The DNA barcode reveals cryptic diversity and a new record for the genus Leporinus (Characiformes, Anostomidae) in the hydrographic basins of central northern Brazil
- Published
- Accepted
- Received
- Academic Editor
- Jörg Oehlmann
- Subject Areas
- Aquaculture, Fisheries and Fish Science, Genetics, Molecular Biology, Zoology, Freshwater Biology
- Keywords
- Molecular identification, Freshwater fish, Leporinus, Neotropical biodiversity, Systematics
- Copyright
- © 2023 Nascimento et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. The DNA barcode reveals cryptic diversity and a new record for the genus Leporinus (Characiformes, Anostomidae) in the hydrographic basins of central northern Brazil. PeerJ 11:e15184 https://doi.org/10.7717/peerj.15184
Abstract
Leporinus is one of the most speciose genera of the order Characiformes, with 81 valid species distributed throughout much of Central and South America. The considerable diversity of this genus has generated extensive debate on its classification and internal arrangement. In the present study, we investigated the species diversity of the genus Leporinus in central northern Brazil, and conclude that six valid species—Leporinus maculatus, Leporinus unitaeniatus, Leporinus affinis, Leporinus venerei, Leporinus cf. friderici, and Leporinus piau—are found in the hydrographic basins of the Brazilian states of Maranhão, Piauí, and Tocantins. We analyzed 182 sequences of the Cytochrome Oxidase subunit I gene, of which, 157 were obtained from Leporinus specimens collected from the basins of the Itapecuru, Mearim, Turiaçu, Pericumã, Periá, Preguiças, Parnaíba, and Tocantins rivers. The species delimitation analyses, based on the ABGD, ASAP, mPTP, bPTP, and GMYC methods, revealed the presence of four distinct molecular operational taxonomic units (MOTUs), identified as L. maculatus, L. unitaeniatus, L. affinis, and L. piau (from the Parnaíba River). The bPTP method restricted L. venerei to a single MOTU, and confirmed the occurrence of this species in the rivers of Maranhão for the first time. The separation of L. cf. friderici into two clades and the subsequent formation of different operational taxonomic units was consistent with polyphyly in this species, which indicates the existence of cryptic diversity. The arrangement of L. cf. friderici and L. piau in two different clades supports the conclusion that the L. piau specimens from Maranhão were misidentified, based on their morphological traits, reflecting the taxonomic inconsistencies that exist among morphologically similar species. Overall, then, the species delimitation methods employed in the present study indicated the presence of six MOTUs—L. maculatus, L. unitaenitus, L. affinis, L. cf. friderici, L. venerei, and L. piau. In the case of two other MOTUs identified in the present study, one (L. venerei) is a new record for the state of Maranhão, and we believe that the other represents a population of L. piau from the basin of the Parnaíba River.
Introduction
The family Anostomidae is a prominent group of Neotropical fish that includes 15 genera and approximately 151 valid species (Ramirez et al., 2016; Britski & Birindelli, 2019; Ramirez et al., 2020). The most speciose genus is Leporinus, which has approximately 81 valid nominal species (Fricke, Eschmeyer & Laan, 2021). Géry (1977) concluded that Leporinus is one of the most diverse genera of the order Characiformes, which is distributed between Central America and southern South America.
The considerable diversity found in the genus Leporinus has led to numerous attempts to classify its species and determine its internal arrangement. A number of studies have proposed subdivisions based on the position of the mouth, and the shape and arrangement of the teeth (Borodin, 1929; Myers, 1950; Garavello, 1979). Britski & Garavello (1978) divided the genus into three groups based on coloration patterns, that is, banding, spots, and longitudinal lines, although these proposals have been contradicted by more comprehensive studies, such as those of Sidlauskas & Vari (2008) and Ramirez et al. (2016). In their cytogenetic study, Galetti, Lima & Venere (1995) confirmed the existence of a well-defined ZZ/ZW sex chromosome system in six Leporinus species. These authors proposed that the presence of the ZW system represents a synapomorphy, and that the six species with this system form a monophyletic group. This conclusion is reinforced by morphological traits, such as coloration patterns, relatively large body sizes, and the number of teeth, as confirmed by Ramirez et al. (2016), which led to the allocation of this group to a new genus, Megaleporinus, by Ramirez, Birindelli & Galetti (2017).
Using osteological markers, Sidlauskas & Vari (2008) evaluated the phylogenetic relationships of the anostomids, and concluded that this family is monophyletic, although they were unable to confirm the monophyly of the genus Leporinus. Ramirez et al. (2016) used nuclear and mitochondrial molecular markers to confirm the paraphyly of the genus Leporinus, and concluded that the recuperation of the monophyly of the group would depend on further taxonomic reviews, including the creation of new genera and the description of new species.
Traditional taxonomic approaches have been essential for the delimitation of anastomid species based on morphological traits, although this does not necessarily resolve some natural groups, given that morphologically similar species may be assigned to the same nominal taxon (Bickford et al., 2007). Deciphering and defining cryptic diversity accurately is fundamental to the understanding of the ecological, biogeographic, and evolutionary patterns of a group of organisms, in addition to its other biological features (Kress et al., 2015).
Hebert et al. (2003) proposed the use of a DNA barcode, based on a standard sequence of the mitochondrial Cytochrome Oxidase subunit I (COI) gene, as the basis for a global species identification system. This approach has been widely-used for the identification of species and the resolution of cryptic diversity within genera and, in particular, in species complexes. A species complex consists of a group of closely-related taxa that have typically undergone recent speciation, which means that their taxonomic differences are still incipient, as observed in the case of the Leporinus cf. friderici species complex, in which Silva-Santos et al. (2018) confirmed the presence of eight distinct Molecular Operational Taxonomic Units (MOTUs) arranged in three clades.
Leporinus is not only one of the most diverse fish genera, but its species also play an important ecological role in many freshwater ecosystems, as well as having considerable economic and social importance for local fisheries. Given this, we compiled a dataset of the mitochondrial COI gene of 182,179 Leporinus specimens, which included specimens from the hydrographic basins of the Brazilian state of Maranhão to verify the potential intrageneric diversity of this genus, i.e., the presence of different putative species for the study region.
Here we present the diversity of Leporinus from hydrographic basins of central northern Brazil. We used integrative taxonomy tools to assess the species diversity of Leporinus based on (i) morphological identification from external characters, (ii) morphological identification from dentary characters, and (iii) molecular identification from COI gene fragment.
Material and Methods
Sampling
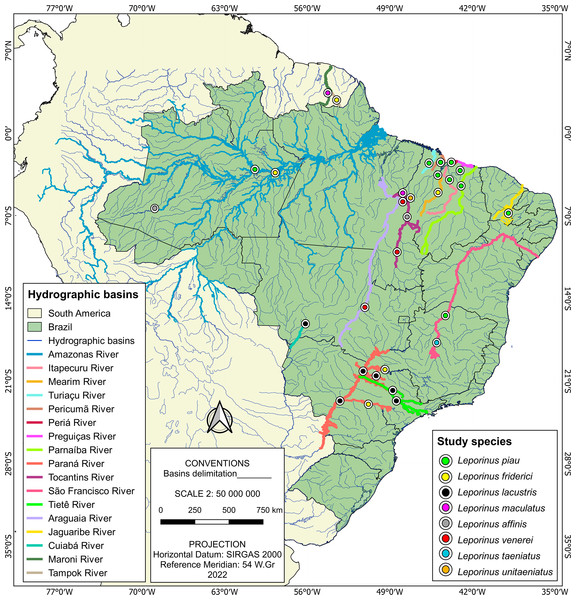
The present study was based on the analysis of a total of 185 sequences, of which 182 were of Leporinus species, with the other three representing the outgroup. The vast majority (157) of these 182 Leporinus sequences were collected during the present study, being extracted from specimens collected from basins in the Brazilian states of Maranhão (Itapecuru, Mearim, Turiaçu, Pericumã, and Periá rivers), Piauí (Parnaíba River), and Tocantins, that is, the Tocantins River (Fig. 1 and Table S1). The other 25 sequences were obtained from GenBank (Table S2).
Figure 1: Sample localities.
Each data point indicates the location where Leporinus samples were collected.The samples from the rivers of Maranhão, Piaui and Tocantins were obtained during extensive fieldwork, which has been ongoing since 2006. This research was authorized by the Brazilian Institute of the Environment and Renewable Natural Resources (IBAMA) through license 02012.004159/2006, and licenses ICMBio/MMA 42119-1/2013, ICMBio/MMA 46367-1/2015, ICMBio/MMA 83138-1/2022, ICMBio/MMA 73790-6/2022 issued by the Chico Mendes Institute for Biodiversity Conservation.
After collection, the specimens were taken to the Genetics and Molecular Biology Laboratory (GENBIMOL) of the Advanced Studies Center of Maranhão State University (CESC/UEMA), where they photographed and registered using a coding system. Samples of muscle tissue were extracted from the specimens for the genetic analyses. The specimens were then fixed em 10% formaldehyde and conserved in 70% alcohol, before being sent to the Museum of Zoology at Londrina State University (MZUEL) in Londrina, Paraná, Brazil, for morphological identification and cataloguing. The study of wild animals was approved by the Regulatory Committee for the Ethical Treatment of Animals of Maranhão State University (protocol 47/2022) and by the Committee for the Ethical Use of Animals of the National Institute for Amazonian Research, registered under protocol number 006/2021, SEI 01280.000116/2021-45.
The total DNA was extracted using the Wizard Genomic DNA Purification kit from Promega, following the maker’s instructions. The genomic region was isolated and amplified by Polymerase Chain Reaction (PCR), using the universal primers COI FishF1 5′-TCAACCAACCACAAAGACATTGCCAC-3′ and COI FishR1 5′- TAGACTTCTGGGTGGCCAAAGAATCA-3′, described by Ward et al. (2005). The samples were sequenced by the Sanger, Nicklen & Coulson (1977) method, using the Big Dye kit in an ABI Prism™ 3500 automatic sequencer (Applied Biosystems, EUA).
The sequences were aligned and edited in the Clustal W (Thompson, Higgins & Gibson, 1994) application of the Bioedit 7.2.5 program (Hall, 1999). All newly generated sequences (175) were deposited in GenBank under accession numbers OP781850–OP781884, OP782222 –OP782283, OP782350–OP7882375 and OP782385–OP782418 (Table S1). The haplotypes were delineated in DnaSP 5.1 (Librado & Rozas, 2009). The mean genetic distances and the Maximum Likelihood (ML) tree were obtained in MEGA X (Kumar et al., 2018), using the Kimura 2-Parameter and Hasegawa-Kishino-Yano (HKY) models, respectively, with the trees being reconstructed using 1,000 bootstrap replicates.
The optimum evolutionary model for the construction of the Bayesian Inference (BI) and Maximum Likelihood (ML) trees was generated in JModelTest2 (Darriba et al., 2012), which is available at CIPRES Science Gateway v3.3 (Miller, Pfeiffer & Schwartz, 2010), using the Hasegawa-Kishino-Yano (HKY+G+I) algorithm. The BI tree was generated in BEAST v.1.10.4 (Drummond et al., 2012; Suchard et al., 2018), using the relaxed lognormal clock (Drummond et al., 2006) and the birth-death speciation model (Gernhard, 2008).
This analysis was based on 40,000,000 generations with the log files being verified in Tracer v1.6 (Rambaut et al., 2014) to evaluate convergence and the most adequate burn-in, with the convergence being considered adequate when the Effective Sample Size (ESS) was over 200. The trees generated in BEAST were summarized in TreeAnnotator v.10.4 (Suchard et al., 2018) to obtain the consensus tree, which was then visualized and edited in Fig Tree v1.4.2 (Rambaut, 2014) and the Inkscape image editing system. Clades with a bootstrap percentage of at least 85% or posterior probability of at least 0.95 were considered to be well supported.
The delimitation analyses of the MOTUs of the COI gene were run using the following models: the Automatic Barcode Gap Discovery (ABGD), Assemble Species by Automatic Partitioning (ASAP), Poisson Tree Process (PTP), and the Generalized Mixed Yule Coalescent (GMYC) model. The ABGD test (Puillandre et al., 2012) was run in https://bioinfo.mnhn.fr/abi/public/abgd/ using the dataset of aligned sequences, while the ASAP test (Puillandre, Brouillet & Achaz, 2020) was implemented in https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html using the matrix of genetic distances, extracted using MEGA X, as the input. The PTP (Zhang et al., 2013) was run on the web server https://species.h-its.org/. In this case, the input was the Maximum Likelihood phylogenetic tree produced in RaxML v.8.29 (Stamatakis, 2014), which is available in the CIPRES Science Gateway v3.3 (Miller, Pfeiffer & Schwartz, 2010). The GMYC (Fujisawa & Barraclough, 2013) was based on the ultrametric consensus tree constructed in BEAST v1.10.1, which was processed in the Ape (Paradis & Schliep, 2019), Splits (Ezard, Fujisawa & Barraclough, 2009), Paran (Dinno, 2009), and Mass (Venables & Ripley, 2002) packages available in the R v. 4.1.0 software (R Core Team, 2021).
Results
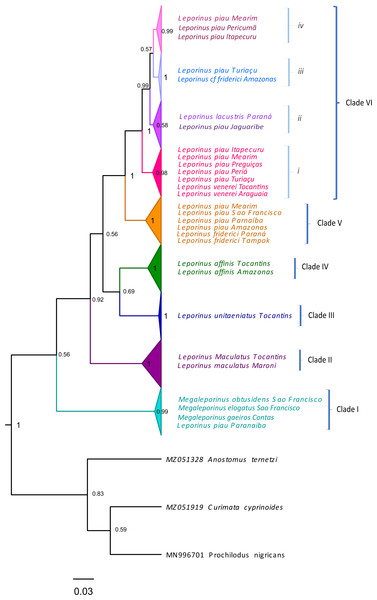
The present study focused on 182 sequences of the COI gene of Leporinus, each consisting of 620 base pairs (bps). The phylogenetic trees generated by the ML and BI analyses were highly congruent and well-supported at both the intra- and interspecific levels (Figs. 2–4), except in the case of Leporinus piau, which grouped with either Leporinus cf. friderici or Leporinus venerei. The ABGD analysis delimited 12 MOTUs, while the ASAP defined 15, the mPTP and bPTP each delimited nine, and the GMYC, six MOTUs (Fig. 2).
Figure 2: Bayesian Inference tree showing the arrangement of the MOTUs of the Leporinus species analyzed in the present study.
This arrangement was obtained using the ABGD, ASAP, mPTP, bPTP, and GMYC species delimitation approaches for the analysis of the mitochondrial COI gene, based on the Hasegawa-Kishino-Yano (HKY+G+I) algorithm, generated in BEAST. The species delimitated by the specific estimates are shown by the vertical bars, with the color representing the current status of the species. The blue bars correspond to valid species, while the gray bars indicate the species delimited diûerently from the current classification.The results of the five delimitation methods applied in the present study had three species in common—L. maculatus, L. unitaeniatus, and L. affinis—as well as differentiating two specimens (PALEP01 and PALEP09) from the basin of the Parnaíba River in a distinct molecular taxonomic unit, which indicates the occurrence of a fourth species, which we believe to be L. piau.
In the present study, the five delimitation methods had three species in common—L. maculatus, L. unitaeniatus, and L. affinis—and differentiated specimens from the basin of the Parnaíba River in a distinct molecular taxonomic unit, which is more basal than the other Leporinus species, and groups with the the Megaleporinus species that were previously assigned to Leporinus.
Clade VI (Fig. 3) was strongly supported, and includes L. venerei, L. lacustris, L. piau, and L. cf. friderici, with L. piau occurring in Maranhão, in the Mearim, Itapecuru, Pericumã, Turiaçu, Preguiças and Periá basins. In this case, the clade was formed by L. venerei from the Tocantins basin, L. lacustris from the basin of the Paraná River, L. piau from the Jaguaribe, Itapecuru, Mearim, Pericumã, Turiaçu, Preguiças and Periá basins, and L. cf. friderici from the Amazon and Mearim basins, which all share a single molecular taxonomic unit. Only the bPTP analysis separated L. venerei from L. lacustris, L. piau, and L. cf. friderici, which together formed a single MOTU in the ABGD, ASAP, mPTP, and GMYC models (Fig. 2).
Figure 3: Maximum Likelihood tree of the Leporinus species.
Maximum Likelihood tree showing the arrangement of the Leporinus species based on the analysis of 185 samples of the mitochondrial COI gene using the Hasegawa-Kishino Yano (HKY+G+I) algorithm, generated in MEGA X. The node support, that is, is given by the Bayesian posterior probability/ML bootstrap values, respectively. Each clade and its subdivisions (when present) are demarcated by the brackets. The Roman numerals in upper case represent the clades, while those in lower case indicate the subclades.The BI and ML analyses identified the formation of subclades within clade VI (Figs. 2 and 3), in which the L. piau from Maranhão, in the Itapecuru, Turiaçu, Mearim, and Periá basins, grouped with L. venerei from the Tocantins basin, with genetic distances ranging from only 0.16% to 1.54% (Table S3). Other L. lacustris and L. piau subclades were identified in the Jaguaribe basin, where the genetic distances ranged from 0.0% to 3.5% (Table S3). The L. piau subclade from Maranhão, found in the Itapecuru, Mearim, Pericumã, and Turiaçu basins, grouped with L. cf. friderici from the Mearim (Maranhão) and Amazon basins (Amazonas state), with genetic distances of between 0.16% and 5.88% (Table S3). All three groups were supported by significant posterior probability (BI) and bootstrap(ML) values (Figs. 2 and 3).
Leporinus cf. friderici, whose type locality is the basin of the Tampok River in French Guiana, formed a group together with L. piau from the basins of the São Francisco, Amazon, and Mearim rivers, an arrangement found in both the species delimitation models and the BI and ML trees. In the ABGD, ASAP mPTP, and bPTP delimitation models, however, L. cf. friderici was differentiated in its own operational unit (Fig. 2).
The genetic distance matrix derived from the molecular taxonomic units revealed relatively high values for both the intra- and inter-MOTU distances. The highest mean intra-MOTU distance was 5.9%, in L. maculatus, while the lowest mean was 0.4%, in L. unitaeniatus, whereas the mean inter-MOTU distances ranged from 7.8% to 17.4% The MOTUs formed by L. venerei, L. lacustris, L. piau, and L. cf. friderici were separated by a mean genetic distance of 2.2% (Table 1). In this context, it is important to note the genetic distance of 7.8% between L. piau (MOTU 1) and L. cf. friderici (MOTU 2), which may be the result of an error in the identification of the species of one of the groups.
Figure 4: Collapsed Bayesian inference tree of the MOTUs of the Leporinus species.
Collapsed Bayesian inference tree showing the arrangement of the MOTUs of the Leporinus species based on 185 samples of the mitochondrial COI gene analyzed using the Hasegawa-Kishino-Yano (HKY+G+I) algorithm, applied in BEAST. The groups were deûned by observing the congruence between the MOTUs generated in the species delimitation analyses based on the ABGD, ASAP, mPTP, bPTP, and GMYC methods.Given the levels of congruence identified in the different delimitation analyses applied in the present study, the ASAP method appeared to be the most effective interpretation, in biological terms, of the dataset considered here, given that it identified 10 MOTUs, which distinguished four of the seven nominal species, including L. venerei, in a distinct MOTU. This confirmed the occurrence of this species the Itapecuru, Mearim, Turiaçu, Preguiças and Periá basins, which constitutes the first record of L. venerei in the Brazilian state of Maranhão.
Discussion
An adequate taxonomic assessment is fundamental for the success of many types of biological research, and DNA data have provided additional insights for the resolution of taxonomic questions in many groups of organisms, including elements of the megadiverse Neotropical fish fauna, such as the anostomids. The COI barcode proved to be an extremely valuable tool for the identification and separation of the species assessed in the present study, based on the analysis of genetic distances and species delimitation, which identified evidence of the potential presence of more than one taxon in some nominal species.
In many previous studies of DNA barcoding and molecular diversity, the number of species or lineages delimited by the analysis has tended to exceed the number of nominal taxa or even the morphospecies analyzed (Carvalho et al., 2018). A similar tendency was observed here, in addition to the opposite pattern, given that, in some species delimitation analyses, more than one valid species was allocated to the same MOTU, as in the case of L. venerei, L. lacustris, L. cf. friderici, and L. piau.
In the present study, the L. venerei, L. lacustris, L. cf. friderici, and L. piau specimens were assigned to a single molecular taxonomic unit by the ABGD, ASAP, mPTP, and GMYC methods, reflecting their similar morphological characteristics, such as their coloration pattern, dental formula, and meristic parameters (Table 2), although the intra-MOTU analyses revealed mean genetic distances of 2.7% (Table 1), ranging from 0.0% to 6.11% (Table S3). This is consistent with the current classification of the valid nominal species (BI and ML analyses: Figs. 2–4; Table 1). All the species of clade VI shared the same morphological pattern, which is considered to be diagnostic of L. friderici, such as the number of spots along the lateral line (1–3) and the 4/4 dental formula, except for L. venerei, which has four teeth in the pre-maxilla and three in the dentary row. Leporinus lacustris and L. venerei are highly similar morphologically, given their relatively tall body, terminal mouth, long, dark anal fin, and three spots on the lateral line (Britski & Birindelli, 2008; Silva-Santos et al., 2018). Leporinus piau presents the D-type coloration pattern described by Garavello (1979), which consists of three well-defined black spots on the lateral line, and four teeth in both rows, with a dental formula of 4/4. The L. venerei, L. lacustris, L. cf. friderici, and L. piau MOTU was subdivided into three subclades (i, ii, iii and iv—Fig. 3). Subclade i includes L. piau from Maranhão and L. venerei from the Tocantins basin, while subclade ii has L. cf. friderici from the Amazon and L. piau from the Turiaçu basins. Subclade iii groups L. lacustris and L. piau from the Jaguaribe basin, and subclade iv groups L. piau from Maranhão and L. cf. friderici from the Mearim. The composition of subclade i (L. piau and L. venerei—Fig. 3), together with the diagnostic morphological features of the species, indicates that the specimens from the basins of Maranhão identified as L. piau may in fact be L. venerei, which would be the first record of this species from this Brazilian state.
| MOTU | Genetic Distance | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 1. Leporinus lacustris+Leporinus venerei+Leporinus piau+Leporinus cf friderici | 2.7 | |||||
| 2. Leporinus piau+Leporinus friderici | 7.91 | 2.2 | ||||
| 3. Leporinus maculatus | 10.89 | 10.58 | 5.9 | |||
| 4. Leporinus affinis | 12.61 | 11.26 | 11.05 | 1.8 | ||
| 5. Leporinus unitaeniatus | 13.04 | 13.49 | 11.97 | 11.27 | 0.4 | |
| 6. Leporinus piau | 15.49 | 16.24 | 15.64 | 17.45 | 14.53 | 0.5 |
| Species | Body | Coloration | Number of scales around the peduncle | Dental formula | Number of scales in the lateral line | Fin coloration |
|---|---|---|---|---|---|---|
| L. affinis Günther (1864) | Pre-dorsal somewhat convex; dorsal inclined slightly between the dorsal and adipose fins, and concave between the adipose and caudal fins. | Body yellowish, with 7 dark transversal stripes on the body and 3–4 on the head. | 16 | 4/4 | * | Peitoral and pelvic fins light yellow; all other fins hyaline. |
| L. friderici Bloch (1794) | Body tall and robust; large, with Standard Length (SL) of ca. 40 cm; body height 26–30% of SL, head length 27–29% of SL; mouth terminal. | Body browny chestnut, with 2–4 dark spots, rounded or oval, on the lateral line. | 16 | 4/4 | 38 to 40 | Anal fin dark gray; all other fins yellowish-gray. |
| L. lacustris Campos (1945) | Body elongated, with a maximum standard length of 20 cm; mouth terminal; incisors truncated. | 2–3 dark, rounded, mediolateral spots on the dorsal fin, the first larger and more conspicuous. | 16 | 4/4 | 33 to 36 | All fins yellowish, except the adipose and anal fins, que which are darkened. |
| L. maculatus Müller and Troschel (1844) | Body small, with Standard Length (SL) of ca. 10 cm; body height 22–26% of SL, head length 23–25% of SL; mouth subterminal. | Body with 4 black, rounded spots connected by 3 transversal stripes, which cross the lateral line. | 16 | 4/4 | 39 to 40 | * |
| L. piau | Body relatively tall. | Body with 3 black spots on the flank, which are elongated horizontally. | 16 | 4/4 | 35 to 37 | * |
| L. unitaeniatusGaravello & Santos (2007) | Body elongated and fusiform; small, with maximum standard length of 12 cm; relatively low body (23% of the standard length); mouth subterminal. | Body yellowish, with a conspicuous black longitudinal streak running along the lateral line; 11–13 dark chestnut transversal stripes separated from the lateral line by two rows of scales. | 16 | 4/4 | 40 to 44 | Hyaline. |
| L. venereiBritski & Birindelli (2008) | Body tall; mouth terminal; anal fin long and dark. | 3 small, dark spots on the lateral line, of which, the last 2, in particular, the last, are typically faded. | 16 | 4/3 | 36 to 37 | * |
Notes:
The L. venerei, L. lacustris, L. cf. friderici, and L. piau MOTU was subdivided into three subclades (i, ii, iii and iv—Fig. 3). Subclade i includes L. piau from Maranhão and L. venerei from the Tocantins basin, while subclade ii has L. cf. friderici from the Amazon and L. piau from the Turiaçu basins. Subclade iii groups L. lacustris and L. piau from the Jaguaribe basin, and subclade iv groups L. piau from Maranhão and L. cf. friderici from the Mearim. The composition of subclade i (L. piau and L. venerei—Fig. 3), together with the diagnostic morphological features of the species, indicates that the specimens from the basins of Maranhão identified as L. piau may in fact be L. venerei, which would be the first record of this species from this Brazilian state.
One other clade, formed by L. friderici from French Guiana and Paraná with L. piau from the Mearim, São Francisco, Parnaíba, and Amazon basins, is also well supported (Fig. 3). This raises two important points: (1) the clear polyphyly of L. friderici and L. piau, which, in the latter case implies a possible error of identification based on the type specimen, and (2) the existence of cryptic diversity in the genus Leporinus, in particular in L. friderici. Silva-Santos et al. (2018) concluded that the samples identified morphologically as L. friderici are in fact a polyphyletic group, given that the specimens collected from the basins of the Brazilian Shield are different from those of L. friderici from the type locality. The polyphyly of L. cf. friderici was also confirmed in the present study, which is consistent with Silva-Santos et al. (2018), in which a species complex is revealed by the genetic differentiation of the populations present in distinct hydrographic basins. In this case, individuals identified consistently as L. cf. friderici may not in fact be conspecific with L. friderici from the type locality, that is, they represent different species. Ramirez, Birindelli & Galetti (2017) confirmed the presence of cryptic diversity in this taxon, which may represent a typical scenario of recent diversification, when closely-related taxa may be poorly-distinguished morphologically, creating predictable taxonomic uncertainties, such as those observed in the populations of L. friderici.
In the present study, the relationship found among L. piau, L. friderici, and L. cf. friderici (Figs. 2–4) alludes to a possible taxonomic inconsistency derived from Fowler’s (1941) description of L. piau, as well as the geographic origin of the specimen analyzed in the present study, which was from the São Francisco basin. Fowler (1941) described Leporinus piau based on a type specimen from the Salgado River in the Jaguaribe basin of the Brazilian state of Ceará, but included a paratype from the Jatobá River, in the São Francisco basin, which led to the subsequent identification of most Leporinus specimens from the São Francisco River as L. piau (Garavello & Britski, 2003; Carvalho et al., 2011). However, Silva-Santos et al. (2018), who analyzed nuclear and mitochondrial genetic markers, including COI, noted that the Leporinus specimens from the São Francisco basin represent a species distinct from L. piau from the type locality in the Jaguaribe basin. Clearly, Fowler’s (1941) inclusion of a paratype from a distinct hydrographic basin have contributed fundamentally to the taxonomic uncertainties surrounding L. piau.
In the present study, L. maculatus, L.unitaeniatus, and L. affinis are valid nominal species, which presented considerable congruence between the traditional and molecular taxonomies. These three species constitute distinct MOTUs, which reflect their arrangement in different clades (BI and ML analyses: Figs. 2–4). All these species present easily distinguished diagnostic traits, such as the numerous spots dotting the body of L. maculatus, the single longitudinal stripe of L. unitaeniatus, and the lateral bands with no subdivisions observed in L. affinis (Britski & Garavello, 2005; Sidlauskas & Vari, 2012).
The samples from the Parnaíba basin identified here as L. piau and defined as a single MOTU by all the species delimitation models were grouped in a single clade with a genetic distance of 0.5%. These samples were delimited clearly as a more basal species separate from all the others, with evidence that they had been wrongly identified, and are in fact representatives of the genus Megaleporinus. This genus was described recently by Ramirez, Birindelli & Galetti (2017), based on a combined morphological, chromosomal, and molecular approach, which assigned the large-bodied Leporinus to a monophyletic clade, which was denominated Megaleporinus. In the present study, these samples were delimited clearly as a single, basal species well separated from all the others, although a more detailed analysis would be necessary to better determine their taxonomic status.
The samples from the basins of Maranhão and Piauí, together with those from the Tocantins River collected for the present study revealed the cryptic diversity found in Leporinus, given that the specimens from the basins of the Itapecuru, Mearim, Pericumã, Turiaçu, Periá, and Preguiças rivers in Maranhão, and the Parnaíba River in Piauí were identified as L. piau based on their morphological traits. The study of the DNA barcode and the analytical tools employed here confirmed that L. friderici likely constitutes a polyphyletic species complex, leading to the frequent misidentification of specimens as L. piau. It will only be possible to resolve this scenario definitively with a systematic re-evaluation of the specimens collected from the hydrographic basins of the states of Maranhão and Piauí.
In the specific case of subclade i (Fig. 3; Tables S3-S4), which groups L. piau from Maranhão with L. venerei from the Tocantins basin, the most parsimonious interpretation of the results of this analysis, together with the diagnostic traits of the two species, would be to consider them to be a single taxon, that is, L. venerei. This would thus be the first record of L. venerei from the basins of the Itapecuru, Mearim, Turiaçu, and Periá rivers, in the state of Maranhão.
Prior to the present study, three Leporinus species were considered to be present in the hydrographic basins of the Brazilian state of Maranhão—L. affinis, in the Itapecuru basin(Abreu et al., 2019), L. friderici in the Itapecuru, Mearim, Maracaçumé, Munim, Periá, and Parnaíba basins (Piorski et al., 1998; Soares, 2005; Ramos, Ramos & Ramos, 2014; Melo et al., 2016; Abreu et al., 2019; Brito et al., 2019; Brito et al., 2020; Guimarães et al., 2021a; Guimarães et al., 2021b; Guimarães et al., 2021c), and L. piau in the basins of the Itapecuru, Mearim, Turiaçu, and Parnaíba rivers (Barros, Fraga & Birindelli, 2011; Ramos, Ramos & Ramos, 2014; Ribeiro et al., 2014; Assega & Birindelli, 2019; Abreu et al., 2019). Based on analyses of molecular data, however, Fraga et al. (2014) and Nascimento et al. (2016) found evidence of two distinct lineages in the L. piau group from the Itapecuru basin, while in the present study, L. piau was assigned to three different clades, being associated strongly with L. cf. friderici in two clades and with L. venerei in one. This leads us to conclude that L. piau is, in fact, absent from the basins of Maranhão, which are instead populated by L. cf. friderici and L. venerei, with the latter being recorded in Maranhão for the first time. This restricts L. piau to the basin of the Parnaíba River.
Conclusions
The molecular analyses presented here, including the different species delimitation approaches, identified the presence of four Leporinus species in the hydrographic basins of central northern Brazil—L. maculatus, L. unitaenitus, L. affinis, and L. venerei. However, the species delimitation analyses also assigned L. cf. friderici and L. piau to two different molecular operational units, which leads us to believe that an additional species, morphologically indistinguishable from L. cf. friderici, may be present. The analyses also revealed a distinct group of two of the specimens, which indicates emphatically the presence of L. piau in the basin of the Parnaíba River, which indicates the presence of a total of six nominal species in the hydrographic basins of central northern Brazil. The confirmation of the presence of L. venerei in the Itapecuru, Mearim, Turiaçu, Preguiças and Periá basins represents a new record for the state of Maranhão, amplifying the known distribution of this species in Brazil.
Supplemental Information
Localities sampled of the hydrographic basins of central northern Brazil.
Localities sampled in the present study of Leporinus reservoirs in hydrographics basins in the state of Maranhao, Piaui and Tocantins.
Localities sampled in the 25 sequences were obtained from GenBank
Genetic distance matrix
Genetic distance, based on the Kimura 2-Parameter algorithm, for the genus Leporinus and Megaleporinus in the hydrographic basins of central northern Brazil.
Arrangement of the specimens in groups and the number of Molecular Operational Units proposed for Leporinus.
ML, Maximum Likelihood. Unnamed, group with more than one valid species.
Sequence Data
The sequence data is also available at GenBank: OP781850 to OP781884, OP782222 to OP782283, OP782350 to OP7882375 and OP782385 to OP782418.