Association between INHA gene polymorphisms and litter size in Hainan black goats
- Published
- Accepted
- Received
- Academic Editor
- Antonio Amorim
- Subject Areas
- Agricultural Science, Genetics, Molecular Biology, Zoology
- Keywords
- Hainan black goats, INHA gene, Polymorphism, Reproductive performance, Litter size
- Copyright
- © 2023 Bian et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Association between INHA gene polymorphisms and litter size in Hainan black goats. PeerJ 11:e15381 https://doi.org/10.7717/peerj.15381
Abstract
Background
The inhibin alpha (INHA) gene is one of the important genes affecting the reproductive traits of animals. Hainan black goats are the main goat breed in Hainan Island (China), whose development is limited by low reproductive performance. However, the relationship between INHA gene and the reproductive performance of Hainan black goats is still unclear. Therefore, the purpose of this work was to explore the effect of INHA gene polymorphisms on the litter size of Hainan black goats.
Methods
Single nucleotide polymorphisms (SNPs) of INHA were detected, and the genetic parameters and haplotype frequency of these SNPs were calculated and association analysis was performed for these SNPs with the litter size. Finally, the SNP with significant correlations to litter size was analyzed by Bioinformatics tools.
Results
The results showed that the litter size of individuals with the AC genotype at loci g.28317663A>C of INHA gene was significantly higher than those with the AA genotype. This SNP changed the amino acid sequence, which may affect the function of INHA protein by affecting its structure. Our results suggest that g.28317663A>C loci may serve as a potential molecular marker for improving the reproductive traits in Hainan black goats.
Introduction
As one of the earliest domesticated livestock species, goats are closely related to human activities (Tabbaa & Al-Atiyat, 2009). Hainan black goats are a unique local breed in Hainan and the only breed on the tropical island in China (Hu et al., 2016). Hainan black goats have a good tolerance to hot and humid environment, and their meat is delicious and popular among people in southern China (Shi et al., 2020). Hainan black goats usually attain sexual maturity at 4–6 months of age, and first breed at 7–8 months of age. They normally produce a single kid per year and sometimes three kids in 2 years (Hua et al., 2019). Due to their poor reproductive performance, their development is currently limited.
Kidding traits are affected by several factors (genetics, environment, nutritional status, etc.). Genetic factors are the most critical factors (González-Rodríguez et al., 2018; Ofori & Hagan, 2020; Zheng et al., 2020). The INHA gene has been shown to be associated with the kidding ability of many animals, such as the Suhuai pigs and Dazu black goats (Liu et al., 2017a; Wang et al., 2021). INHA gene is the main gene controlling the biological activity of inhibin. Inhibin is mainly secreted by male testicular cells and female ovarian granulosa cells (Cai et al., 2011; Cui et al., 2020). Inhibin regulates reproductive functions through endocrine, paracrine, and autocrine modes, specifically acting on the pituitary gland. It inhibits the secretion of follicle-stimulating hormone (FSH), and locally regulates the production of estrogen (E) (Burger, 1988). High levels of FSH and E will lead to greater ovulation in animals and thus more chances of multiple births, which may be an important reason why INHA affects litter size. However, INHA gene polymorphism in Hainan black goats has not been studied.
When considering candidate genes involved in reproductive performance, it is important to detect single nucleotide polymorphisms (SNPs) and to analyze the associations between these SNPs and reproductive traits. Therefore, this study aims to detect INHA gene polymorphisms in Hainan black goats by sequencing and analyzing the relationship between INHA gene variants and litter size.
Materials and Methods
Animals, sample collection, and DNA preparation
All of the conducted procedures were approved by the Hainan University Institutional Animal Use and Care Committee (No. HNUAUCC-2022-000121). Jugular blood samples of 211 Hainan black does were collected from Hainan Chuxin Animal Husbandry Co,. Ltd. (Ding’an, Hainan, China) in two different locations. There were 90 Hainan black does with records of kidding, of which 37 had two kids per litter and 53 had single kid per litter. The remaining 121 Hainan black does were randomly selected in the farms. The genomic DNA of these ewes was extracted with a blood genomic DNA extraction kit (Tiangen Biotech Co., Ltd., Beijing, China).
Amplification of the INHA coding sequence
The full-length exon sequence (accession number: NC_030809.1) of the INHA gene of Hainan black goats was used for designing four pairs of primers using the Primer 3Plus web-site according to the INHA gene sequence (GENE ID: 100861261) in NCBI (Table 1). We divided 211 DNA samples of Hainan black does into three groups consisting of 70/70/71 samples. DNA samples in each group were pooled and PCR conducted using primers 1, 2, 3, 4. Primer 1 is used to amplify exon 1, and primer 2–4 is used to amplify exon 2.
| Primer name | Primer sequence (5′–3′) | Position in goat sequence | Annealing temperture/°C |
Amplified fragment/bp |
|---|---|---|---|---|
| INHA-1-F | AGAGATAGGAGGTCTCAATG | INHA-exon 1 (−198…406) | 56 | 605 |
| INHA-1-R | AGGAAGGTTTAGTAACTGGT | |||
| INHA-2-F | AGAGGGGTCCCAGGTTTT | INHA-exon 2 (-59…540) | 56 | 600 |
| INHA-2-R | GCTCCTGGAAAGAGATATTGA | |||
| INHA-3-F | GTTGTCCTCTCTGTTCCTG | INHA-exon 2 (316…821) | 56 | 506 |
| INHA-3-R | TTAGATGCAAGCACAGTGC | |||
| INHA-4-F | ATCTTCCACTACTGTCAC | INHA-exon 2 (579…1010) | 56 | 432 |
| INHA-4-R | CACTTATCAGAGAAGCTTG | |||
| INHA-5-F | GGACAGACAGGAGACCACT | INHA-exon 2 (131…910) | 59 | 780 |
| INHA-5-R | GCCTCTGAGCAGAGAGGAG |
Note:
The numbers in parentheses start with the first base of the corresponding CDS region of INHA gene.
Genotyping of INHA polymorphism
The gene sequence was detected by Shanghai Shengong Bioengineering Co., LTD., and then visualized using SnapGene software. The genotype was determined by the visual sequencing peak. In pooled sequencing, we found the SNPs in both exon 1 and 2. For the one SNP on exon 1, we used primer INHA-1 for amplification. The multiple SNPs on exon 2 spanned the fragment of primers INHA-2, 3, and 4, so we redesigned primer INHA-5 to amplify these SNPs (Table 1). Finally, the distribution of the SNP loci was compared and analyzed. The sequencing data of INHA gene has been uploaded in the GenBank database with the accession number (OQ461736–OQ462157).
Statistical analysis
Popgen32 software was used to count the genotype frequency, allele frequency, observed heterozygosity (Ho), expected heterozygosity (He), effective allele number (Ne), and polymorphism information content (PIC), and to conduct a Hardy Weinberg equilibrium test. Haploview4.2 software was used to analyze the haplotypes and linkage disequilibrium (LD) of the SNPs. At the same time, the same general linear model (GLM) program of SPSS 19.0 software was used to analyze the influence of genotypes and haplotypes on the litter size of Hainan black goats. The models was as follows:
where Yij is the litter size phenotype of the individual Hainan black goats, μ is the population mean, Gi is the effect of genotype or haplotype, and eij is the random error effect.
Bioinformatics analysis
First, the integral and coding sequences of gene were obtained from NCBI (https://www.ncbi.nlm.nih.gov/). Then the secondary and the tertiary structures of the INHA protein and its variant, which was significantly associated with litter size, were predicted by the online websites PRABI (http://www.prabi.fr/) (Combet et al., 2000) and SWISS (https://swissmodel.expasy.org/), respectively (Waterhouse et al., 2018).
Results
INHA gene specific fragment
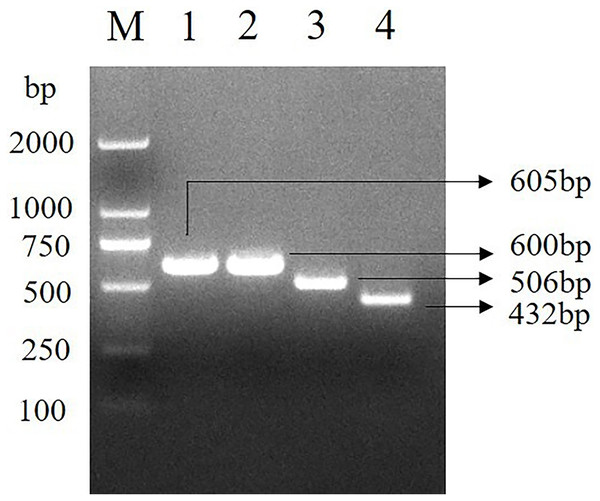
Four pairs of primers were used to amplify the INHA gene of Hainan black goats, and the four DNA sequence fragments of 605, 600, 506 and 432 bp were obtained (Fig. 1). The amplified fragment was consistent with the target fragment and could be analyzed directly by sequencing.
Figure 1: INHA gene specific fragment.
The electrophoresis results of PCR amplified fragments of the INHA gene. M. D2000 DNA Marker; 1. INHA-1-F/INHA-1-R primer amplified fragment; 2. INHA-2-F/INHA-2-R primer amplified fragment; 3. INHA-3-F/INHA-3-R primer amplified fragment; 4. INHA-4-F/INHA-4-R primer amplified fragment.SNPs identified by sequencing
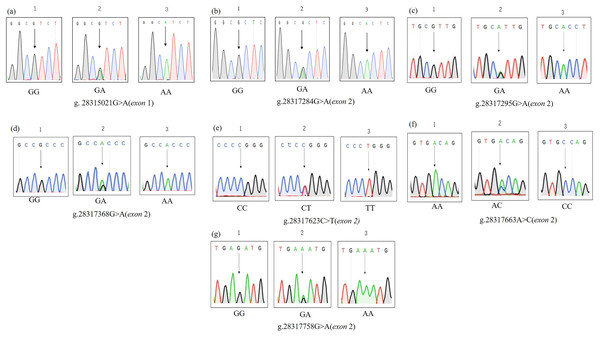
According to the pooled sequencing results, we found seven SNPs. One SNP was located in exon 1, and the other six SNPs were located in exon 2. Since the six SNPs in exon 2 span fragments of primers INHA-2, 3, and 4, we redesigned primer INHA-5 (Material S1) to amplify these SNPs. The PCR products were sequenced, and the sequences of different genotypes are shown (Fig. 2). Among them, g.28315021G>A, g.28317295G>A and g.28317663A>C resulted in amino acid sequence changes (Table 2).
Figure 2: (A–G) SNPs identified by sequencing.
SNPs distribution of the INHA gene in Hainan black goats.| Mutation type | Mutation location | Mutation region | Amino acid change |
|---|---|---|---|
| G/A | g.28315021 | Exon 1 | (Arg→His) |
| G/A | g.28317284 | Exon 2 | (Ala→Ala) |
| G/A | g.28317295 | Exon 2 | (Arg→His) |
| G/A | g.28317368 | Exon 2 | (Pro→Pro) |
| C/T | g.28317623 | Exon 2 | (Pro→Pro) |
| A/C | g.28317663 | Exon 2 | (Thr→Pro) |
| G/A | g.28317758 | Exon 2 | (Glu→Glu) |
Polymorphism analysis of INHA gene in Hainan black goats
Genetic analysis of Hainan black goats showed that all SNPs loci were in Hardy-Weinberg equilibrium (p > 0.05). The g.28315021G>A, g.28317284G>A, g.28317295G>A, g.28317623C>T, and g.28317663A>C sites were in a low polymorphic information content state (PIC < 0.25). The g.28317368G>A and g.28317758G>A sites were in a moderate polymorphic information content state (0.25 < PIC < 0.50). The results show that these two loci have strong selection potential in Hainan black goats, and their genetic diversity is rich. The homozygosity of all loci was higher than heterozygosity (Table 3).
| Locus | Genotype | Allele frequency | He | Ne | PIC | test | |||
|---|---|---|---|---|---|---|---|---|---|
| g.28315021G>A | GG | GA | AA | G | A | 0.244 | 1.323 | 0.241 | 0.013 |
| 155 | 52 | 4 | 0.86 | 0.14 | |||||
| g.28317284G>A | GG | GA | AA | G | A | 0.156 | 1.185 | 0.166 | 0.196 |
| 176 | 34 | 1 | 0.91 | 0.09 | |||||
| g.28317295G>A | GG | GA | AA | G | A | 0.202 | 1.253 | 0.196 | 0.048 |
| 166 | 42 | 3 | 0.89 | 0.11 | |||||
| g.28317368G>A | GG | GA | AA | G | A | 0.470 | 1.886 | 0.471 | 2.293 |
| 35 | 89 | 87 | 0.38 | 0.62 | |||||
| g.28317623C>T | CC | CT | TT | C | T | 0.095 | 1.105 | 0.095 | 0.555 |
| 191 | 19 | 1 | 0.95 | 0.05 | |||||
| g.28317663A>C | AA | AC | CC | A | C | 0.234 | 1.305 | 0.241 | 1.126 |
| 156 | 53 | 2 | 0.86 | 0.14 | |||||
| g.28317758G>A | GG | GA | AA | G | A | 0.283 | 1.395 | 0.282 | 3.68 |
| 149 | 52 | 10 | 0.83 | 0.17 | |||||
Note:
He is the expected heterozygosity; Ne is the number of effective alleles. 0.05 (df = 1) = 3.84.
Association of INHA gene polymorphism and litter size in Hainan black goats
The results showed that the genotypes of 90 Hainan black goats at locus g.28317663 were significantly correlated with litter size. In the Hainan black goat breed, the does with AC genotype had greater litter size than those with AA genotypes (p = 0.046) (Table 4).
| Locus | Genotype | Number of samples | Litter size (Mean ± SD) |
|---|---|---|---|
| g.28315021G>A | GG | 73 | 1.397 ± 0.492 |
| GA | 17 | 1.470 ± 0.515 | |
| g.28317284G>A | GG | 75 | 1.427 ± 0.498 |
| GA | 14 | 1.357 ± 0.497 | |
| AA | 1 | 1.000 ± 0.000 | |
| g.28317295G>A | GG | 74 | 1.419 ± 0.467 |
| GA | 16 | 1.375 ± 0.500 | |
| g.28317368G>A | GG | 12 | 1.500 ± 0.522 |
| AA | 46 | 1.435 ± 0.501 | |
| GA | 32 | 1.344 ± 0.483 | |
| g.28317623C>T | CC | 80 | 1.388 ± 0.490 |
| CT | 10 | 1.600 ± 0.516 | |
| g.28317663A>C | AA | 66 | 1.349 ± 0.480b |
| AC | 24 | 1.583 ± 0.504a | |
| g.28317758G>A | GG | 65 | 1.354 ± 0.482 |
| GA | 22 | 1.500 ± 0.512 | |
| AA | 3 | 2.000 ± 0.000 |
Note:
Results are expressed as mean ± standard deviation. Different letters indicate significant differences (p < 0.05).
Bioinformatics analysis of the INHA g.28317663A>C
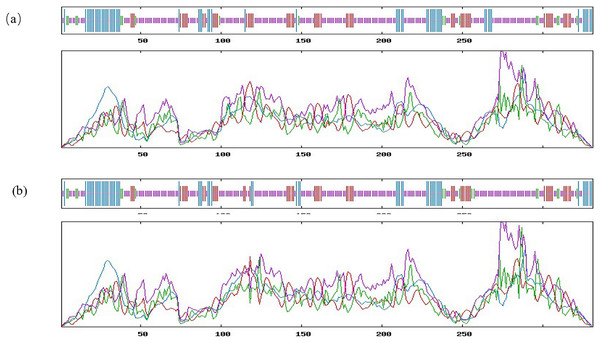
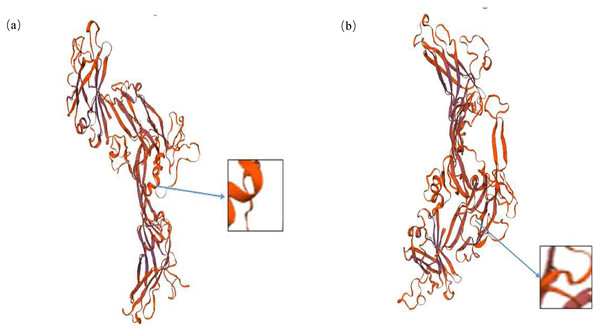
It is noteworthy that the mutation at g.28317663 loci leads to the change of amino acid sequence at the 316 site of inhibin A protein, which is Thr change Pro. PRABI was used to predict the protein secondary structure of alleles A and C at locus 316. The secondary structure of the alpha helix of allele A inhibin protein was 18.13%, that of the extended strand was 16.62%, that of the beta turn was 5.44%, and that of the random coil was 59.82%. The secondary structure of the alpha helix of allele C inhibin protein was 17.52%, that of the extended strand was 16.01%, that of the beta turn was 5.44%, and that of the Random coil was 61.03%. It was found that the missense mutation site changed the alpha helix, random coil, and extended strand in the secondary structure of the protein (Fig. 3). Using SWISS to predict the changes in the tertiary structure of the proteins of alleles A and allele C at locus 316, significant changes with g.28317663A>C mutation were observed (Fig. 4).
Figure 3: Bioinformatics analysis of the INHA gene with g.28317663A>C.
Prediction of the secondary structure of the INHA protein of alleles A (A) and allele C (B). Blue, alpha helix; purple, extended strand; green, beta turn; orange, random coil.Figure 4: Bioinformatics analysis of the INHA gene with g.28317663A>C.
Prediction of the tertiary structure of the INHA protein for alleles A (A) and allele C (B).Linkage disequilibrium and haplotype frequency
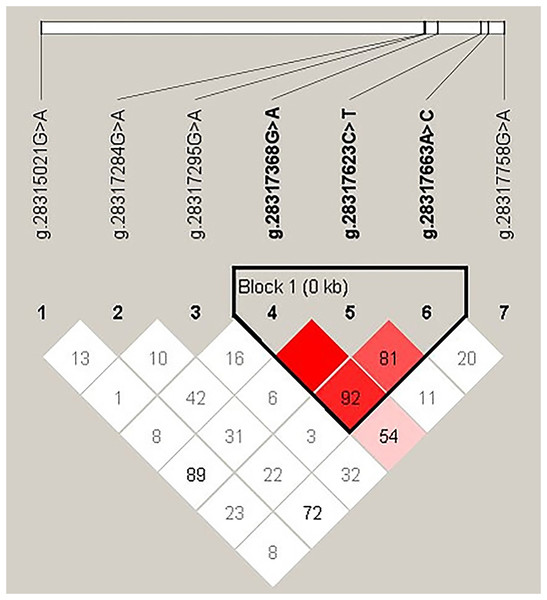
In order to explore the linkage relationship among the seven SNPs in the INHA gene of Hainan black goats (Fig. 5), linkage disequilibrium parameters (D′ and r2) were used. The results of the linkage disequilibrium are showed in Table 5. Linkage disequilibrium was found between g.28317368G>A, g.28317623C>T, and g.28317663A>C of the INHA gene. In these three SNPs, the D′ values ranged from 0.817 to 1.000 and the r2 values from 0.079 to 0.264. When subject to a 1% haplotype frequency threshold (Haploview4.2), a total of four haplotypes were identified within three SNPs of the Hainan black goat INHA gene. With regard to haplotype frequency, the ACA haplotype was found to be the most common, observed at a frequency of 0.595, while the GTC haplotype displayed the lowest frequency, occurring at a frequency of 0.042 (Table 6).
Figure 5: Disequilibrium of linkage among seven SNPS of INHA gene in Hainan black goats.
The color of the square indicates the degree of linkage, the darker the color, the higher the degree of linkage; The value represents the strength of the correlation between sites (percentage).| D′ | r2 | |
|---|---|---|
| g.28317368G>A/g.28317623C>T | 1.000 | 0.079 |
| g.28317368G>A/g.28317663A>C | 0.921 | 0.264 |
| g.28317623C>T/g.28317663A>C | 0.817 | 0.220 |
| Haplotype | g.28317368G>A | g.28317623C>T | g.28317663A>C | Haplotype frequency |
|---|---|---|---|---|
| Hap1 (ACA) | A | C | A | 0.595 |
| Hap2 (GCA) | G | C | A | 0.260 |
| Hap3 (GCC) | G | C | C | 0.089 |
| Hap4 (GTC) | G | T | C | 0.042 |
Association analysis of INHA gene haplotype and litter size in Hainan black goats
We analyzed the association between INHA gene haplotypes and litter size in Hainan black goats (Table 7). The results showed that there was no significant association between haplotypes and litter size.
| Haplotype | Number of samples | Litter size (Mean ± SD) |
|---|---|---|
| Hap1 (ACA) | 77 | 1.407 ± 0.493 |
| Hap2 (GCA) | 58 | 1.448 ± 0.501 |
| Hap3 (GCC) | 23 | 1.608 ± 0.499 |
| Hap4 (GTC) | 7 | 1.714 ± 0.487 |
| p value | 0.135 |
Discussion
Inhibin is an important inhibitor of pituitary gonadotropin secretion of FSH (Cai et al., 2015). Inhibin can competitively bind activing (ACT) receptor with ACT to weaken the action of ACT, thereby reducing the effect of ACT on the positive feedback regulation of pituitary FSH synthesis and secretion, and achieving the effect of negative regulation of FSH secretion (Gaccioli et al., 2018). FSH can stimulate the growth and development of follicles (Casarini & Crepieux, 2019). Decreased inhibin induces multiple ovulation and increase pituitary secretion of FSH, which appears to be the primary mechanism for stimulating additional follicle growth (Wang et al., 2012). Hence, the polymorphism of INHA gene involving the functional center of inhibin has the potential to affect the reproductive performance of animals. Previous studies have found that a large number of INHA gene mutations are closely related to reproductive system problems such as male infertility (Li et al., 2015), premature ovarian failure (Yoon et al., 2012), superovulation (Luo et al., 2019), follicular cysts (Li et al., 2016), and sperm quality (Rafaqat et al., 2020). Considering the above research results, we believe that the INHA gene is an important candidate gene for improving kidding in does. However, it is still unknown whether INHA gene polymorphism has an effect on the reproductive traits of Hainan black goats. Therefore, the relationship between SNPs of the INHA gene and the kidding of Hainan black goats was analyzed, helping to find useful molecular markers for breeding selection Hainan black goats.
Polymorphisms of the INHA gene have been verified in different animals. In this study, we found seven SNPs in this gene, including three new SNPs, which were g.28315021G>A, g.28317368G>A, and g.28317758G>A. Wu et al. (2009) identified 12 SNPs of the INHA gene in Boer goats, four of which were consistent with our study. In addition, studies in other animals have also found that this gene has higher polymorphisms, such as 12 SNPs being detected in the INHA gene of chickens (Cui et al., 2019) and three SNPs in the INHA gene in Murrah bulls (Chandra et al., 2020). Combined with our results, the INHA gene has abundant polymorphisms.
Polymorphisms of the INHA gene are related to reproductive traits in different animals. Studies have found that polymorphisms in the INHA gene are closely related to the reproductive traits of Holstein cows (Sang et al., 2011; Tang et al., 2011). INHA gene polymorphisms have been shown to be significantly correlated with litter size in several goat breeds. For example, Jining Grey goat does with the genotype GA having 0.79 (p < 0.01) more kids than those with the genotype GG in the g.28315680 locus of the INHA gene (Liu et al., 2017b). For g.28318073C>T of the INHA gene, Nigerian goat does with the genotype CT had 0.33 (p < 0.05) more kids than those with the genotype CC (Isa et al., 2017). These results are similar to those in the present study. In our study, for g.28317663A>C of the INHA gene, the litter size of the AC genotype was significantly higher than that of the AA genotype.
In this study, g.28317663A>C, which is a missense mutation, was significantly associated with litter size in Hainan black goats. INHA missense mutations have also been found to be significantly associated with litter size in Malabari goats (Pillai & Venkatachalapathy, 2020). Missense mutations cause amino acid changes that can alter protein properties (e.g., binding, expression, and protein stability) (Norn, André & Theobald, 2021). For example, studies have found that mutations in the ASMT and ADAMTS1 genes can cause changes in protein structure and affect protein function, which may affect the litter size of goats (Hu et al., 2020). We speculate that the change of amino acids from Thr to Pro may affect the function of INHA protein by impacting the structure of secondary and tertiary proteins. This change may affect the secretion of pituitary FSH, and then affect the concentration of FSH, which may ultimately increase the litter size of Hainan black goats. However, the effect still needs further verification.
In this study, three SNPs of the INHA gene were found to be in linkage disequilibrium in Hainan black goats (Table 4) which include g.28317368G>A, g.28317623C>T, and g.28317663A>C of INHA genes. To the best of our knowledge, our study is the first to analyze the linkage disequilibrium of the SNPs of the INHA gene in Hainan black goats. In addition, linkage disequilibrium in the SNPs of this gene has also been found in Luhua chickens (Cui et al., 2019). If these polymorphisms are in linkage disequilibrium with genes that influence variation in reproductive traits, segregation based on marker alleles will result in phenotypic differences. Therefore, the linkage disequilibrium phenomenon needs to be considered when this gene polymorphism is applied to the breeding of goats with a high litter size.
Due to the ancestral structure captured in the distribution of haplotypes, the haplotypes show more influence than the single SNPs for important trait associations (Akey, Jin & Xiong, 2001). Although no significant correlation was found between haplotype and litter size in the haplotype analysis, we found that H3 and H4 enriched with the g.28317663 C allele had a higher litter size than H1 and H2. The g.28317663 C allele may be related to litter size, which supports g.28317663A>C being a useful genetic marker affecting the litter size of Hainan black goats.
Conclusions
In this study, a total of seven SNPs loci were found in the INHA gene of Hainan black goats, among which g.28317663A>C was significantly associated with litter size. Mutation at this site will cause changes in amino acids from Thr to Pro, affecting the secondary and tertiary structure of INHA protein. The INHA gene is one of the genes that affect the reproductive capacity of Hainan black goats, and the g.28317663A>C loci may be a potential genetic marker for future breeding. It will be nevertheless necessary to expand the sample size to further study the impact of INHA gene polymorphism on the litter size of Hainan black goats.
Supplemental Information
The electrophoresis result of PCR amplified by primer INHA-5.
The primer INHA-5 was used to amplify 6 SNPs in exon 2 of the INHA gene