Genome-wide identification of DUF506 gene family in Oryza sativa and expression profiling under abiotic stresses
- Published
- Accepted
- Received
- Academic Editor
- Elsayed Mansour
- Subject Areas
- Agricultural Science, Bioinformatics, Genetics, Genomics, Plant Science
- Keywords
- DUF506, Gene family, Identification, Abiotic stress, Expression profile
- Copyright
- © 2023 Dong et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Genome-wide identification of DUF506 gene family in Oryza sativa and expression profiling under abiotic stresses. PeerJ 11:e16168 https://doi.org/10.7717/peerj.16168
Abstract
The domain of unknown function 560 (DUF560), also known as the PDDEXK_6 family, is a ubiquitous plant protein that has been confirmed to play critical roles in Arabidopsis root development as well as ABA and abiotic responses. However, genome-wide identification and expression pattern analysis in rice (Oryza sativa) still need to be improved. Based on the phylogenetic relationship, 10 OsDUF506 genes were identified and classified into four subfamilies. Segmental duplication was essential to the expansion of OsDUF506s, which were subjected to purifying selective pressure. Except for OsDUF50609 and OsDUF50610, the OsDUF506s shared colinear gene pairs with five monocot species, showing that they were conserved in evolution. Furthermore, the conserved domains, gene structures, SNPs distribution, and targeting miRNAs were systematically investigated. Massive cis-regulatory elements were discovered in promoter regions, implying that OsDUF506s may be important in hormone regulation and abiotic stress response. Therefore, we analyzed plant hormone-induced transcriptome data and performed qRT-PCR on eight OsDUF506s under drought, cold, and phosphorus-deficient stresses. The results revealed that most OsDUF506s respond to ABA and JA treatment, as well as drought and cold conditions. In conclusion, our findings provided insights into the evolution and function of OsDUF506s, which could benefit crop breeding in the future.
Introduction
Domains of unknown functions (DUFs) are batches of gene families with conserved domains but unknown functions that are common in eukaryotes (Bateman, Coggill & Finn, 2010). The Pfam database contains 4,716 DUF families (https://www.ebi.ac.uk/interpro/entry/pfam/). Although the majority of DUF families remain unknown, some have been studied. In Oryza sativa, OsDUF1618 (Wang et al., 2014), OsDUF221 (Ganie, Pani & Mondal, 2017), OsDUF1110 (Harada et al., 2016), OsDUF810 (Li et al., 2018), OsDUF668 (Zhong et al., 2019a), OsDUF231 (Zhong, Cui & Ye, 2019b), OsDUS936 (Li et al., 2017) have been characterized. Previous studies showed that DUF genes were engaged in different biological functions in Oryza sativa. For instance, SWOLLEN TAPETUM AND STERILITY 1 (STS1), which contained a DUF726 domain, interacted with Polyketide Synthase 2 (OsPKS2) and Acyl-CoA Synthetase 12 (OsACOS12) to contribute to sporopollenin biosynthesis (Yuan et al., 2022). Another DUF726 protein, encoded by Leaked and Delayed Degraded Tapetum 1 (OsLDDT1), was involved in fatty acid synthesis and anther epidermis formation (Sun et al., 2023). ROLLED and ERECT LEAF 2 (REL2), which contained DUF630 and DUF632 conserved domains, was involved in regulating leaf morphology, its functional loss resulted in rolling leaves (Yang et al., 2016). DUF genes have also been implicated in various biotic and abiotic responses. For example, the Oryza sativa Stress Responsive DUF740 Protein (OsSRDP) gene belonged to the DUF740 family, and its overexpressed transgenic plants, driven by promoter AtRd29A, revealed increased resistance to drought, salinity, and cold stresses as well as rice blast fungus (Jayaraman et al., 2022). The DUF966-stress repressive gene 2 (OsDSR2) gene was involved in the negative regulation of salt and drought stress responses (Luo et al., 2014).
The DUF506 family, also called the PDDEXK_6 family, is a group of plant proteins that are distant homologs of the PD-(D/E)XK nuclease superfamily. The nuclear structure is retained as α-β-β-α-β and includes the typical PDDEXK motifs II and III in modified forms as xDxxx motif located in the second core beta-strand, where x is any hydrophobic residue, and a (D/E)X(D/N/S/C/G) pattern. The missing positively charged residue of motif III may be replaced by a conserved arginine in motif IV, which is located in the proceeding alpha-helix (Knizewski et al., 2007). So far, DUF506 proteins in Oryza sativa have not been characterized systemically and functionally. Previous research merely identified that the expressions of 13 AtDUF506s were ubiquitous in organs and associated with abiotic stresses and ABA response (Ying, 2021). The comparative microarray data showed that AT2G20670 was inhibited by B. cinereal, heat, salinity, and osmotic stress (Sham et al., 2019). Recent studies revealed that REPRESSOR OF EXCESSIVE ROOT HAIR ELONGATION 1 (AtRXR1) gene encoded AT3G25240 protein, which was strongly induced by phosphorus limitation and suppressed root hairs (RHs) extension by interacting with RabD2c GTPase. Moreover, its function under phosphorus limitation was conserved in both monocot and dicot, as proven by the analogous function of Brachypodium distachyon DUF506 (Ying et al., 2022). AtRXR3 (AT1G62420), another P-inducible AtDUF506 gene, inhibited RHs elongation by a distinct mechanism. AtRXR3 was transactivated by ROOT HAIR DEFECTIVE6-LIKE4 (RSL4) and interacted with cytosolic calmodulins to repress RHs elongation (Ying & Scheible, 2022). Current studies on DUF506s in Arabidopsis suggested that DUF506 family members were significant in plant growth and abiotic resistance, but DUF506s functions in Oryza sativa have rarely been investigated. LOC_Os01g68650, the closest homologous gene of AtRXR1, was upregulated under drought stress, and its expression in drought tolerance 11 (OsDT11) overexpression line was higher than in the wild line, indicating that this gene may participate in drought response and be enhanced by OsDT11 (Zhao et al., 2020). LOC_Os01g54340 was identified as a nitrogen-sensitive gene that was rapidly repressed by nitrogen starvation (Hsieh et al., 2018). Until now, the OsDUF506 family has not been genome-wide identified and the functions are still unknown.
In this study, we identified all OsDUF506 family members in Oryza sativa by bioinformatic methods. The phylogeny, conserved motifs, cis-acting regulatory elements, distribution of non-synonymous SNPs, target miRNA, synteny, and tissue expression specificity were analyzed. The expression pattern of OsDUF506s under plant hormones treatments and drought, cold, and phosphorus-deficient stresses was investigated using transcriptome and qRT-PCR. Our study provided a more comprehensive identification and classification of OsDUF506s, broadened our recognition of the functions under abiotic stresses, and laid the groundwork for molecular breeding in Oryza sativa.
Materials and Methods
Identification of DUF506 members in 10 plant species
All the genome databases were downloaded from the EnsemblPlants database (http://plants.ensembl.org). Thirteen Arabidopsis DUF506 protein sequences were obtained from UniProtKB/Swiss-Prot (SwissProt) database (https://www.uniprot.org) and used as queries to perform protein blast search in 10 plant species (Oryza sativa, Arabidopsis thaliana, Glycine max, Solanum tuberosum, Gossypium raimondii, Zea mays, Hordeum vulgare, Triticum aestivum, Sorghum bicolor and Ananas comosus) by using TBtools (Chen et al., 2020). Meantime, the typical domain of DUF506 (PF04720, PDDEXK_6) was downloaded from the PFAM database (http://pfam.xfam.org) and was used to search for DUF506 with the HMMER tool (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). All the candidates were merged and edited to eliminate redundancies. The candidates were then submitted to NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd/) to test for the existence of the complete DUF506 conserved domain and those with an incomplete N end or C end were eliminated. The molecular weight (Mw), isoelectric point (pI), instability index, aliphatic index, and grand average of hydropathicity (GRAVY) of DUF506 members were predicted with ExPASy (http://web.expasy.org/protparam). The subcellular localizations were predicted with WoLF PSORT (https://wolfpsort.hgc.jp).
Phylogenetic relationship, structure, and conserved motifs analysis of OsDUF506 members
A total of 130 DUF506s was used for aligning with the MUSCLE method (default parameters) and construction of an ML phylogenetic tree (default parameters) using MEGA 11 software by setting bootstrap to 1,000 and JTT+G model (Tamura, Stecher & Kumar, 2021). The result was displayed by ChiPlot (https://www.chiplot.online). The conserved motifs were predicted with MEME (https://meme-suite.org/meme/doc/meme.html) and visualized by TBtools (Chen et al., 2020).
Prediction of CREs of OsDUF506 members
The 2,000 bp upstream sequences of promoters were used to search for CREs with PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) and visualized by TBtools (Chen et al., 2020).
Analysis of SNPs of OsDUF506 members
The OsDUF506 sequences were used to query the SNP-Seek database for nsSNPs against Nipponbare reference (https://snp-seek.irri.org). SNP-index was used to assess the subpopulation specificity of nsSNPs. The SNP-index was calculated as SNPGJ-index = Nref/Nall × 100%, SNPH-index = Nh/Nall × 100%, SNPXI-index = 1-SNPGJ-index-SNPH-index, Nref represented the number of varieties sharing the same allele with reference, Nh represented the number of varieties with heterozygous allele, Nall represented the total number of varieties with determined alleles at the SNP locus.
Syntenic analysis of OsDUF506 members
The duplication events and syntenic relationship of DUF506s between Oryza sativa and other plants were obtained by MCScanX (Wang et al., 2012). The results were visualized by TBtools (Chen et al., 2020). The nonsynonymous (Ka) and synonymous (Ks) calculations were performed by the simple Ka/Ks calculator kit of TBtools (Chen et al., 2020).
Predict analysis of miRNAs interacting with OsDUF506 members
The miRNAs targeting OsDUF506s were predicted by psRNATarget (https://www.zhaolab.org/psRNATarget/analysis?function=2) and visualized by ChiPlot (https://www.chiplot.online). The expressions of the predicted miRNAs were obtained from the PmiRExAt database (http://pmirexat.nabi.res.in/searchdb.html) and the normalized TPM values (log2) were used for constructing a heatmap, the scale method of normalized was used to intuitively reflect the expressing differences between tissues and treatments by TBtools (Chen et al., 2020).
Expression analysis of OsDUF506 members in different tissues and induced by plant hormones
The expression data in various tissues and induced by 50 μM abscisic acid (ABA), 10 μM gibberellin 3 (GA3), 10 μM auxin (IAA), 100 μM jasmonic acid (JA), 1 μM brassinolide (BL), and 1μM trans-Zeatin (tZ) were obtained from RiceXPro database (https://ricexpro.dna.affrc.go.jp/quick-guide.html), the normalized signal intensity values (log2) were used for constructing heatmap, and the scale method of normalized was used to intuitively reflect the expressing changes of particular genes at different treating time points by TBtools (Chen et al., 2020).
Plant growth conditions and abiotic stresses treatments
Japonica rice variety Yunkegeng 5 was used in the expression analysis. The plants were cultivated in a climate chamber (160 μmol−2 s−1 light intensity, 14 h-light and 10 h-dark a cycle, 28 °C, 45% RH) for 14 days in Yoshida rice nutrient solution (NS1040; Coolaber Technology Co., Ltd., Beijing, China). For drought stress, plants were transferred to a nutrient solution of 20% (w/v) polyethylene glycol (PEG-6000) for 3 h. For cold stress, plants were transferred to the climate chamber under 4 °C treatment for 3 h. For phosphorus deficiency stress, plants were transferred to phosphorus-deficient Yoshida nutrient solution (NSP1040-P; Coolaber Technology Co., Ltd., Beijing, China) for 7 days. Three biological replicates of the treated and control plants were harvested and stored at −80 °C.
Expression analysis of OsDUF506 members by qRT-PCR
Primer design and specificity check was performed by Primer-BLAST of NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi). The total RNA was extracted by TaKaRa MiniBEST Plant RNA Extraction Kit, cDNA was synthesized using Vazyme HiSript III 1st Strand cDNA Synthesis Kit (+gDNA wiper). The qRT-PCR was accomplished by Vazyme ChamQ SYBR Color qPCR Master Mix (Without ROX) in LightCycler96 system under the PCR condition of 95 °C for 60 s, 45 cycles of 95 °C for 10 s, 54 to 60 °C for 20 s and 72 °C for 20 s. The relative expressions data were calculated by 2−ΔΔCT method, and the reference used in this study was OsActin. The primer sequences were listed in Table S9. The significant differences level was analyzed by unpaired t-test with GraphPad Prism 8.0 software.
Results
Identification and phylogenetic relationship of DUF506 members in Oryza sativa and nine additional plant species
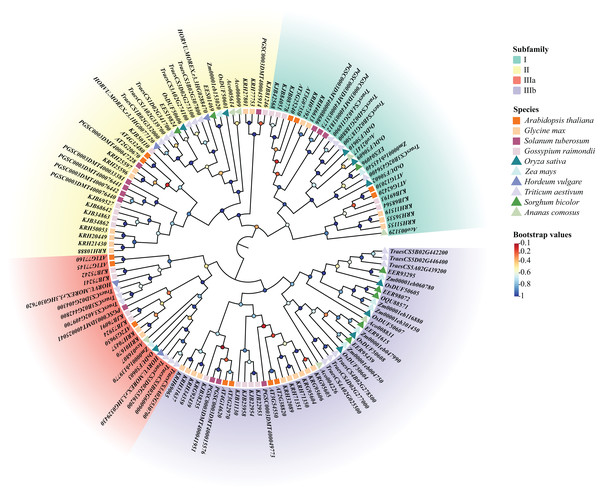
Using the previous method, we identified 10 DUF506s in Oryza sativa and named them OsDUF50601 to OsDUF50610. Moreover, 13, 25, 13, 20, 8, 4, 22, 8, 7 DUF506s were identified in Arabidopsis thaliana, Glycine max, Solanum tuberosum, Gossypium raimondii, Zea mays, Hordeum vulgare, Triticum aestivum, Sorghum bicolor and Ananas comosus, respectively (Table S1). A total of 130 DUF506s were used to construct an ML phylogenetic tree and divided into four subfamilies based on the evolutionary distance referred to previous literature (Ying, 2021). The IIIb subfamily contained the most DUF506s, while the IIIa subfamily had the fewest. Three members belonged to subfamily I (OsDUF50602, OsDUF50609, and OsDUF50610), two belonged to subfamily II (OsDUF50601 and OsDUF50606), only OsDUF50603 belonged to subfamily IIIa, while the rest four members composed the largest subfamily IIIb (Fig. 1).
Figure 1: Phylogenetic tree of DUF506 members identified in ten plant species.
The members were divided into four subfamilies shown in different colors. Circles in different colors represent different bootstrap values. Bootstrap values were calculated from 1,000 replications and only the values over 0.7 bootstrapping were considered significant.Characterization and conserved motifs of OsDUF506 members
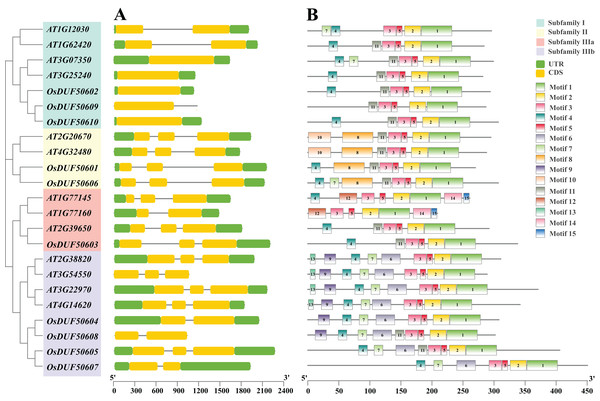
The 10 OsDUF506s were located on chromosome 1, 3, 5, 7, 10, 11. The summary of characteristics was shown in Table 1. Their protein lengths ranged from 269 to 507. The molecular weight, theoretical isoelectric points, and aliphatic index were predicted between 29,861.74 to 54,428.55 Da, 5.58 to 9.12, and 62.39 to 86.85, respectively. The instability index of proteins exceeded 40, and their GRAVY values were negative, suggesting that they were unstable hydrophilic proteins. The subcellular localization of subfamily II members (OsDUF50601 and OsDUF50606) was predicted to be in the nucleus, while the rest were predicted to be in the chloroplast. OsDUF506s possessed 1 to 3 exons, and the exon/intron structure of genes within a subfamily was comparable (Fig. 2A). The OsDUF506s from subfamily I had only one CDS region, while those from subfamilies II and IIIa had three CDS regions. The number of CDS regions led to the division of subfamily IIIb into two branches. The result suggested that members of different subfamilies might have distinct functions.
| Gene ID | Subfamily | Locus ID | Size(aa) | Protein MW (Da) | pI | Instability index | Aliphatic index | GRAVY | Chromosome | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| OsDUF50601 | II | LOC_Os01g54340 | 269 | 29,861.74 | 7.61 | 47.02 | 65.76 | −0.460 | 1 | Nucleus |
| OsDUF50602 | I | LOC_Os01g68650 | 293 | 30,429.34 | 8.97 | 52.76 | 75.22 | −0.134 | 1 | Chloroplast |
| OsDUF50603 | IIIa | LOC_Os01g74250 | 337 | 37,616.96 | 7.25 | 60.92 | 86.85 | −0.325 | 1 | Chloroplast |
| OsDUF50604 | IIIb | LOC_Os03g06680 | 307 | 33,011.21 | 6.68 | 45.12 | 80.59 | −0.416 | 3 | Chloroplast |
| OsDUF50605 | IIIb | LOC_Os03g58230 | 405 | 43,737.47 | 9.12 | 50.54 | 68.52 | −0.489 | 3 | Chloroplast |
| OsDUF50606 | II | LOC_Os05g44300 | 306 | 32,169.30 | 6.76 | 56.42 | 62.39 | −0.267 | 5 | Nucleus |
| OsDUF50607 | IIIb | LOC_Os07g08390 | 507 | 54,428.55 | 9.07 | 59.08 | 66.07 | −0.481 | 7 | Chloroplast |
| OsDUF50608 | IIIb | LOC_Os10g28210 | 301 | 32,041.06 | 6.52 | 50.73 | 79.60 | −0.347 | 10 | Chloroplast |
| OsDUF50609 | I | LOC_Os11g25020 | 286 | 30,211.19 | 9.03 | 42.62 | 76.71 | −0.108 | 11 | Chloroplast |
| OsDUF50610 | I | LOC_Os11g25040 | 306 | 32,464.54 | 5.58 | 50.32 | 82.06 | −0.176 | 11 | Chloroplast |
Figure 2: Gene structure, and conserved motifs of DUF506 in Oryza sativa and Arabidopsis thaliana.
Four background colors indicate four subfamilies. (A) Exon/intron structures. Green bars, yellow bars, and lines indicate UTRs, exons, and introns, respectively. (B) Distributions of conserved motifs. Motifs are shown by 15 different color bars.Fifteen conserved protein motifs of OsUDF506s were identified (Fig. 2B). All Oryza sativa and Arabidopsis DUF506 members shared motifs 1, 2, 3, and 5. In subfamily I, the most conserved motif was 4 and 11. Ultimately, OsDUF50602 and OsDUF50610 shared the same motifs with AT1G62420 and AT3G25240, indicating they may have the same biological function. The subfamily II members shared motifs 8 and 11. The motif construction of OsDUF50602 from subfamily IIIa was identical to that of AT2G39650. All subfamily IIIb members shared motifs 4, 6, and 7. Three-quarters possessed motif 9, but motif 13 was exclusive to genes from Arabidopsis. The result showed that DUF506s belonging to the same subfamily shared similar motif characteristics, indicating that they have a similar function.
Analysis of cis-acting regulatory elements (CREs) of OsDUF506 members
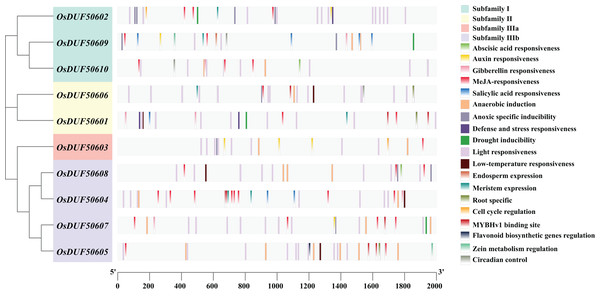
To analyze the prospective function of OsDUF506s, we searched the promoter regions (2,000 bp upstream of the start codon) and predicted 321 potential CREs (Fig. 3). OsDUF50604 possessed the most CREs, while OsDUF50603 and OsDUF50609 possessed the fewest (Table S2). Nineteen types of CREs were identified and grouped into three functional categories: hormone response, abiotic stress response, and plant growth and metabolism. Hormone response-related CREs included MeJA-response elements (TGACG-motif and CGTCA-motif), abscisic acid response elements (ABRE), auxin response elements (TGA and AuxRR-core), salicylic acid response elements (TCA) and gibberellin response elements (GARE-motif and P-box). Each member of OsDUF506s contained at least two types of hormone response elements, MeJA-response elements and abscisic acid response elements. Regarding abiotic stress response, light response elements, anaerobic induction elements, and anoxic specific inducible elements were the most prevalent. In subfamily II and IIIb, there were low-temperature response (LTR) elements. Drought inducible elements (MSB) existed in OsDUF50601, OsDUF50602, OsDUF50607, and OsDUF50609, indicating that they may be associated with drought stress. Fewer CREs were related to plant growth and metabolism. CAT-boxes relating to meristem expression were found in half of OsDUF506s, while Motif I in OsDUF50601, OsDUF50605, and OsDUF50606 was involved in root specificity. Only a few OsDUF506 members possessed CREs associated with circadian control, cell cycle regulation, zein metabolism regulation, endosperm expression, and flavonoid biosynthetic genes regulation. These results suggested that OsDUF506s might be essential in hormone regulation and abiotic stress response.
Figure 3: Distribution of CREs in OsDUF506s promoters.
Predicted CREs with different functions are displayed with boxes in different colors. The light gray bars represent the 2,000 bp sequence upstream of OsDUF506s.Analysis of SNPs in OsDUF506 members
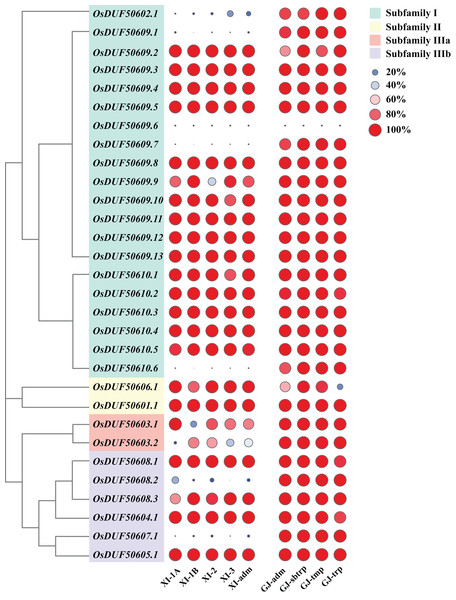
According to the Rice SNP-Seek Database, 78 SNPs were identified in OsDUF506s, of which 30 were non-synonymous SNP (nsSNP) (Table 2). OsDUF50609 and OsDUF50610 possessed 13 and six nsSNPs, respectively, while the other members possessed between one to three nsSNP, indicating they were relatively conserved. To explore the subpopulation-specific variants, 30 nsSNP genotyping data of 2,644 Oryza sativa varieties from nine subpopulations (five Xian/indicia (XI) subpopulations and four Geng/japonica (GJ) subpopulations) were analyzed. SNPGJ-index represented the proportion of varieties that shared the same allele as the reference Nipponbare. Three nsSNPs (OsDUF50609.1, OsDUF50609.7, and OsDUF50610.6) from subfamily I and one nsSNP (OsDUF50607.1) from subfamily IIIb were specific between Indica and Japonica varieties, because the SNPGJ-index values in Indica subpopulations were less than 5%, while the values in the Japonica subpopulations were greater than 85% (Fig. 4 and Table S3).
| Subfamily | Gene ID | No. total SNP | nsSNP ID | Ref allele | Alt allele | Position |
|---|---|---|---|---|---|---|
| I | OSDUF50602 | 2 | OSDUF50602.1 | C | T | Chr1:39867256 |
| OsDUF50609 | 23 | OsDUF50609.1 | G | T | Chr11:14248387 | |
| OsDUF50609.2 | C | A | Chr11:14248411 | |||
| OsDUF50609.3 | C | T | Chr11:14248432 | |||
| OsDUF50609.4 | A | G | Chr11:14248446 | |||
| OsDUF50609.5 | C | T | Chr11:14248498 | |||
| OsDUF50609.6 | A | G | Chr11:14248539 | |||
| OsDUF50609.7 | C | G | Chr11:14248616 | |||
| OsDUF50609.8 | C | G | Chr11:14248636 | |||
| OsDUF50609.9 | C | T | Chr11:14248665 | |||
| OsDUF50609.10 | G | T | Chr11:14248737 | |||
| OsDUF50609.11 | C | T | Chr11:14248861 | |||
| OsDUF50609.12 | A | C | Chr11:14248932 | |||
| OsDUF50609.13 | T | C | Chr11:14249448 | |||
| OsDUF50610 | 9 | OsDUF50610.1 | G | T | Chr11:14267840 | |
| OsDUF50610.2 | C | T | Chr11:14267960 | |||
| OsDUF50610.3 | G | A | Chr11:14268083 | |||
| OsDUF50610.4 | C | G | Chr11:14268298 | |||
| OsDUF50610.5 | C | T | Chr11:14268593 | |||
| OsDUF50610.6 | T | C | Chr11:14268614 | |||
| II | OsDUF50606 | 3 | OsDUF50606.1 | G | A | Chr5:25778321 |
| OSDUF50601 | 9 | OSDUF50601.1 | A | G | Chr1:31272839 | |
| IIIa | OSDUF50603 | 12 | OSDUF50603.1 | G | A | Chr1:43017098 |
| OSDUF50603.2 | C | A | Chr1:43018592 | |||
| IIIb | OsDUF50608 | 7 | OsDUF50608.1 | G | C | Chr10:14658953 |
| OsDUF50608.2 | G | A | Chr10:14658996 | |||
| OsDUF50608.3 | C | G | Chr10:14659216 | |||
| OsDUF50604 | 3 | OsDUF50604.1 | T | C | Chr3:3381185 | |
| OsDUF50607 | 3 | OsDUF50607.1 | G | C | Chr7:4305870 | |
| OsDUF50605 | 7 | OsDUF50605.1 | T | A | Chr3:33171316 |
Figure 4: SNPGJ-index values of OsDUF506s nsSNPs in nine subpopulations.
Circles in different colors and sizes represent different SNPGJ-index values.Synteny analysis of OsDUF506 members
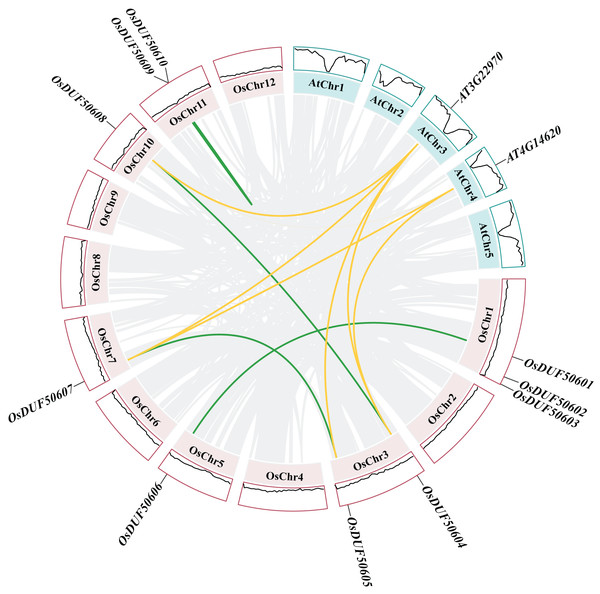
Gene duplications are significant for gene family evolution. The synteny analysis result showed three segmental duplications (OsDUF50601-OsDUF50606, OsDUF50604-OsDUF50608, and OsDUF50605-OsDUF50607) and one tandem duplication (OsDUF50609-OsDUF50610) in Oryza sativa (Fig. 5), indicating that segmental duplication was the core expansion dynamic of OsDUF506s evolution. All duplications had Ka/Ks ratios below 0.5 (Table S4), demonstrating that OsDUF506s have undergone purifying selective pressure during evolution. In addition, these duplicated events were speculated to occurred at least 20.48 million years ago (Table S4).
Figure 5 : Synteny analysis of DUF506s in Oryza sativa and Arabidopsis.
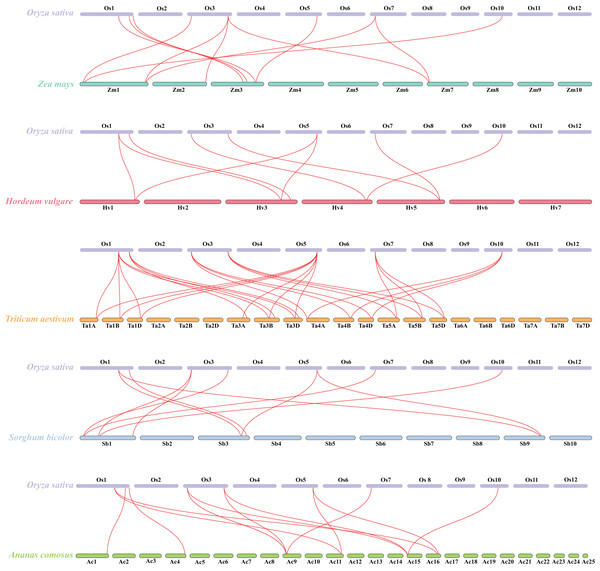
Pink and green bars represent the chromosomes of Oryza sativa and Arabidopsis respectively. The black lines in the colored box show the gene density of the chromosomes. The green lines suggest duplicated gene pairs in Oryza sativa, the yellow lines indicate the collinearity between Oryza sativa and Arabidopsis thaliana.Besides, duplicated events of DUF506s in Oryza sativa, the dicot model plant (Arabidopsis thaliana), and other monocotyledonous species (Zea mays, Hordeum vulgare, Triticum aestivum, Sorghum bicolor, and Ananas comosus) were also identified. OsDUF50604, OsDUF50605, OsDUF50607, and OsDUF50608 in Oryza sativa and AT3G22970 and AT4G14620 in Arabidopsis from subfamily IIIb formed six homologous gene pairs and showed multiple collinearities (Fig. 5). In five monocotyledonous species, other than OsDUF50609 and OsDUF50610 on chromosome 11, the other OsDUF506s all had homologous genes. A total of 10 colinear gene pairs were found between Oryza sativa and Hordeum vulgare, 11 pairs with Zea mays and Sorghum bicolor, respectively, 12 pairs with Ananas comosus, and 30 pairs with Triticum aestivum (Fig. 6 and Table S5). The results suggested that these DUF506s might be derived from the same ancestral type and function similarly. The OsDUF506s from subfamily IIIb were involved in gene pairs with Arabidopsis thaliana and those five monocots, respectively, indicating that they might be critical to the evolution of the DUF506 family.
Figure 6: Synteny analysis of DUF506s between Oryza sativa and five monocot species.
Red lines suggest syntenic gene pairs.Predicted miRNAs analysis
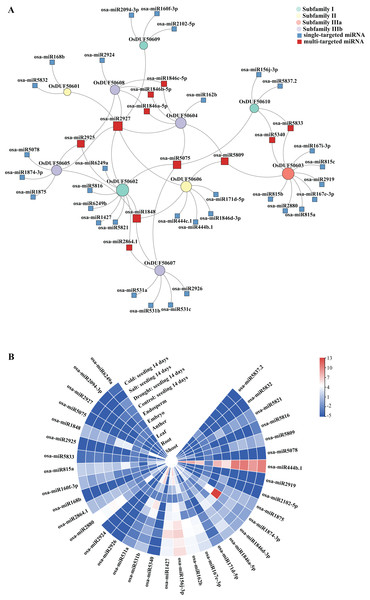
Sixty-nine predicted miRNAs that target OsDUF506s and might be involved in expression regulations were identified (Fig. 7A). All OsDUF506s were targeted by multiple miRNAs, suggesting these genes were strictly regulated by the combination of multiple miRNAs. Multiple OsDUF506s were targeted by osa-miRNA2927, osa-miRNA5075, osa-miRNA1848, osa-miRNA2925, and osa-miRNA5809, indicating that these miRNAs were essential to OsDUF506s. The lengths of matured miRNAs ranged between 19 and 24 nucleotides (Table S6). Most of the miRNAs inhibited OsDUF506 expressions by cleavage, only osa-miR2880, osa-miR5340, osa-miR2926, osa-miR156j-3p, osa-miR1875, osa-miR444b.1, osa-miR444c.1, and osa-miR5832 inhibited OsDUF506 expressions by translation repression.
Figure 7: Analysis of predicted miRNA targeting OsDUF506s.
(A) Identified miRNAs targeting OsDUF506s. Circles represent OsDUF506s, squares represent the related miRNAs. (B) Expressions of predicted miRNAs in different tissues and under abiotic stresses. The heatmap demonstrates the expression level, the color gradient from blue to red presents increasing expression values.The expression analysis revealed that the majority of miRNAs were expressed at a modest level in Oryza sativa tissues (Fig. 7B). The osa-miR1874-3p, which targeted OsDUF50605 specifically, showed the highest expression level in embryo. The osa-miR444b.1 targeting OsDUF50606 was highly expressed in all tissues except anther, indicating that OsDUF50606 expression was limited in these tissues, which may inhibit the function of OsDUF50606 in plant growth. The expression levels of miRNAs under drought, salt, and cold stress showed no significant changes, suggesting they did not participate in abiotic stress responses.
Expression analysis of OsDUF506 members in different tissues and induced by plant hormones
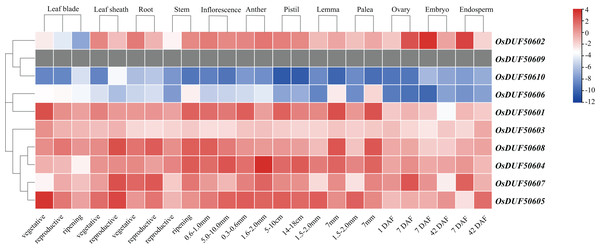
To investigate the expression specificities of OsDUF506s, the expression values in the leaf blade, leaf sheath, root, stem, inflorescence, anther, pistil, lemma, palea, ovary, embryo, and endosperm were analyzed (Fig. 8; Table S7). OsDUF50602 from subfamily I expressed in all tissues, with the highest expressions in the embryo and endosperm 7 days after flowering, whereas OsDUF506010 was expressed at an exceedingly low level in all tissues. The expression modes of the segment duplication gene pair OsDUF50601-OsDUF50606 from subfamily II were wholly dissimilar, suggesting that they might be involved in functional redundancy. Compared to the genes from subfamily IIIa, the expression levels of four genes from subfamily IIIb were higher in most tissues. The segment duplication gene pair OsDUF50604-OsDUF50608 was highly expressed in anther and leaf sheath, respectively. In contrast, another segment pair, OsDUF50605-OsDUF50607, was highly expressed in the leaf blade and leaf sheath, revealing that they might be involved in functional differentiation.
Figure 8: Expressions of OsDUF506 members in different tissues.
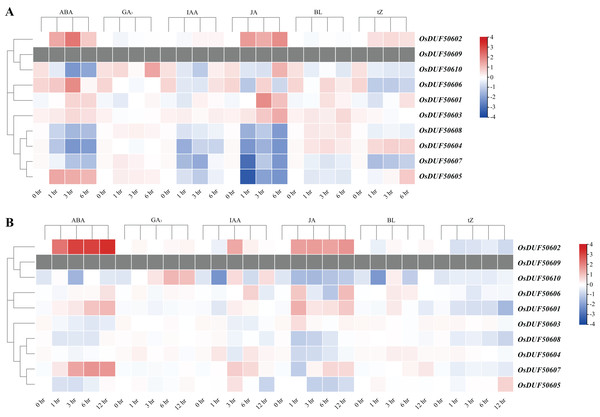
The heatmap demonstrates the expression level, the color gradient from blue to red presents increasing expression values. Grey presents missing data.The CREs analysis of OsDUF506s suggested that they were widely involved in hormonal regulation, thus we analyzed their expression profiles in root and shoot treated with six plant hormones, including abscisic acid (ABA), gibberellic acid (GA3), indole-3-acetic acid (IAA), jasmonic acid (JA), brassinolide (BL), and trans-zeatin (tZ). OsDUF506s showed distinct regulating modes (Fig. 9 and Table S8). Significant upregulation of OsDUF50602 was induced by ABA and JA, whereas opposite regulation was induced by tZ in root and shoot. Subfamily II members, OsDUF50601 and OsDUF50606 exhibited identical regulation under ABA treatment. In the root, except OsDUF50605 was upregulated by ABA induction, the other members from subfamily IIIb were downregulated by ABA, IAA, and JA. Under BL treatment, OsDUF50604 and OsDUF50608 were upregulated, whereas OsDUF50605 and OsDUF50607 were downregulated in root. Interestingly, OsDUF50607 was significantly upregulated by ABA induction in shoot, while the opposite was observed in root. OsDUF50605 also showed opposite effects on shoot and root regulations by ABA induction.
Figure 9: Expressions of OsDUF506s induced by plant hormones in root (A) and shoot (B).
The heatmap demonstrates the expression levels, the color gradient from blue to red presents increasing expression values. Grey presents missing data.Expression analysis of OsDUF506 members under drought stress, cold stress, and phosphorus-deficient stress
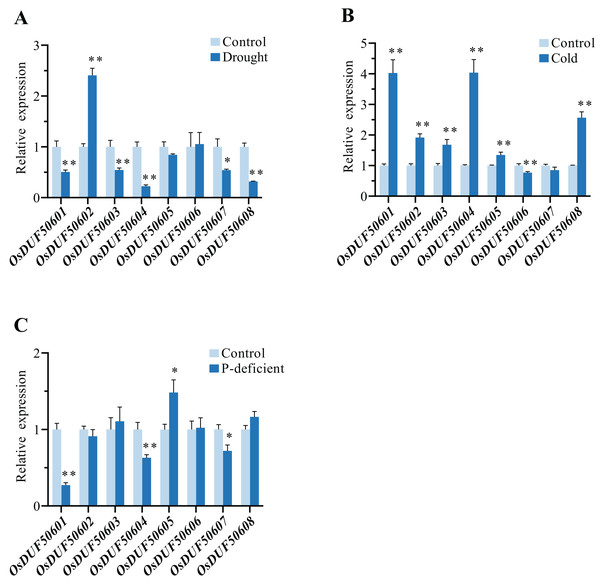
To explore the responses of OsDUF506s under abiotic stresses, the relative expressions of eight OsDUF506s under drought, cold, and phosphorus-deficient stresses were analyzed by qRT-PCR (Fig. 10). Under drought condition, the expressions of OsDUF50601, OsDUF50603, OsDUF50604, OsDUF50607, and OsDUF50608 were significantly downregulated, whereas only OsDUF50602 was significantly upregulated with a more than 2-fold increase. By contrast, OsDUF506s were more sensitive to cold stress. Under cold treatment, six OsDUF506s were upregulated with the exception of OsDUF50606 and OsDUF50607, OsDUF50601, and OsDUF50504, which showed a 4-fold increase in gene expression. Under phosphorus-deficient condition within a week, OsDUF50601, OsDUF50604, and OsDUF50607 were significantly downregulated, whereas only OsDUF50605 was significantly upregulated but by less than 2-fold. The result demonstrated the majority of OsDUF506s were induced by drought and cold stresses, suggesting these genes might be implicated in these stress responses.
Figure 10: Relative gene expressions of OsDUF506s under drought (A), cold (B) and phosphorus-deficient (C) stresses.
Data represent the mean ± SE of three biological and technical replicates. The significant differences level was analyzed by unpaired t-test (*p < 0.05, **p < 0.01).Discussion
DUFs are a specific type of genes with conserved domains but unknown functions. DUF506 family belongs to the PD-(D/E)XK nuclease superfamily, which consists of numerous enzymes involved in significant cellular processes (Knizewski et al., 2007). Most families are certified to be distinct restriction endonucleases with functions including repair of damaged DNA, resolution of holliday junctions, and excessive cleaving in DNA recombination (Bujnicki, 2003). The family retains the characteristic motif II of PD-(D/E)XK, but lacks a functionally-characterized domain (Knizewski et al., 2007). Until now, DUF506 family research has only performed in Arabidopsis (Ying, 2021). In this study, we identified 10 OsDUF506s with an intact DUF506 domain in Oryza sativa and categorized them into four subfamilies based on the previous study in Arabidopsis (Table 1). Additionally, 120 DUF506 genes from five other monocots and four dicots were also identified and their phylogenetic relationship with OsDUF506s was analyzed. In contrast to DUF1618s, which only existed in monocot, DUF506s were present in both monocots and dicots (Fig. 1), indicating that DUF506 was an ancient gene family that originated prior to the dicotyledon-monocotyledon divergence (Wang et al., 2014). DUF506 members from monocots or dicots preferred to congregate on the same branch (Fig. 1), indicating a substantial divergence in DUF506s between monocots and dicots. Moreover, genome size did not correlate with the number of DUF506 genes. In dicots, for example, Arabidopsis thaliana and Solanum tuberosum possessed the same quantity of DUF506 genes, but their genome sizes differed significantly.
Gene duplication is widespread in plants and contributes to genome expansion and the evolution of new functions. The majority of gene duplications consist of whole-genome duplications and tandem duplications (Panchy, Lehti-Shiu & Shiu, 2016). However, in OsDUF506 duplication events, segmental duplication accounted for 75%, while tandem duplication only accounted for 25% (Table S4), suggesting that segmental duplication was critical for expanding OsDUF506 family members. The dynamic processes between gene duplications and gene losses contribute to genomes differences (Holland et al., 2017). Research on Oryza sativa duplications revealed that 85% of duplicates underwent loss, subfunctionalization, or neofunctionalization during 50-70 million years of evolution (Throude et al., 2009). OsDUF50602 and OsDUF50603 did not form duplicate gene pairs (Fig. 5), indicating they could undergo gene losses. The mechanisms of duplicate gene loss include the deletion of duplicate sequence and pseudogenization, the latter of which refers to gene silence and genetic redundancy (Ho-Huu et al., 2012; Lynch & Conery, 2000; Thibaud-Nissen, Ouyang & Buell, 2009). The duplicated gene with low expression may experience pseudogenization, and pseudogenes typically originate from tandem duplications (Yang et al., 2011). This viewpoint is supported by the tandem duplicate pair OsDUF50609-OsDUF50610, which originated 58.69 million years ago. According to the gene expression profiles of the RiceXPro database (Fig. 8) and Rice Genome Annotation Project database (http://rice.uga.edu/expression.shtml), they were barely expressed in any tissue. Besides, compared to other members, OsDUF50609 and OsDUF50610 contained the most nsSNP mutations (Table 2), and excessive nonsynonymous nucleotide mutants are a characteristic of pseudogenes (Balakirev & Ayala, 2003). DUF family appears to be abundant in pseudogenes, such as DUF1311, DUF 1124, and DUF 3054 (Thibaud-Nissen, Ouyang & Buell, 2009). On the other side, OsDUF50609 and OsDUF50610 exhibited a general loss bias in Gramineae species, including Zea mays, Hordeum vulgare, Triticum aestivum, and Sorghum bicolor (Fig. 6). Except for the presumed pseudogenes, the other OsDUF506s all have colinear gene pairs with the other five monocots (Fig. 6), revealing that they were conserved in the evolution and expansion of the DUF506 family in plants and they might have originated from the same ancestor. Previous studies suggested that more than half of duplicated genes have diverged in gene expression in both Arabidopsis and Oryza sativa (Blanc & Wolfe, 2004; Yim, Lee & Jang, 2009). Two duplicated gene pairs from subfamily IIIb (OsDUF50604-OsDUF50608 and OsDUF50605-OsDUF50607) revealed different tissue specificity (Fig. 8), and duplicates respond conversely under phosphorus-deficient conditions (Fig. 10), suggesting that they might be neofunctionalized.
MicroRNAs (miRNAs) are fundamental noncoding riboregulators for gene expression. In plants, miRNA silences genes by guiding RNA cleavage or translation inhibition (Song et al., 2019). They cooperate closely with target genes and transcription factors to regulate plant growth and resistance; it could be an effective strategy for precisely improving Oryza sativa varieties by derepressing specific genes using CRISPR/Cas9 (Lin et al., 2021; Nadarajah & Kumar, 2019). We predicted the miRNAs targeting OsDUF506s and analyzed their expression profiles in tissues and under stresses of drought, cold, and salt (Fig. 7). The osa-miR444b.1, which specifically targeted OsDUF50606, was the only one highly expressed in tissues excluding the anthers and under the four stresses. It was functionally unknown. One copy of the segmental duplicates from subfamily II (OsDUF50606) expressed extremely low in all tissues, it showed no significant changes in expression level under those abiotic stresses. In contrast, the other copy (OsDUF50601) was constitutively expressed and was strongly induced by drought, cold, and phosphorus-deficient stresses (Figs. 8 and 10). Further verification is required to ascertain if the expression difference originates from the translation inhibition by osa-miR444b.1.
The results of expression induced by plant hormones showed that OsDUF506s were more sensitive to ABA and JA treatments than IAA and GA3 treatments, which was consistent with the presence of hormone-responsive CREs observed in their promoters (Figs. 3 and 9). Although the promoter region of some OsDUF506s contained a few GA3 and IAA CREs, they show no significant and specific response trend under corresponding hormone treatment.
The CREs analysis also showed that OsDUF50601, OsDUF50602, OsDUF50607, and OsDUF50609 each contained one MBS, which was a MYB transcription factor binding site responsive to drought. However, only OsDUF50602 was upregulated under drought treatment (Figs. 3 and 10), indicating that their promotors might recruit different MYBs to active OsDUF50602 expression and inhibit OsDUF50601 and OsDUF50607 expressions to regulate drought tolerance. Perhaps this is due to the vital function of hormones under the drought condition. ABA is an important plant hormone regulating water status and stomatal movement. When plants suffer from a drought environment, they synthesize ABA. Increasing ABA could induce plants to close their stomatal and retain water (Lim et al., 2015). Seven copies of ABRE(ABA-responsive element)existed in the promotor region of OsDUF50602. Its expression significantly increased in both shoot and root under ABA treatment, suggesting that it might also be involved in the ABA-related signaling pathway of drought responses (Fig. 9). Moreover, a previous study revealed that the expression of OsDUF50602 was higher in OsDT11 OE lines than in the wild line under drought treatment, indicating that the drought response of OsDUF50602 might be enhanced in the particular genetic background (Zhao et al., 2020). ABRE has been proved to be the most conserved drought-inducible promoter in Arabidopsis, Oryza sativa, and soybean, indicating that the transcriptional regulation of drought-inducible genes like OsDUF50602 is similar across these species (Maruyama et al., 2012). Therefore, we analyzed the expressions of its homologous genes under drought stress in the eplant database (http://bar.utoronto.ca/). In Triticum aestivum, TraesCS3A02G420900 gene expression increased 3.34-fold under drought stress. On the other hand, JA and its derivatives, which occur at low levels under normal condition, accumulate to high levels and are transmitted over long distances under abiotic stress (Wang et al., 2021a). The promoter of OsDUF50602 contained six copies of the CGTCA/TGACG-motif (JA-responsive element). With JA treatment, the expression of OsDUF50602 was rapidly and significantly upregulated in both the shoot and root (Fig. 9). However, further verification is required to determine whether these two hormones collaboratively induce the drought responses of OsDUF50602.
The OsDUF506s were more sensitive to cold stress than to drought stress. Under cold stress, except for the slight downregulation of OsDUF50606 and OsDUF50607, the other OsDUF506s were significantly upregulated, and three genes containing LTR elements (OsDUF5060601, OsDUF5060604, and OsDUF50608) showed the highest level (Fig. 10). DUF506s from other species also revealed an active response to cold stress. For instance, At1g62420, At3g25240, Bradi2g58590, and Bradi2g62310 were strongly induced by cold stress in Arabidopsis and Brachypodium (Ying, 2021). The above results demonstrated that DUF506 genes play a crucial role in cold response with different mechanisms.
Recent studies showed five AtDUF506 genes belonging to subfamilies I and II (At1g62420, At3g07350, At3g25240, At2g20670, and At4g32480) were strongly upregulated by P-limitation (Ying et al., 2022; Ying & Scheible, 2022). However, although the OsDUF506s, which belong to subfamilies I and II, shared a highly similar exon/intron structure and conserved motifs (Fig. 3), only OsDUF50601 was significantly downregulated by P-limitation, indicating that monocot and dicot species differed in phosphorus responding signal pathway.
Rice (Oryza sativa) is an essential staple food for approximately half of the world’s population, and stable rice production is crucial for food security, especially in Asia (Zhang, 2007). However, extreme weather and climate change, such as drought, flooding, salinity, low temperature, high temperature, and mineral deficiency, seriously affect crop productivity and sustainability. Among the abiotic stress, drought is the most destructive threat. Half of the world’s arable land will suffer from drought in the next three decades (Singhal et al., 2016). Hence, it is urgent to breed rice varieties with superior drought resistance. Drought resistance is a complex agronomic trait regulated by multiple genes. Exploring drought resistance genes through linkage analysis or GWAS is inefficient and genes with minor effects are hard to clone (Sun et al., 2022). With the development of bioinformatics and gene editing technology, the reverse genetics method can be combined to identify and characterize abiotic resistance-related genes such as OsDUF506s. Abiotic resistance improvements usually require increased expression of related genes, including DROUGHT1 (DROT1), ONAC066, OsMADS23, DROUGHT-INDUCED BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE (OsDIAT), CHILLING-TOLERANCE DIVERGENCE 1 (COLD1), and Cyclic Nucleotide-gated Channels 9 (OsCNGC9) (Sun et al., 2022; Yuan et al., 2007; Li et al., 2021; Shim et al., 2023; Ma et al., 2015; Wang et al., 2021b). A donor-DNA-free CRISPR/Cas-based approach to knock-up genes in rice could be helpful for rice breeding practices and simplify regulatory approval of the edited plants (Lu et al., 2021).
Conclusions
In this study, 10 OsDUF506 family members in Oryza sativa were identified and classified into four subfamilies. We analyzed the phylogenetic relationship, gene structures, conserved motif, CREs, SNP distribution, and targeting miRNA, which filled the gap of DUF506 family in rice. The results of public expression profiles and RT-qPCR data in tissues and under plant hormones and abiotic stresses demonstrated that OsDUF506s were actively involved in ABA and JA response and had different expression patterns under drought and cold, which laid the foundation for further functional analysis of OsDUF506 family.