
| 83 | 191 | Actin-42A | P02572 | 41797 | 5.3 | 41 | 2 | 7 |
| 84 | 194 | Tubulin α-3 chain | P06605 | 49859 | 5 | 78 | 1 | 3 |
| 85 | 198 | Actin-79B | P83967 | 41760 | 5.29 | 118 | 5 | 20 |
| 86 | 200 | Actin-87E | P10981 | 41775 | 5.3 | 194 | 5 | 20 |
| 87 | 201 | Actin-57B | P53501 | 41808 | 5.23 | 112 | 3 | 10 |
| 88 | 202 | Paramyosin | P35415 | 102277 | 5.47 | 36 | 5 | 7 |
| 89 | 203 | Actin-42A | P02572 | 41797 | 5.3 | 83 | 2 | 6 |
| 90 | 208 | Actin-5C | P10987 | 41795 | 5.3 | 45 | 3 | 9 |
| Heat shock proteins | ||||||||
| 91 | 31 | Heat shock 82 kDa protein | P02828 | 81814 | 4.91 | 23 | 4 | 9 |
| 92 | 42 | Heat shock 70 kDa protein | P29844 | 72216 | 5.22 | 84 | 6 | 12 |
| 93 | 43 | Heat shock 70 kDa protein | P11147 | 71087 | 5.36 | 188 | 8 | 16 |
| 94 | 46 | Heat shock 60 kDa protein | O02649 | 60771 | 5.38 | 26 | 1 | 1 |
| 95 | 180 | Heat shock factor protein | P22813 | 76886 | 4.87 | 27 | 1 | 3 |
| Ion-channel/cellular transport | ||||||||
| 96 | 54 | Calreticulin | P29413 | 46779 | 4.4 | 89 | 2 | 5 |
| 97 | 55 | Tubulin β-1 chain | Q24560 | 50115 | 4.76 | 149 | 9 | 19 |
| 98 | 78 | Voltage-dependent anion-selective channel (Porin) | Q94920 | 30531 | 7.74 | 387 | 7 | 36 |
| 99 | 96 | Calcium-transporting ATPase | P22700 | 111630 | 5.28 | 27 | 1 | 1 |
| 100 | 160 | ADP/ATP translocase | Q26365 | 34193 | 9.82 | 247 | 6 | 14 |
| 101 | 181 | Voltage-dependent anion-selective channel (Porin) | Q94920 | 30531 | 6.44 | 195 | 4 | 20 |
| 102 | 205 | Voltage-dependent anion-selective channel (Porin) | Q94920 | 30531 | 6.44 | 22 | 1 | 3 |
| 103 | 206 | Voltage-dependent anion-selective channel (Porin) | Q94920 | 30531 | 6.44 | 27 | 1 | 3 |
| 104 | 207 | Transient receptor potential locus C protein | P36951 | 29075 | 6.07 | 24 | 2 | 11 |
| Redox homeostasis | ||||||||
| 105 | 85 | Capon-like protein | Q8SXX4 | 77233 | 8.98 | 22 | 1 | 1 |
| 106 | 86 | Superoxide dismutase [Cu–Zn] | P61851 | 15689 | 5.67 | 162 | 8 | 62 |
| 107 | 120 | Glutathione-S-transferase | P41043 | 27596 | 4.57 | 224 | 5 | 19 |
| 108 | 134 | Catalase | ACP17336 | 57113 | 8.39 | 54 | 4 | 9 |
| 109 | 175 | Superoxide dismutase [Cu–Zn] | P61851 | 15689 | 5.67 | 56 | 3 | 24 |
| 110 | 197 | Glutathione-S-transferase | P41043 | 27596 | 4.57 | 74 | 3 | 9 |
| 111 | 204 | Glutathione-S-transferase | P20432 | 23851 | 6.75 | 61 | 1 | 4 |
| Protein synthesis/folding/degradation | ||||||||
| 112 | 50 | Protein disulfide isomerase | P54399 | 55746 | 4.72 | 65 | 2 | 5 |
| 113 | 99 | Furin-like protease | P30430 | 120917 | 6.25 | 22 | 1 | 1 |
| 114 | 121 | 40S ribosomal protein SA | P38979 | 30209 | 4.76 | 50 | 2 | 9 |
| 115 | 164 | Elongation factor 1α (EF-1α) | P08736 | 59384 | 9.09 | 52 | 4 | 9 |
| 116 | 167 | 40S ribosomal protein SA | P38979 | 30209 | 4.76 | 38 | 2 | 9 |
| 117 | 179 | 40S ribosomal protein S12 | P80455 | 15159 | 5.93 | 32 | 1 | 10 |
| Miscellaneous | ||||||||
| 118 | 100 | Thioredoxin peroxidase (apoptosis) | Q9V3P0 | 21724 | 5.52 | 122 | 4 | 26 |
| 119 | 176 | Caspase-8 precursor (apoptosis) | Q29IM7 | 57539 | 6.31 | 22 | 1 | 3 |
| 120 | 62 | Vitellogenin-2 precursor (development) | ACP02844 | 49630 | 7.74 | 56 | 6 | 12 |
| 121 | 87 | Protein stand still (development) | P92189 | 35753 | 9.28 | 26 | 1 | 2 |
| 122 | 190 | DNA polymerase α-catalytic subunit (replication) | P26019 | 169796 | 8.28 | 33 | 1 | 0 |
| 123 | 130 | RNA helicase (transcription) | Q6J5K9 | 144797 | 5.67 | 30 | 1 | 0 |
| 124 | 38 | Mitosis initiation protein fs(1)Ya (cell division) | P25028 | 77677 | 9.54 | 21 | 1 | 2 |
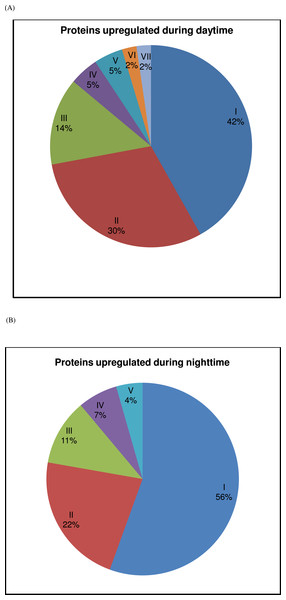
Figure 2: Contribution of protein groups over the 24-h period.
(A) The contribution of upregulated proteins of each group (I—metabolism, II—muscle activity, III—ion-channel/cellular transport, IV—protein synthesis/folding/degradation, V—redox homeostasis, VI—development and VII—transcription) during daytime (representing 08:00, 12:00 and 16:00) is represented. (B) The contribution of upregulated proteins of each group (I—metabolism, II—muscle activity, III—heat shock proteins, IV—protein synthesis/folding/degradation and V—apoptosis) during nighttime (representing 20:00, 00:00 and 04:00) is represented. See Table 2 for further details.Among the protein spots which showed significant variations over the 24 h period are various enzymes involved in metabolism ((6-phosphofructokinase, succinate dehydrogenase, N-glycanase, maltase H, vacuolar ATP synthase catalytic subunits A and B, enolase, ATP synthase subunits (α, β and D chain), fructose bisphosphate aldolase, arginine kinase, stellate protein CG33247 (protein kinase regulator), triosephosphate isomerase, protein l(2)37Cc (DOPA decarboxylase), inorganic pyrophosphatase, pyruvate kinase, phosphoglycerate kinase, glycerol-3-phosphate dehydrogenase, isocitrate dehydrogenase, thioredoxin reductase, alcohol dehydrogenase and cytochrome P450)). Others which showed significant variation of expression at different time points include: (i) proteins involved in muscular activities (various types of actin (88F, 57B, 5C, 79B, 87E and 42A), myosin regulatory light chain-2, synapse associated protein, ADP-ribosylation factor-8, tubulin α-3 chain and paramyosin), (ii) ion-channel/proteins involved in transport processes (calreticulin, tubulin β-1 chain, porin, ADP/ATP translocase and transient receptor potential locus C protein), (iii) different types of heat shock proteins (82 kDa, 70 kDa, 60 kDa and heat shock factor protein), (iv) proteins associated with synthesis/folding/degradation (protein disulfide isomerase, furin-like protease, 40 S ribosomal protein SA and S12 and elongation fator 1α (EF-1α), (v) redox homeostasis protein (glutathione-S-transferase) (vi) apoptosis proteins (caspase-8 precursor and thioredoxin peroxidase (also as antioxidant)), (vii) proteins involved in development (vitellogenin-2 precursor) and transcription (RNA helicase) (Table 2 and Figs. 3B–3I).
| S. No. | Spot/ Cluster ID/No. | Protein name | Significance level of % volumea | |||||
|---|---|---|---|---|---|---|---|---|
| 04:00 | 08:00 | 12:00 | 16:00 | 20:00 | 00:00 | |||
| Metabolism | ||||||||
| 1 | 12 | 6-phosphofructokinase | 0.0168 ± 0.080 | 0.013871 ± 0.0093 p = 0.00017c | 0.00355 ± 0.00336 p = 0.00026d | |||
| 2 | 25 | Succinate dehydrogenase | 0.05666 ± 0.03670 | 0.01156 ± 0.01067 p = 6.8929E–05b | 0.02717 ± 0.01942 p = 0.00980d | |||
| 3 | 32 | N-glycanase | 0.04920 ± 0.02726 | 0.02594 ± 0.01233 p = 0.00870b | 0.00113 ± 0.00013 p = 2.5021E–05c | 0.00715 ± 0.00467 p = 6.7294E–07d | ||
| 4 | 41 | Maltase H | 0.03721 ± 0.02519 | 0.04786 ± 0.06067 | 0.03036 ± 0.01416 | 0.01673 ± 0.01416 | 0.0195 ± 0.01690 | 0.000583 ± 0.00065 p = 0.00036bp = 0.00233c |
| 5 | 44 | Vacuolar ATP synthase catalytic subunit A | 0.00312 ± 0.02442 | 0.03587 ± 0.06408 | 0.04102 ± 0.03517 | 0.01178 ± 0.01333 | 0.02862 ± 0.01673 | 0.0080 ± 0.0015 p = 0.00771bp = 0.00177c |
| 6 | 51 | Vacuolar ATP synthase catalytic subunit B | 0.08049 ± 0.05789 | 0.06707 ± 0.03213 p = 0.00198c | 0.04721 ± 0.01989 | 0.01295 ± 0.00224 p = 7.1834E–05bp = 0.00198c | ||
| 7 | 61 | Enolase | 0.03289 ± 0.00521 | 0.05179 ± 0.07539 | 0.09340 ± 0.08054 p = 0.00139b | 0.12059 ± 0.11632 p = 0.00239b | 0.0561 ± 0.07604 | 0.00145 ± 0.00177 |
| 8 | 64 | ATP synthase subunit α | 0.01221 ± 0.01438 | 0.07412 ± 0.00754 p = 7.3863E–08bp = 5.7179E–06c | 0.01719 ± 0.0472 | 0.02236 ± 0.04303 | 0.01559 ± 0.00272 | 0.0345 ± 0.02371 |
| 9 | 69 | Fructose bisphosphate aldolase | 0.02904 ± 0.00464 | 0.05306 ± 0.03179 | 0.10312 ± 0.00128 p = 0.00186bp = 0.00093c | 0.0154 ± 0.0300 | 0.02551 ± 0.00406 | 0.05898 ± 0.02373 |
| 10 | 71 | Arginine kinase | 0.12971 ± 0.07577 | 0.13765 ± 0.02036 p = 0.00838c | 0.04893 ± 0.04735 | 0.00027 ± 0.00018 p = 0.0010b | ||
| 11 | 80 | Stellate protein CG33247 | 0.01139 ± 0.00267 | 0.0197 ± 0.0438 | 0.05522 ± 0.01000 p = 0.00483b | 0.0296 ± 0.0644 | 0.00743 ± 0.01502 | 0.00266 ± 0.00303 |
| 12 | 81 | Stellate protein CG33247 | 0.00784 ± 0.00121 | 0.02921 ± 0.0239 | 0.05163 ± 0.00106 p = 0.00507b | 0.01766 ± 0.0354 | 0.0129 ± 0.02987 | 0.00360 ± 0.00349 |
| 13 | 83 | Triosephosphate isomerase | 0.01722 ± 0.07347 | 0.01545 ± 0.06742 | 0.10425 ± 0.08459 | 0.11867 ± 0.08459 | 0.12435 ± 0.02904 | 0.02646 ± 0.01363 p = 0.00061c |
| 14 | 90 | ATP synthase subunit β | 0.0121 ± 0.00155 | 0.00539 ± 0.00112 p = 0.00247b | 0.019 ± 0.03214 p = 0.00024d | |||
| 15 | 97 | ATP synthase D chain | 0.08024 ± 0.01850 | 0.04083 ± 0.0364 | 0.05072 ± 0.04827 | 0.03286 ± 0.02797 | 0.06776 ± 0.06139 | 0.01847 ± 0.00439 p = 0.00263b |
| 16 | 105 | ATP synthase subunit β | 0.06354 ± 0.05091 | 0.01256 ± 0.03672 | 0.04666 ± 0.05516 | 0.0397 ± 0.02289 | 0.02342 ± 0.03046 | 0.00179 ± 0.00281 p = 0.00151b |
| 17 | 111 | Protein l(2)37Cc | 0.06736 ± 0.04438 | 0.018423 ± 0.00270 p = 0.00809b | 0.05768 ± 0.02585 | 0.07649 ± 0.10300 | 0.0401 ± 0.03811 | 0.00559 ± 0.00881 p = 0.00022bp = 0.00787c |
| 18 | 117 | ATP synthase subunit β | 0.05724 ± 0.00662 | 0.02367 ± 0.0637 | 0.07744 ± 0.09473 | 0.0326 ± 0.06556 | 0.0476 ± 0.0820 | 0.00466 ± 0.00134 p = 0.00096b |
| 19 | 123 | ATP synthase subunit β | 0.01906 ± 0.01613 | 0.00331 ± 0.00433 | 0.02062 ± 0.0099 | 0.01013 ± 0.0012 | 0.03737 ± 0.02032 | 0.00281 ± 0.00336 p = 0.00080c |
| 20 | 124 | Inorganic pyrophosphatase | 0.03386 ± 0.02401 | 0.0325 ± 0.01715 | 0.04636 ± 0.02574 | 0.02309 ± 0.02131 | 0.05833 ± 0.00977 | 0.00568 ± 0.00112 p = 0.00721bp = 1.5608E–06c |
| 21 | 131 | Fructose bisphosphate aldolase | 0.05415 ± 0.05416 | 0.01170 ± 0.02879 p = 0.00780c | 0.03636 ± 0.01339 | 0.0522 ± 0.07434 | 0.0694 ± 0.00939 | 0.054637 ± 0.00665 |
| 22 | 133 | Pyruvate kinase | 0.05199 ± 0.03744 | 0.0145 ± 0.01690 | 0.0377 ± 0.0492 | 0.01838 ± 0.01070 p = 0.00287b | ||
| 23 | 152 | Phosphoglycerate kinase | 0.08607 ± 0.05137 | 0.17323 ± 0.03216 | 0.14976 ± 0.02226 | 0.34022 ± 0.07880 p = 0.00945b | 0.13099 ± 0.00249 | 0.01186 ± 0.00197 p = 0.00016c |
| 24 | 155 | Glycerol-3-phosphate dehydrogenase | 0.03523 ± 0.01054 | 0.0993 ± 0.05331 | 0.03185 ± 0.01802 | 0.01244 ± 0.00796 | 0.00780 ± 0.00056 p = 0.00706d | 0.00171 ± 0.00257 p = 1.6430E–05bp = 0.00440c |
| 25 | 156 | Isocitrate dehydrogenase | 0.09404 ± 0.06044 | 0.02818 ± 0.00736 | 0.03816 ± 0.0096 | 0.02102 ± 0.06067 | 0.06589 ± 0.10657 | 0.00184 ± 0.00023 p = 5.3619E–05b |
| 26 | 157 | Glyceraldehyde-3-phosphate dehydrogenase | 0.03387 ± 0.00190 | 0.17997 ± 0.03068 p = 0.00119b | 0.11537 ± 0.00203 p = 9.0253E–07d | |||
| 27 | 161 | Thioredoxin reductase I | 0.06182 ± 0.03565 | 0.01353 ± 0.01329 | 0.15359 ± 0.05446 p = 0.00536c | 0.04874 ± 0.01566 | 0.01517 ± 0.00499 | 0.00059 ± 0.00048 p = 0.00265bp = 0.00012c |
| 28 | 162 | Fructose bisphosphate aldolase | 0.03210 ± 0.01170 | 0.02430 ± 0.01086 p = 0.00021c | 0.06483 ± 0.01934 p = 0.00315bp = 0.00938c | 0.07216 ± 0.02846 p = 0.00248b | 0.12061 ± 0.00710 p = 4.1997E–08d | 0.01689 ± 0.00260 p = 0.00263c |
| 29 | 163 | Alcohol dehydrogenase | 0.04532 ± 0.01706 p = 0.00964c | 0.00838 ± 0.00187 | ||||
| 30 | 165 | ATP synthase subunit α | 0.06259 ± 0.02558 | 0.05752 ± 0.04504 | 0.01409 ± 0.01 | 0.09364 ± 0.01434 | 0.0328 ± 0.00413 | 0.00082 ± 0.00054 p = 0.00123b |
| 31 | 166 | Maltase H | 0.00333 ± 0.00325 | 0.08944 ± 0.09212 | 0.00040 ± 0.00019 p = 0.00216c | 0.00617 ± 0.00298 | 0.00480 ± 0.00155 | 0.04754 ± 0.03454 |
| 32 | 170 | Pyruvate kinase | 0.00480 ± 0.00251 | 0.02907 ± 0.00478 p = 0.00839d | ||||
| 33 | 171 | Glycerol-3-phosphate dehydrogenase | 0.01344 ± 0.00408 | 0.00372 ± 0.00255 p = 0.00016c | 0.00526 ± 0.00287 p = 0.00020c | 0.00482 ± 0.00032 p = 0.00736b | 0.05168 ± 0.00546 p = 0.00063d | 0.00688 ± 0.00578 p = 0.00063b |
| 34 | 173 | ATP synthase subunit β | 0.00046 ± 0.00031 p = 0.00025c | 0.15433 ± 0.02157 p = 0.00126c | 0.04854 ± 0.00678 | |||
| 35 | 177 | Fructose bisphosphate aldolase | 0.00406 ± 0.00075 | 0.00050 ± 0.00027 p = 0.00153bp = 0.00030c | 0.04982 ± 0.00730 p = 0.00041d | |||
| 36 | 178 | ATP synthase subunit α | 0.00288 ± 0.00394 | 0.00459 ± 0.00615 p = 0.00025c | 0.05003 ± 0.00671 p = 3.2629E–06d | |||
| 37 | 185 | ATP synthase subunit β | 0.00871 ± 0.00543 | 0.01777 ± 0.00956 p = 1.7782E–06c | 0.04401 ± 0.01014 p = 0.00020bp = 0.00029c | 0.09301 ± 0.00422 p = 5.0701E–09d | 0.00877 ± 0.00187 p = 1.0732E–05c | |
| 38 | 186 | Fructose bisphosphate aldolase | 0.10034 ± 0.05736 | 0.00754 ± 0.01134 p = 2.2442E–07b | 0.06666 ± 0.02744 p = 0.00087c | 0.05637 ± 0.02768 p = 0.00536c | 0.00989 ± 0.02621 p = 5.4053E–06d | |
| 39 | 187 | Fructose bisphosphate aldolase | 0.01225 ± 0.01942 | 0.09305 ± 0.03952 p = 0.00727b | 0.02897 ± 0.01348 p = 0.00241c | 0.02634 ± 0.04315 p = 0.00226c | 0.15707 ± 0.06963 p = 0.00277d | 0.00230 ± 0.00212 p = 2.0640E–05c |
| 40 | 193 | Cytochrome P450 | 0.00662 ± 0.00730 | 0.00985 ± 0.00105 p = 0.00174bp = 0.00028c | 0.02990 ± 0.00493 p = 0.00012c | 0.05572 ± 0.00066 p = 1.0684E–06d | ||
| 41 | 195 | Fructose bisphosphate aldolase | 0.00581 ± 0.00388 | 0.03529 ± 0.05554 | 0.02590 ± 0.00795 p = 0.00391b | 0.04218 ± 0.00960 p = 0.00090b | 0.02665 ± 0.02258 | 0.00053 ± 0.00047 p = 0.00139bp = 0.00307c |
| 42 | 199 | Inorganic pyrophosphatase | 0.02621 ± 0.01431 | 0.04274 ± 0.01619 | 0.00144 ± 0.00105 p = 1.0825E–06bp = 5.2773E–09c | |||
| 43 | 209 | Arginine kinase | 0.01591 ± 0.00931 | 0.04940 ± 0.01749 p = 0.00610b | 0.07943 ± 0.01468 p = 2.8958E–05b | 0.079384 ± 0.03733 p = 0.00411b | 0.05711 ± 0.00992 p = 0.00047d | 0.00402 ± 0.00257 p = 0.00013c |
| Muscle activity | ||||||||
| 44 | 59 | Actin-88F | 0.02046 ± 0.00313 | 0.06569 ± 0.00868 p = 5.8021E–05b | 0.0269 ± 0.07303 | 0.01924 ± 0.04082 | 0.01591 ± 0.02909 | 0.0039 ± 0.00216 p = 1.1928E–06bp = 1.1046E–05c |
| 45 | 102 | Actin-57B | 0.07233 ± 0.01558 | 0.02407 ± 0.01990 p = 0.00101b | 0.05179 ± 0.02690 | 0.00096 ± 0.00085 p = 5.0262E–07bp = 0.00038c | ||
| 46 | 104 | Actin-88F | 0.11418 ± 0.01078 | 0.12801 ± 0.01392 | 0.14641 ± 0.11335 | 0.0227 ± 0.00535 p = 0.00738bp = 0.00032c | 0.18435 ± 0.17218 | 0.04722 ± 0.03402 |
| 47 | 114 | Myosin regulatory light chain-2 | 0.03078 ± 0.00338 | 0.00768 ± 0.02413 | 0.02481 ± 0.08161 | 0.02176 ± 0.05122 | 0.0174 ± 0.0255 | 0.00723 ± 0.00110 p = 0.00103b |
| 48 | 115 | Actin-5C | 0.02452 ± 0.00371 | 0.00461 ± 0.00065 p = 0.00750b | 0.02079 ± 0.0455 | 0.00308 ± 0.00072 p = 4.9214E–05b | 0.00398 ± 0.00122 p = 0.00549d | 0.00535 ± 0.00147 p = 0.00911b |
| 49 | 116 | Actin-79B | 0.06538 ± 0.00565 | 0.00187 ± 0.00272 p = 7.981E–06bp = 0.00435c | 0.02916 ± 0.08442 | 0.00724 ± 0.00169 p = 1.6044E–07b | 0.01044 ± 0.00186 p = 2.6139E–06d | 0.00615 ± 0.00132 p = 3.6987E–07b |
| 50 | 149 | Actin-5C | 0.05339 ± 0.03502 | 0.04162 ± 0.0414 | 0.05021 ± 0.04663 | 0.15640 ± 0.19589 | 0.0328 ± 0.04848 | 0.00433 ± 0.00909 p = 0.00601b |
| 51 | 151 | Synapse associated protein | 0.01538 ± 0.0055 p = 0.00118c | 0.00974 ± 0.00762 p = 0.00132c | 0.03087 ± 0.00508 | |||
| 52 | 158 | ADP-ribosylation factor-8 | 0.06229 ± 0.03123 | 0.01840 ± 0.00531 p = 0.00325c | 0.10726 ± 0.01584 | 0.16042 ± 0.06361 | 0.09388 ± 0.02007 | 0.00920 ± 0.00118 p = 0.00746bp = 0.00025c |
| 53 | 168 | Actin-79B | 0.00114 ± 0.00188 | 0.0201 ± 0.02050 p = 0.00921b | 0.02964 ± 0.01304 p = 2.1258E–05d | |||
| 54 | 169 | Actin-88F | 0.00256 ± 0.00179 | 0.04922 ± 0.02446 p = 0.00191d | ||||
| 55 | 174 | Actin-57B | 0.00193 ± 0.00344 | 0.04580 ± 0.01204 p = 0.00085b | 0.03273 ± 0.00470 p = 0.00016d | 0.00812 ± 0.00773 p = 0.00102c | ||
| 56 | 182 | Actin-87E | 0.00355 ± 0.00395 | 0.05350 ± 0.03246 p = 0.00520b | 0.07146 ± 0.01176 p = 9.2309E–07d | |||
| 57 | 184 | Actin-57B | 0.00062 ± 0.00095 | 0.12755 ± 0.00720 p = 1.3775E–07bp = 3.7756E–05c | 0.0122 ± 0.02055 | |||
| 58 | 188 | Actin-57B | 0.05530 ± 0.01465 | 0.14250 ± 0.00663 p = 0.00071bp = 4.5473E–05c | 0.08487 ± 0.05086 | 0.09601 ± 0.01685 p = 0.00666c | 0.04235 ± 0.00629 | 0.0421 ± 0.07126 |
| 59 | 189 | Actin-5C | 0.05170 ± 0.00922 | 0.27330 ± 0.07415 p = 0.00680bp = 0.00632c | 0.11201 ± 0.05377 | 0.06672 ± 0.04820 | 0.0506 ± 0.0797 | 0.07088 ± 0.0666 |
| 60 | 194 | Tubulin α-3 chain | 0.00143 ± 0.00293 | 0.02587 ± 0.01795 p = 0.00082bp = 0.00204c | 0.00259 ± 0.00353 p = 1.8164E–08c | 0.07357 ± 0.00403 p = 8.0941E–09d | ||
| 61 | 198 | Actin-79B | 0.0135 ± 0.0029 | 0.05796 ± 0.00308 p = 5.6253E–05b | 0.08917 ± 0.00759 p = 8.7657E–05b | |||
| 62 | 200 | Actin-87E | 0.0110 ± 0.01541 | 0.02004 ± 0.00506 | 0.05987 ± 0.00764 p = 0.00248b | 0.06444 ± 0.05897 | 0.03010 ± 0.02593 | 0.00197 ± 0.00100 p = 0.00156c |
| 63 | 201 | Actin-57B | 0.06669 ± 0.02571 | 0.00313 ± 0.00456 p = 4.3266E–05c | ||||
| 64 | 202 | Paramyosin | 0.01868 ± 0.00829 | 0.08292 ± 0.05254 | 0.05174 ± 0.00992 p = 0.00556b | 0.15828 ± 0.04194 p = 0.00481b | 0.04854 ± 0.0097 | 0.01727 ± 0.01595 |
| 65 | 203 | Actin-42A | 0.07337 ± 0.01193 p = 0.00736c | 0.02297 ± 0.01262 | ||||
| 66 | 208 | Actin-5C | 0.03647 ± 0.01753 | 0.01795 ± 0.01412 | 0.04708 ± 0.01341 | 0.08915 ± 0.02907 | 0.03239 ± 0.00920 | 0.00092 ± 0.00019 p = 0.00407c |
| Ion-channel/cellular transport | ||||||||
| 67 | 54 | Calreticulin | 0.06318 ± 0.072311 | 0.07632 ± 0.01609 | 0.05745 ± 0.01702 | 0.02592 ± 0.01448 p = 0.00454c | 0.12495 ± 0.02609 | 0.0131 ± 0.00333 p = 0.00012c |
| 68 | 55 | Tubulin β-1 chain | 0.0158 ± 0.00237 | 0.00237 ± 0.0025 | 0.03131 ± 0.05206 | 0.06529 ± 0.00427 p = 0.00053bp = 0.00834c | 0.0227 ± 0.02573 | 0.00163 ± 0.00122 |
| 69 | 78 | Voltage-dependent anion-selective channel (Porin) | 0.0109 ± 0.01073 | 0.2164 ± 0.0563 | 0.1006 ± 0.1049 | 0.0921 ± 0.01156 p = 0.00140b | 0.13172 ± 0.11014 | 0.03589 ± 0.00776 |
| 70 | 160 | ADP/ATP translocase | 0.00215 ± 0.00150 | 0.0302 ± 0.02585 p = 0.00012bp = 9.6783E–07c | 0.00391 ± 0.00784 | |||
| 71 | 206 | Voltage-dependent anion-selective channel (Porin) | 0.07494 ± 0.06443 | 0.0116 ± 0.04136 | 0.06563 ± 0.00627 | 0.076146 ± 0.02511 | 0.04800 ± 0.10300 | 0.00101 ± 0.00116 p = 0.00581b |
| 72 | 207 | Transient receptor potential locus C protein | 0.07783 ± 0.01334 p = 0.00394c | 0.02824 ± 0.00534 | 0.00036 ± 0.00019 p = 4.0791E–08c | |||
| Heat shock proteins | ||||||||
| 73 | 31 | Heat shock 82 kDa protein | 0.02077 ± 0.01520 | 0.012483 ± 0.0060 p = 1.7087E–05c | 0.00111 ± 0.00014 p = 0.00101d | |||
| 74 | 42 | Heat shock 70 kDa protein | 0.05238 ± 0.01099 | 0.00207 ± 0.00078 p = 0.00012b | 0.0305 ± 0.00145 p = 0.00939b | 0.00959 ± 0.00818 p = 5.2107E–05b | 0.02227 ± 0.01378 p = 0.00187d | 0.00374 ± 0.00089 p = 1.62E–08bp = 0.00028c |
| 75 | 43 | Heat shock 70 kDa protein | 0.05023 ± 0.04775 | 0.03493 ± 0.00646 | 0.04306 ± 0.02875 p = 0.00157c | 0.01095 ± 0.00139 | 0.01596 ± 0.01483 p = 0.00948d | 0.00101 ± 0.00228 p = 8.70E–03bp = 8.95E–06c |
| 76 | 46 | Heat shock 60 kDa protein | 0.05832 ± 0.03522 | 0.01751 ± 0.01643 p = 0.00400b | 0.02924 ± 0.01869 | 0.00205 ± 0.02335 p = 0.00819b | 0.00164 ± 0.00208 p = 0.00080d | 0.01227 ± 0.00143 p = 0.00048b |
| 77 | 180 | Heat shock factor protein | 0.01626 ± 0.00932 | 0.04055 ± 0.00227 p = 0.00755d | ||||
| Protein synthesis/folding/ degradation | ||||||||
| 78 | 50 | Protein disulfide isomerase | 0.05850 ± 0.00582 | 0.00525 ± 0.000696 | 0.01486 ± 0.00272 | 0.01374 ± 0.01066 | 0.01429 ± 0.02467 | 0.00699 ± 0.00148 p = 0.00237b |
| 79 | 99 | Furin-like protease | 0.02738 ± 0.01222 | 0.04072 ± 0.01344 | 0.04672 ± 0.0098 | 0.05014 ± 0.00689 p = 0.00352c | 0.03017 ± 0.00327 | 0.2291 ± 0.00774 |
| 80 | 121 | 40S ribosomal protein SA | 0.13683 ± 0.01787 | 0.02732 ± 0.01356 p = 0.00107bp = 0.00908c | 0.06246 ± 0.02463 p = 0.00708b | 0.00885 ± 0.00078 p = 2.5433E–05bp = 2.0399E–08c | 0.05494 ± 0.00225 p = 0.00023d | 0.00305 ± 0.00589 p = 1.61E–06bp = 0.00022c |
| 81 | 164 | Elongation factor 1α (EF-1α) | 0.01402 ± 0.01559 | 0.04487 ± 0.04267 | 0.03916 ± 0.03485 | 0.05944 ± 0.00664 p = 0.00147b | 0.06306 ± 0.00781 p = 0.00098d | 0.00452 ± 0.00442 p = 2.9002E–09c |
| 82 | 179 | 40S ribosomal protein S12 | 0.00028 ± 0.00028 | 0.08388 ± 0.00936 p = 2.3206E–05d | ||||
| Miscellaneous | ||||||||
| 83 | 197 | Glutathione-S-transferase (redox homeostasis) | 0.01685 ± 0.00487 | 0.02996 ± 0.01557 | 0.01077 ± 0.00323 | 0.08026 ± 0.02642 | 0.04748 ± 0.03732 | 0.00108 ± 0.00099 p = 3.3424E–08bp = 0.00028c |
| 84 | 204 | Glutathione-S-transferase (redox homeostasis) | 0.05135 ± 0.03322 | 0.09755 ± 0.05312 | 0.02023 ± 0.02641 | 0.09738 ± 0.04084 | 0.04298 ± 0.01096 | 0.00243 ± 0.00236 p = 0.00030bp = 1.1889E–07c |
| 85 | 100 | Thioredoxin peroxidase (apoptosis) | 0.09598 ± 0.01127 | 0.09064 ± 0.10128 | 0.10623 ± 0.0569 | 0.03505 ± 0.01026 p = 0.00228bp = 8.085E–06c | 0.12797 ± 0.00070 p = 0.00205b | 0.00244 ± 0.00142 p = 1.2261E–06bp = 1.00E–07c |
| 86 | 176 | Caspase-8 precursor (apoptosis) | 0.00092 ± 0.00013 | 0.00133 ± 0.00074 p = 4.1969E–06c | 0.00473 ± 0.00655 p = 1.77E–06c | 0.04464 ± 0.00337 p = 0.00012d | ||
| 87 | 62 | Vitellogenin-2 precursor (development) | 0.02446 ± 0.00269 | 0.0436 ± 0.0398 | 0.04901 ± 0.02959 p = 0.00238bp = 0.00149c | 0.0417 ± 0.05467 | 0.01699 ± 0.00314 | 0.00053 ± 0.00009 |
| 88 | 130 | RNA helicase (transcription) | 0.08621 ± 0.03634 | 0.10347 ± 0.0988 | 0.09000 ± 0.01469 | 0.04088 ± 0.01000 p = 0.00092c | 0.07469 ± 0.00027 | 0.00160 ± 0.00133 p = 0.00046bp = 0.00071c |
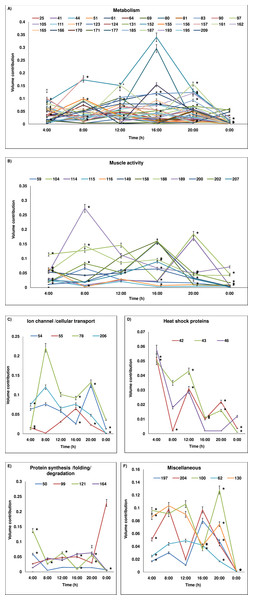
Figure 3: Temporal variation in expression level of proteins.
Protein level variations (as percentage of volume contribution) of the groups across 24-h period are shown. (A) metabolism, (B) muscle activity, (C) ion-channel/cellular transport, (D) heat shock proteins, (E) protein synthesis/folding/degradation, (F) miscellaneous. The proteins which show expression at all time points are represented and protein spot/cluster number is given in the figure. The mean ± SD values of percentage of volume contribution are plotted. At some time points, the SD values are nearly 0 and hence the values may not be visible in the reduced scale. The time points at which the expression is significantly different are marked with *. In (A) as numerous temporal variations are plotted, the SD values and * marks are plotted only for certain proteins for easy visualization. The SD values and significant variations of all proteins (A–F) are given in Table 2.Protein spot IDs which showed different levels of expression at a minimum of any four time points were: 32, 42, 43, 46, 51, 59, 71, 100, 102, 115, 116, 121, 130, 152, 158, 161, 164, 171, 174, 176, 185, 187, 193, 194, 200 and 209. The enzymes involved in metabolism and muscular activities—fructose bisphosphate aldolase (187), arginine kinase (209), thioredoxin reductase I (161) and actin-87E (200) were upregulated during daytime and showed a lowest level of expression at 00:00. In addition, proteins/enzymes involved in various active cellular processes (i.e., (40S ribosomal protein SA (121), ATP synthase subunit β (185), heat shock protein 70 kDa (43), vacuolar ATP synthase subunit B (51), actin-88F (59), 57B (174) and 87E (200), thioredoxin peroxidase (100), RNA helicase (130), phosphoglycerate kinase (152), ADP-ribosylation factor-8 (158), and elongation factor-1α (164)) were apparently upregulated during the daytime but showed lowest level of expression at midnight (00:00). Conversely, proteins involved in apoptosis and toxin metabolism (caspase-8 precursor (176) and cytochrome P450 (193)) appeared upregulated at 20:00 compared to the daytime points (Figs. 3B–3I).
Discussion
Living organisms perform their functions according to the light-dark cycle. Since, proteins and enzymes play vital roles in almost all the physiological functions of the body, a proper control of protein expression in a temporal manner is a crucial aspect of an organism. As complex interplay of multiple processes involved in the generation of overt rhythms of multiple biological functions, it is obvious that numerous proteins vary their expression in a temporal manner in the body of the fly. Studies on overall assessment of fly’s circadian proteome have confirmed the rhythmic nature of translation in D. melanogaster (Huang et al., 2013). However, it could be hypothesized that the circadian proteome is an outcome of regulatory stages at multiple steps of transcription and translation (RNA processing, posttranscriptional and posttranslational modifications). The conservation of circadian and clock controlled genes regulating similar pathways across various species, including Drosophila and mammals, is also well known (Akhtar et al., 2002). Since the majority of the regulatory mechanisms and signaling pathways are known to be conserved between Drosophila and humans (Rodriguez et al., 2013) the data generated in Drosophila may be applied to humans as well.
Our proteomics investigation demonstrated identification of 124 protein spots in the whole body of the fruit fly, Drosophila melanogaster. Eighty-eight of these proteins apparently showed temporal variation in expression. Analogous to our study, Rodriguez et al. (2013) had recognized more than 130 cycling transcriptional units in the heads of D. melanogaster, of which nearly one-third (44) cycled significantly. Among the 44, the peak times of mRNAs of cytochrome P450 (20:00 h) and glutathione-S-transferease (16:00) appeared synchronous with accumulation of their proteins that was observed in our study. However, the significant oscillations of several other proteins involved in metabolism, muscle activity, cellular transport, redox homeostasis, protein synthesis/folding/degradation, cell division and transcription that were also reported by Rodriguez et al. (2013) were not seen in our proteomics investigation. This may be partly due to the mRNA levels being investigated in heads of the fly by Rodriguez et al. (2013), whilst our analysis was performed on the whole body proteome.
The data of our study, when compared to other reports, demonstrated higher percentage of the fruit fly proteins under the clock control. For example, Reddy et al. (2006) had reported that only about 20% of soluble proteins in the proteome profile of whole mouse liver were under the circadian regulation. In addition, the proteome of suprachiasmatic nuclei (mammalaian circadian pacemaker) showed roughly 13% of soluble proteins demonstrated robust oscillations, with 53 of the protein spots in the 2DE proteome profile (Deery et al., 2009). In their study, more protein spots showed maximum expression during the day (65%) than night (35%). However, our study on the whole fly proteome showed slightly higher numbers of protein spots that were upregulated during nighttime than daytime.
The rhythmic protein abundance observed in the study may be caused by differences in the synthesis and/or half-life of proteins. In addition, it could also be attributed to (i) circadian transcriptional regulation by clock transcriptional factors and co-regulators which act on a wide array of circadian clock-controlled genes (ccgs) (Asher & Schibler, 2011), (ii) circadian hormonal signaling to various types of cells (Asher & Schibler, 2011; Lück et al., 2014) and (iii) rhythmically distinct feeding patterns (Vodala et al., 2012). Determination of the composition of proteins in the whole body of fruit fly is an essential step towards understanding the regulation of various proteins as an integrated system. In the whole body of D. melanogaster many genes showed coordinated circadian oscillations of expression but there were significant disconnections between the processes of transcription, post-transcriptional processing and protein synthesis (Beckwith & Yanovsky, 2014). Thus, the analysis of circadian pattern of proteome is useful in analyzing how many fold transcriptional and translational steps vary to maximize organismal adjustments over day and night.
Our study revealed an integrated pattern/regulation of proteome in the body of the fly, which could be necessary for optimizing growth and fitness. In this study, observation of multiple hits for single protein spot could be due to (i) very close-localization of two different spots of proteins, (ii) isoforms of the same proteins with a very close mass and pI and (iii) post-translational modifications. In some cases, a considerable variability in volume contribution of protein spots was observed (e.g., 6-phosphofructokinase at 04:00—0.0168 ± 0.080 or enolase at 16:00—0.1206 ± 0.1163). This could be owing to minuscule variations in the pI of proteins, their posttranslational modifications (like phosphorylation) and presence of isoforms. The missing spots at certain time points (Figs. 1A–1F) could indicate a circadian variation in the expression of proteins.
The gene ontology (GO) analysis proves beneficial in the identification of most meaningful functional aspects occurring in a given set of related gene products or biological insights into the system being studied especially when involving large proteome catalogs, like those that were generated via quantitative proteomics (e.g., ITRAQ and SILAC). Since, the focus of the current study was to investigate the overall pattern of temporal variation as well as to document the rhythmic build-up of proteins in D. melanogaster via 2-DE/MS (qualitative proteomics), the study greatly emphasizes on the correctness and the depth of analysis. We have therefore, presented the results of the experiment typically as a list of proteins (Al-Obaidi et al., 2014; Jessie et al., 2014). Generic biological processes annotated in this study (Table 1) were categorized based on the GO annotation found at the Uniprot Knowledgebase (UniProtKB).
The PERIOD protein is expressed only during daytime in neurons and tissues and is absent during nighttime (Plautz et al., 1997a; Plautz et al., 1997b). In our study, in the whole fly homogenate, we could not identify the protein. The reason may owe to the sensitivity of the techniques employed (2DE/MS/MS). We think that a combination of high-performance liquid chromatography and 2DE/MS/MS would have higher sensitivity to analyse the temporal variation of the PERIOD protein in the whole fly homogenate. As our temporally oscillating proteins are involved in metabolism, muscle activity, cellular transport, protein synthesis, apoptosis and development, a clock regulated release of various neurotransmitters regulating these functions (Moller et al., 2010) could also be suggested. Numerous genes involved in carbohydrate and amino acid metabolism including enzymes and membrane transporters were reported to be rhythmic (Akhtar et al., 2002). The present results showing diurnal upregulation of main enzymes of carbohydrate/amino acid metabolism (fructose bisphosphate aldolase, arginine kinase, ATP synthase subunit β, vacuolar ATP synthase subunit B, arginine kinase, phosphoglycerate kinase and thioredoxin reductase I) demonstrate synchronous activation and inhibition of the pathways involved. In addition, the minimal levels of expression of numerous proteins at midnight suggest that several cellular, biochemical and physiological activities were low at night in the fruit fly. These 24-h variations may be an indirect outcome of circadian control of ingestion or under a direct circadian control, mediated by neural and endocrine entities from the master clock that is located in lateral neurons of the fly (Akhtar et al., 2002).
Of late, an extensive interconnection has been documented between the molecular circadian clock and the underlying biochemical pathways that regulate the bioenergetics of the organism. The scope includes the regulatory role played by coenzymes (NAD(P)+/NAD(P)H), reactive oxygen species (superoxide anion and hydrogen peroxide), antioxidants, and physiological events that modulate the redox state (feeding condition and circadian rhythms) in determining the timing capacity of the molecular circadian clock. Both the circadian timing system and the metabolic network are tightly interlinked (Mendez et al., 2015). In addition, circadian clock gene transcription factors in metabolic tissues synchronize metabolic fuel utilization and storage with alternating durations of feeding and fasting parallel to the rest–activity cycle (Peek et al., 2015).
Recent evidences suggest the temporal accrual of yolk proteins in the seminal vesicles of D. melanogaster (Majewska et al., 2014). While the mechanisms of input pathways to the central circadian clock and the core circadian clock (lateral neurons in D. melanogaster) are extensively known, the processes that regulate the circadian output pathways (which result in the circadian proteome profile) are poorly understood. Recent genome-wide studies in many organisms suggested extensive translational regulation by the circadian clock could mainly contribute to the temporal protein profile, despite the robust mRNA rhythms observed (Montenegro-Montero & Larrondo, 2016).
Previous studies, demonstrated via distinct oscillations of mRNA and protein synthesis of genes, have shown that several genes encoding the cytoskeleton components are under clock control (Akhtar et al., 2002). Among the 23 fast skeletal muscle myosin genes, myh_tc, myh_n1, myh_n4, myo18a_2, and myo18b_2 showed circadian rhythmic expression and possess many circadian-related transcription factor-binding sites (Creb, Mef2 and E-box motifs) within their recognized promoter regions. In addition, the circadian expression of these 5 myosin genes was robustly correlated with the transcription pattern of clock genes in fast skeletal muscle (Lazado et al., 2014). Murphy et al. (2014) reported significant interaction between circadian time and exercise for muscle genes MYF6, UCP3, MYOD1 and PDK4. Hence, the circadian expression of a set of muscle-related proteins in D. melanogaster is expected.
As the proteome of the whole fly temporally vary in multiple biological processes including metabolism, muscle activities, cellular transport, apoptosis etc., our results generally indicate that a wide range of physiological/cellular processes are fine-tuned by the rhythmic expression of protein profiles. Although, the tissue specific expression of proteins and the coordination of protein regulation in various tissues of the fly could not be analyzed in this study, potential avenues of future research in the temporal regulation of intracellular localization of proteins and, exploration of rhythmically varying proteins in specific tissue types are wide open.
Supplemental Information
Supplemental Table 1
Percentage of volume contribution of protein spots/clusters of Drosophila melanogaster.