Occurrence of two myxosporean parasites in the gall bladder of white seabream Diplodus sargus (L.) (Teleostei, Sparidae), with the morphological and molecular description of Ceratomyxa sargus n. sp.
- Published
- Accepted
- Received
- Academic Editor
- Bishoy Kamel
- Subject Areas
- Marine Biology, Parasitology, Taxonomy
- Keywords
- Phylogeny, SSU rRNA gene, Aquaculture, Ultrastructure, Taxonomy, Ceratomyxa sargus n. sp., Zschokkella auratis, Myxosporean parasites
- Copyright
- © 2023 Rocha et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Occurrence of two myxosporean parasites in the gall bladder of white seabream Diplodus sargus (L.) (Teleostei, Sparidae), with the morphological and molecular description of Ceratomyxa sargus n. sp. PeerJ 11:e14599 https://doi.org/10.7717/peerj.14599
Abstract
Myxosporeans are widespread cnidarian parasites that usually parasitize fish as part of their complex life cycle, thus constituting a potential threat for the aquaculture industry. White seabream Diplodus sargus (L.) is a commercially valuable sparid fish reared in Southern European aquacultures. Nonetheless, knowledge on myxosporean infections potentially harming the sustainable production of this fish is extremely limited. In this study, a myxosporean survey was conducted on D. sargus specimens reared in two Southern Portuguese fish farms. Two coelozoic myxosporeans were detected infecting the gall bladder, and are herein reported based on microscopic and molecular procedures: Ceratomyxa sargus n. sp. and Zschokkella auratis Rocha et al., 2013, previously described from reared stocks of gilthead seabream Sparus aurata in the same geographic locality. Ceratomyxa sargus n. sp. is the 12th species of the genus to be reported from Southern European sparids, reinforcing a substantial radiation of Ceratomyxa within this fish family and geographic region. SSU rRNA-based Bayesian inference and maximum likelihood analyses revealed C. sargus n. sp. positioned separately from other sparid-infecting Ceratomyxa spp. reported from Southern European countries, demonstrating that this species does not share a more immediate common ancestor with its closest relatives based on host affinity and geography. The recognition of a novel sparid-infecting lineage within the Ceratomyxa clade strengthens the contention that this genus entered sparid fish multiple times, namely in the Southern European region. The identification of Zschokkella auratis infections in D. sargus demonstrates that host shift has occurred among sparids reared in the Southern Portuguese coast. This agrees with the broad host specificity that is usually attributed to this genus, and that may be suggested to be the outcome of the capacity of the Zschokkella morphotype to undergo host shift/switch based on our findings and the limited molecular data available for this genus. Thus, a better understanding of Zschokkella host-associated diversification and dispersal mechanisms requires the increasing availability of molecular data from infections of the same species occurring in multiple hosts and geographical locations.
Introduction
Myxosporeans are obligate cnidarian parasites with a complex life cycle that usually involves fish as vertebrate hosts. They constitute a diverse and widespread group with more than 2,400 species described (Okamura, Gruhl & Bartholomew, 2015). Ceratomyxa Thélohan, 1892 is the second most speciose myxosporean genus, comprising near 300 species that represent ca. 13% of the taxa currently known. This genus is typically coelozoic, parasitizing the gall bladder of marine teleosts, and exceptionally occurring in elasmobranchs (Eiras, 2006; Lom & Dyková, 2006; Eiras, Cruz & Saraiva, 2018). In turn, the genus Zschokkella Auerbach, 1909 comprises ca. 97 species, most of which are coelozoic and, less frequently, histozoic in freshwater and marine fishes worldwide, including three representatives described from amphibians and reptiles (Lom & Dyková, 2006; Matsche et al., 2021). Several studies have shown that species identification within these genera based on myxospore morphology is artificial (Kent et al., 2001; Holzer, Sommerville & Wootten, 2004; Fiala, 2006; Bartošová, Fiala & Hypša, 2009). However, the lack of a specified threshold for myxosporean interspecific variability makes the usage of morphological features a requirement for differentiating between species (Atkinson et al., 2015; Gunter, Whipps & Adlard, 2009; Bartošová-Sojková et al., 2018). As such, reports of both novel and known species are presently based on the comprehensive analysis of several characters that include myxospore morphology, sequencing of selected molecular markers (usually the SSU rRNA gene), host species, tissue tropism, and geographic locality (Atkinson et al., 2015).
Despite the relative abundance of Ceratomyxa sequences presently available in GenBank, the interrelationships and evolutionary drivers of these parasites remain unknown, as geography and host affinity do not appear to correlate with relatedness of species within the Ceratomyxa clade (Gunter & Adlard, 2008; Heiniger, Gunter & Adlard, 2008; Gunter, Whipps & Adlard, 2009; Fiala et al., 2015; Rocha et al., 2015; Bartošová-Sojková et al., 2018; Alama-Bermejo et al., 2021). In turn, the molecular data currently available for Zschokkella is relatively scarce, with phylogenetic analyses retrieving the genus as polyphyletic (Fiala, 2006; Rocha et al., 2013; Fiala et al., 2015). The progressive expansion of the data available on genetic databases is therefore a key objective of myxosporean research, helping unequivocal species identification, but also the recognition of evolutionary patterns.
Several sparid fish are commercially important for Southern European aquaculture industries. Cultured stocks mainly comprise the gilthead seabream Sparus aurata L., but also sharpsnout seabream Diplodus puntazzo (Walbaum, 1792), white seabream Diplodus sargus (L.), common dentex Dentex dentex (L.), and blackspot seabream Pagellus bogaraveo (Brünnich, 1768). Parasitological studies have shown that the sustainable production of sparids in this geographical region is threatened by myxosporean infections, several of which caused by Ceratomyxa spp. (Georgévitch, 1916; Lubat et al., 1989; Diamant, Lom & Dyková, 1994; Sitjà-Bobadilla, Palenzuela & Álvarez-Pellitero, 1995; Sitjà-Bobadilla & Alvarez-Pellitero, 1995, 2001; Palenzuela, Sitjà-Bobadilla & Alvarez-Pellitero, 1997; Athanassopoulou, Prapas & Rodger, 1999; Kalavati & MacKenzie, 1999; Caffara et al., 2003; Diamant et al., 2005; Golomazou et al., 2009; Alama-Bermejo, Raga & Holzer, 2011; Rocha et al., 2013, 2015; Rangel et al., 2014; Thabet et al., 2019). Myxosporean surveys conducted on Portuguese fish farms have mostly focused on S. aurata and European seabass Dicentrarchus labrax (L.), which are known to host species belonging to the genera Ceratomyxa, Kudoa Meglitsch, 1947, Ortholinea Shulman, 1962, Zschokkella Auerbach, 1909 and Sphaerospora Thélohan, 1892 in this geographic location (Santos, 1996; Costa et al., 1998; Rocha et al., 2013, 2015, 2016; Rangel et al., 2014, 2016, 2017). Despite these fishes being commonly reared together with D. sargus in semi-intensive polyculture regimes established in the Portuguese Southern coast, myxosporean surveys have not been performed in specimens of D. sargus. Worldwide, a single study by Golomazou et al. (2006) reported the occurrence of myxosporean infections in commercial stocks of this fish, including a Myxobolus sp. in the kidney, a Kudoa sp. in the musculature, and Enteromyxum leei in the intestine of specimens reared in cages off Greece. Another study by Sirin, Santos & Rangel (2018) further described the myxosporean Bipteria lusitanica from wild specimens captured off the North Atlantic Portuguese coast.
Considering the above, this study aimed to provide knowledge regarding the diversity of myxosporean parasites occurring in reared populations of D. sargus. To pursue this objective, specimens were sampled from two Southern Portuguese fish farms and their external and internal tissues examined for the presence of myxosporean infection. Herein, we describe the occurrence of two myxosporeans in the gall bladder of D. sargus based on myxospore morphology, sequencing of the SSU rRNA gene, and host and tissue data.
Materials and Methods
Fish sampling and myxosporean survey
Specimens of reared white seabream Diplodus sargus (L.) (Teleostei, Sparidae) were collected from earth ponds located in the Algarve Atlantic coast in southern Portugal. Fifty-four specimens were collected between April 2014 and March 2015 from a fish farm (37°08′N/08°37′W) near the city of Portimão, with further 48 specimens collected between March 2021 and March 2022 from the IPMA’s Aquaculture Research Station (EPPO—Estação Piloto de Piscicultura de Olhão) (37°01′N 7°49′W), near the city of Olhão. All the people involved in animal handling and experimentation received proper training (category B courses accredited by FELASA, the Federation of Laboratory Animal Science Associations) and all fish facilities were accredited by the Portuguese National Authority for Animal Health (DGAV) with the Number 0421/2018. All experimental procedures involving animals followed the European Directive 2010/63/EU and the related guidelines and Portuguese legislation (Decreto-Lei 113/2013) for animal experimentation and welfare. Freshly caught specimens were dissected for performing a myxosporean survey of several organs and tissues, including brain, eye, muscle, gills, heart, spleen, liver, gall bladder, gonads, swim bladder, urinary bladder, kidney, and digestive tube. Fresh pseudoplasmodia and myxospores were observed and photographed using an Olympus CX23 light microscope (Olympus, Tokyo, Japan), equipped with the Olympus EP50 digital camera (Olympus, Japan). Myxospore morphometry was determined following the guidelines of Lom & Arthur (1989). All measurements herein provided were determined from a minimum of 25 myxospores, and include the mean value ± standard deviation, and range of variation.
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/. The LSID for this publication is: urn:lsid:zoobank.org:pub:E70B1814-0651-4396-81B4-0CFECEC525E2. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central SCIE and CLOCKSS.
Electron microscopy
Myxospores and pseudoplasmodia obtained from infected gall bladders were fixed in 5% glutaraldehyde buffered in 0.2 M sodium cacodylate (pH 7.4) for 20–24 h at 4 °C, rinsed in buffer, post-fixed in 2% osmium tetroxide in 0.2 M sodium cacodylate buffer (pH 7.4) for 3–4 h, rinsed in buffer, and dehydrated in a graded series of ethanol. For transmission electron microscopy (TEM), samples of the Ceratomyxa species were then embedded using ascending mixtures of EPON in oxide propylene, ending in EPON. Semithin sections cut from EPON blocks were stained with methylene blue-Azure II, and ultrathin sections double contrasted with uranyl acetate and lead citrate. Ultrathin sections were then observed and photographed using a JEOL 100 CXII TEM (JEOL Optical, Tokyo, Japan), operated at 60 kV. For scanning electron microscopy (SEM), dehydrated samples of the Zschokkella species were critical point dried, coated with a gold-palladium alloy (60%), and observed and photographed with a JSM-6301F SEM (JEOL Optical, Tokyo, Japan), operated at 15 kV.
DNA extraction, amplification, and sequencing
Myxospores belonging to the Ceratomyxa and Zschokkella morphotypes were obtained from bile, each from three separate individuals. Samples were fixed and preserved in absolute ethanol at 4 °C. Genomic DNA extraction was performed using a GenElute™ Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich, St Louis, MI, USA), following the manufacturer’s instructions.
The SSU rRNA gene of both myxosporeans was amplified and sequenced using the universal primers and myxosporean-specific primers shown in Table 1. PCR reactions and cycling conditions were performed as in Rocha et al. (2015). Electrophoresis of the obtained PCR products was performed in a 1% agarose 1× Tris-acetate-EDTA buffer (TAE) gel stained with ethidium bromide. PCR products were purified using the ExoFast method and sequenced directly using a BigDye Terminator v1.1 from the Applied Biosystems kit (Applied Biosystems, Carlsbad, CA, USA), and ABI3700 DNA analyzer (Perkin-Elmer, Waltham, MA, USA; Applied Biosystems, Carlsbad, CA, USA; Stabvida, Oeiras, Portugal).
| Name | Sequence (5′–3′) | Paired with | Source |
|---|---|---|---|
| 18E | CTG GTT GAT CCT GCC AGT | MyxospecR, MYX4R | Hillis & Dixon (1991) |
| MyxospecF | TTC TGC CCT ATC AAC TTG TTG | MYX4R, 18R | Fiala (2006) |
| MYX4F | GTT CGT GGA GTG ATC TGT CAG | 18R | Hallett & Diamant (2001) |
| MyxospecR | CAA CAA GTT GAT AGG GCA GAA | 18E | Fiala (2006) |
| MYX4R | CTG ACA GAT CAC TCC ACG AAC | 18E, MyxospecF | Hallett & Diamant (2001) |
| 18R | CTA CGG AAA CCT TGT TAC G | MyxospecF, MYX4F | Whipps et al. (2003) |
Sequence assembly, distance estimation, and phylogenetic analysis
Forward and reverse sequence segments of the Ceratomyxa and Zschokkella isolates were manually aligned with ClustalW in MEGA X software (Nei & Kumar, 2000; Kumar et al., 2018), with ambiguous bases clarified using corresponding ABI chromatograms. For performing distance estimation analysis, a dataset comprising 33 sequences was constructed and included the new Ceratomyxa isolate, all highly similar putative Ceratomyxa species (above a 90% cut-off) determined by BLASTn, and all sequences available for congeners that infect sparids or that share similar myxospore morphology and geographic location. In turn, the dataset used for distance estimation of the new Zschokkella isolate comprised all SSU rRNA sequences available in GenBank for this genus. Datasets were aligned using the software MAFFT version 7 available online, and distance estimation was performed in MEGA X, with the p-distance model selected, and all ambiguous positions removed for each sequence pair.
For the phylogenetic analysis of the new Ceratomyxa, the dataset used for distance estimation was broadened to encompass other representative Ceratomyxa sequences, Palliatus indecorus (DQ377712), Myxodavisia bulani (KM273030), Unicapsulocaudum mugilum (KP091845), and the outgroup sequences of Kudoa thyrsites (AY542482), and Ellipsomyxa mugilis (AF411336). Alignments were performed using the software MAFFT version 7 available online. Ambiguous characters were removed using Gblocks v0.91b with less stringent parameters (Castresana, 2000; Dereeper et al., 2008). Phylogenetic trees were calculated using maximum likelihood (ML) and Bayesian inference (BI). ML analyses were conducted in MEGA X using the general time reversible model with gamma distributed rate and invariant sites (GTR + G + I), selected based on the lowest score of Bayesian information criterion (BIC) and corrected Akaike information criterion (AIC), and with bootstrap confidence values calculated from 1,000 replicates. BI analyses were conducted in MrBayes v.3.2.6 (Ronquist & Huelsenbeck, 2003), with the general time reversible model with gamma-shaped rate variations across sites (Invgamma) (GTR + I + G) selected. Posterior probability distributions were generated using the Markov chain Monte Carlo method. Four chains were run simultaneously for 2 million generations, with burn-in set at 25%, and trees sampled every 500 generations.
Results
Analyzed fish specimens did not present external signs of infection or disease, and neither was morbidity or mortality reported from the sampled fish stocks. The macro- and microscopic analysis of 13 different organs revealed the occurrence of myxosporean infection in the gall bladder by Ceratomyxa Thélohan, 1892 and Zschokkella Auerbach, 1909, in the kidney by Sphaerospora Thélohan, 1892, and in the urinary bladder by Ortholinea Shulman, 1962. Co-infection in the gall bladder was determined based on myxospores observation and could be detected in four out of the 54 specimens collected from the fish farm near the city of Portimão. Infections by Zschokkella were not observed at the EPPO sampling location. The microscopic and molecular procedures performed identified the ceratomyxid as new species that is herein described as Ceratomyxa sargus n. sp., and the second isolate as Zschokkella auratis Rocha et al., 2013.
Ceratomyxa sargus n. sp. urn:lsid:zoobank.org:act:3103EB6E-000F-445C-AA10-F129C0509EFC
Taxonomic placement
Phylum Cnidaria Hatschek, 1888
Sub-phylum Myxozoa Grassé, 1970
Class Myxosporea Bütschli, 1881
Order Bivalvulida Shulman, 1959
Family Ceratomyxidae Doflein, 1899
Genus Ceratomyxa Thélohan, 1892
Morphological description and taxonomic summary
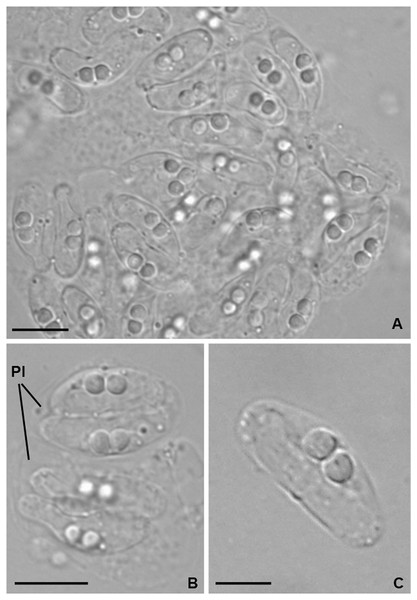
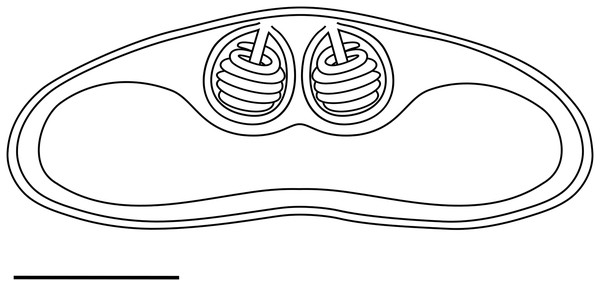
Light microscopy. Pseudoplasmodia at different stages of maturation, and mature myxospores, observed free in the bile (Fig. 1). Young pseudoplasmodia spherical; mature pseudoplasmodia subspherical to elliptical, mostly disporic (Figs. 1A and 1B). Mature myxospores crescent-shaped with convex anterior margin and slightly concave to straight posterior margin; valves with rounded ends. Myxospores 4.9 ± 0.4 (4.2–5.7) µm long and 14.5 ± 1.1 (12.4–16.9) µm thick. Polar capsules located at the same level at the myxospores anterior pole, equally-sized and subspherical, 2.2 ± 0.2 (1.9–2.5) µm long and 1.9 ± 0.2 (1.5–2.3) µm wide (Fig. 1C). A schematic drawing of a myxospore in valvular view is provided in Fig. 2.
Figure 1: Light micrographs of Ceratomyxa sargus n. sp. from the gall bladder of Diplodus sargus.
(A) Cluster of plasmodia and free myxospores. Scale bar = 10 µm. (B) Disporic pseudoplasmodia (Pl). Scale bar = 10 µm. (C) Free crescent-shaped myxospore. Scale bar = 5 µm.Figure 2: Schematic drawing of a mature myxospore of Ceratomyxa sargus n. sp. from the gall bladder of Diplodus sargus
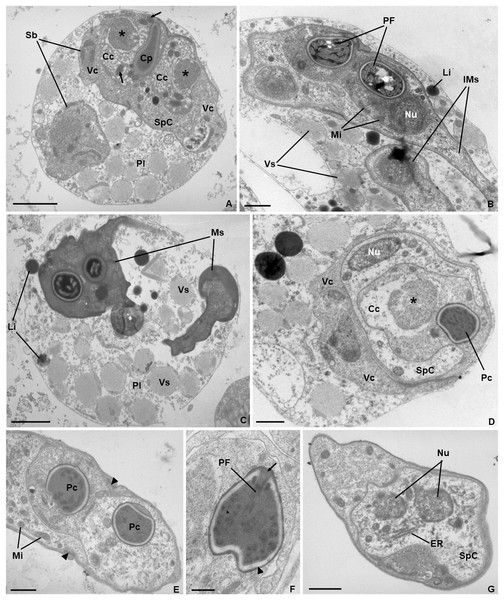
Scale bar = 5 µm.Ultrastructure. Young and mature pseudoplasmodia with smooth surface membrane and displaying numerous vesicles containing granular material of variable electronic density, as well as lipidic droplets, distributed in the cytoplasm (Figs. 3A–3D). More mature pseudoplasmodia containing two sporoblasts in equivalent developmental stages (Figs. 3A–3C). Younger sporoblasts comprising two large and single nucleated valvogenic cells that surround two capsulogenic cells and one sporoplasmogenic cell (Fig. 3A). Capsulogenic cells uninucleate, initially with a globular capsular primordium extending into an external tubule (Fig. 3A). In more advanced stages of sporogenesis, the tubule is coiled within the inner wall of the capsular primordium, forming a young polar capsule and its internal polar tubule (Figs. 3B–3D). Myxospores constituted by two symmetrical valves, thick and smooth, united along a curved suture line (Fig. 3E). Polar capsules with a double-layered wall (outer layer thinner and electron-dense; inner layer thicker and electron-lucent), and a homogenous dense matrix. Polar tubule coiled in two rows, forming ca. five turns in the outer row, and two turns in the inner row (Fig. 3F). Sporoplasm binucleate, displaying endoplasmic reticulum and several mitochondria (Fig. 3G).
Figure 3: Transmission electron micrographs of Ceratomyxa sargus n. sp. from the gall bladder of Diplodus sargus.
(A) Pseudoplasmodium (Pl) containing two developing sporoblasts (Sb), one of which displaying all sporogenic cells: two valvogenic cells (VC) surrounding two capsulogenic cells (CC) and one sporoplasmogenic cell (SpC). Each capsulogenic cell presents a single nucleus (*), and a globular capsular primordium (Cp) that extends into an external tubule (arrows). Scale bar = 2 µm. (B) Pseudoplasmodium containing two immature myxospores (IMs), numerous vesicles (Vs), and some lipidic globules (Li). Notice that the polar capsules are completely formed, with the polar tubule coiling within, but the cytoplasmic content of the capsulogenic cells remains, inclusive the nuclei (Nu) and several mitochondria (Mi). Scale bar = 1 µm. (C) Pseudoplasmodium (Pl) containing two fully matured myxospores (Ms), numerous vesicles, and some lipidic globules. Scale bar = 2 µm. (D) Transverse section of an immature myxospore showing its valvogenic cells (VC), one of the two capsulogenic cells (CC) displaying its degrading nucleus (*) and an almost completely matured polar capsule (PC), and the sporoplasmogenic cell (SpC) also displaying one of its two nuclei (Nu). Scale bar = 1 µm. (E) Oblique section of a myxospore showing its almost matured polar capsules (PC) located in the proximity of the curved suture line (arrowheads) that unites the valves (V). The sporoplasm is riddled with mitochondria (Mi). Scale bar = 1 µm. (F) Longitudinal section of a polar capsule displaying the polar tubule (PT) coiled internally to the double-layered wall (arrowhead) and capped at the apex by a stopper (arrow). Scale bar = 0.5 µm. (G) Oblique section of a myxospore, evidencing the cytoplasmic content of the sporoplasmogenic cell (SpC), in which two nuclei (Nu), and endoplasmic reticulum (ER) can be observed. Scale bar = 1 µm.Type host. Diplodus sargus (L.) (Eupercaria incertae sedis, Sparidae) (common name: white seabream).
Type locality. The Algarve Atlantic coast in southern Portugal, from a fish farm (37°08′N/08°37′W) near the city of Portimão, and another (37°01′N 7°49′W) near the city of Olhão.
Site of infection. Gall bladder.
Prevalence of infection. 11.8% (13.0%, 7 infected in 54 specimens analysed from the fish farm near the city of Portimão; 10.0%, 5 infected in 48 specimens analysed from the EPPO).
Pathogenicity. Analysed fish did not present external signs of infection or disease.
Type material. Series of phototypes of the hapantotype, deposited together with a representative DNA sample in the Natural History and Science Museum of the University of Porto, Portugal, reference CIIMAR 2022.66.
Molecular data. Partial SSU rRNA gene sequence with 1,849 bp and GenBank accession number ON123709.
Etymology. The specific epithet “sargus” refers to the specific epithet of the host species.
Differential diagnosis
The myxospores of Ceratomyxa sargus n. sp. closely resemble those of C. bartholomewae Gunter, Burger & Adlard, 2010, C. cardinalis Heiniger & Adlard, 2013, C. cribbi Gunter & Adlard, 2008, C. cyanosomae Heiniger & Adlard, 2013, C. dehoopi Reed et al., 2007, C. moseri Gunter & Adlard, 2008, C. puntazzi Alama-Bermejo, Raga & Holzer, 2011, C. rueppelli Heiniger & Adlard, 2013 and C. siganicola Zhang et al., 2019 in terms of shape, but can be readily distinguished based on molecular data. Ceratomyxa cardinalis (JX971431) and C. cribbi (EU440367) shared only 97.0% and 96.5% of similarity with C. sargus n. sp., respectively, with all others displaying similarity values lower than 90.0%. Distance estimation determined highest similarity to Ceratomyxa sp. SAR (DQ333430) (99.4%), Ceratomyxa gunterae (JX971422) (98.2%), and Ceratomyxa archamiae (JX971428) (98.1%). Other congeners having similar myxospores, but lacking molecular data allowing prompt differentiation, are C. australis Gaevskaya & Kovaleva, 1979, C. arripica Su & White, 1994, C. castigatoides Meglitsch, 1960, C. declivis Meglitsch, 1960, C. simplex Zhao et al., 2015, and C. sprenti Moser, Kent & Dennis, 1989. However, the myxospores of C. australis have smaller thickness range and thinner polar capsules (Eiras, 2006). Ceratomyxa arripica myxospores are narrower, with spherical polar capsules displaying a lower number of polar tubule coils (3–4) in a single row (Su & White, 1994). The myxospores of C. castigatoides, C. declivis and C. simplex have similar morphometry to those of C. sargus n. sp. but differ in terms of shape. Ceratomyxa castigatoides by having a posterior margin varying from convex to concave and spherical polar capsules, in addition to being slightly longer than the myxospores of C. sargus n. sp.; C. declivis by having a plump crescent shape with nearly truncated ends; and C. simplex by being strongly arcuate, with the anterior edge parallel to and almost symmetrical with the posterior edge, with both ends truncated (Eiras, 2006; Zhao et al., 2015). Finally, the myxospores of C. sprenti differ from those of C. sargus n. sp. by having both the anterior and posterior margins straight, with spherical polar capsules, and 5–6 coils of the polar tubule in a single row. They are also slightly thicker than the myxospores of C. sargus n. sp. (Moser, Kent & Dennis, 1989).
Phylogenetic analysis
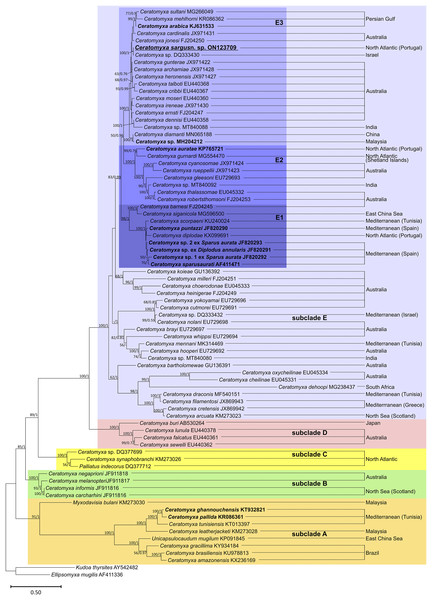
Phylogenetic analysis based on SSU rRNA gene sequences showed the sequences available for sparid-infecting Ceratomyxa, including C. sargus n. sp., positioned within subclades A and E (Fig. 4). Ceratomyxa pallida (KR086361) and C. ghannouchensis (KT932821) from the sparid Boops boops in Tunisia cluster in the basal subclade A together with C. tunisiensis (KT013097) from Carangiformes also in Tunisia, and C. leatherjacketi (KM273028) from Aluterus monoceros (L.) (Tetraodontiformes) in Malaysia. The remaining sequences of sparid-infecting Ceratomyxa spp., including C. sargus n. sp., all clustered within subclade E, which is the most recent and taxon-rich subclade with unresolved deeper nodes. Ceratomyxa sparusaurati (AF411471), Ceratomyxa sp. 1 ex S. aurata (JF820292), Ceratomyxa sp. 2 ex S. aurata (JF820293), C. puntazzi (JF820290), and Ceratomyxa sp. ex D. annularis (JF820291) from South European S. aurata all cluster together to form the subclade E1, which further includes perciform-infecting species, namely C. scorpaeni (KU240024) from Tunisia, C. siganicola (MG596500) from the East China Sea, and C. barnesi (FJ204245) from Australia. The allegedly sparid-infecting species Ceratomyxa diplodae (KX099691) also clusters within subclade E1, despite its only available sequence having been obtained from infections in the gall bladder of European seabass Dicentrarchus labrax (Eupercaria incertae sedis). Ceratomyxa auratae (KP765721) from South European S. aurata clusters with C. gurnardi (MG554470) from the Atlantic Ocean off Scotland, in close relationship with several Ceratomyxa spp. from Kurtiformes and Perciformes in Australian waters, forming the subclade E2. Lastly, C. sargus n. sp. (ON123709) clusters within the subclade E3, which further comprises C. arabica (KJ631533) and a Ceratomyxa sp. (MH204212), also from sparid hosts in the Persian Gulf and Malaysia, respectively, plus an array of Ceratomyxa spp. mostly from Australian Pomacentridae (Ovalentaria incertae sedis) and Kurtiformes. Although the formation of these three subclades is well-supported in both maximum likelihood and Bayesian inference analyses, their exact relationship within subclade E remains uncertain. The inner topology of subclades E1 and E2 further display soft polytomies congruent with highly unresolved inner nodes.
Figure 4: SSU rRNA-based maximum likelihood phylogenetic tree showing the position of sparid-infecting ceratomyxids (marked in bold) within the Ceratomyxa clade.
Nodal supports are maximum likelihood bootstrap values and Bayesian inference probabilities; dashes represent poorly resolved nodes or a different branching pattern in the BI phylogenetic tree; nodes without values were poorly resolved in both trees.Zschokkella auratis Rocha et al., 2013
Fifteen out of the 54 specimens analysed (27.8%) from the fish farm located near the city of Portimão presented infection by this species, which was co-infective with C. sargus n. sp. in four specimens. Infections by Z. auratis were not detected in samples obtained from the EPPO. Species identification was based on the morphological and morphometric aspects of the myxospores, and on molecular data of the SSU rRNA gene.
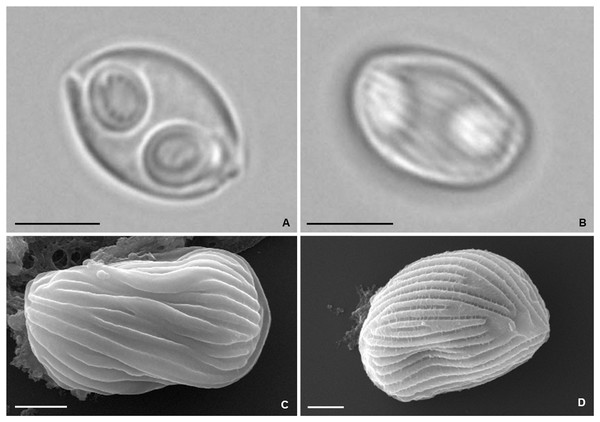
Mature myxospores observed isolated in the bile were ovoidal in sutural and valvular views, with rounded opposite sides, measuring 9.6 ± 0.5 (9.1−10.3) μm in length and 6.7 ± 0.2 (6.4−7.0) μm in width. Myxospore wall thick, composed of two symmetrical valves united along a curved suture line (Figs. 5A and 5B). Light microscopy and SEM observations revealed the presence of numerous surface ridges organized parallel to the suture line and forming a pattern along the entire myxospore body (Figs. 5B–5D). Two symmetrical subspherical polar capsules, 3.7 ± 0.4 (3.0−4.2) μm long and 3.0 ± 0.2 (2.7−3.3) μm wide, were located sub terminally at the same level within the myxospores and opened at nearly opposite positions. Each polar capsule contained a polar tubule coiled in four to five turns (Fig. 5A). These morphological and morphometric features are largely congruent with those reported in the original description of Z. auratis (Rocha et al., 2013).
Figure 5: Photomicrographs of Zschokkella auratis from the gall bladder of Diplodus sargus.
(A) Light micrograph showing a myxospore ovoidal in sutural view, and containing two subspherical polar capsules located subterminally and opening at nearly opposite positions. Scale bar = 5 µm. (B) Light micrograph of a myxospore evidencing the presence of surface ridges. Scale bar = 5 µm. (C) SEM micrograph of a myxospore in sutural view, showing the surface ridges organized parallel to the suture line. Scale bar = 2 µm. (D) SEM micrograph allowing recognition of the surface pattern formed by the ridges that extend throughout the myxospore body. Scale bar = 2 µm.The partial SSU rRNA gene sequence obtained from the isolate comprised 2,055 bp and is deposited in GenBank under the accession no. ON054249. Distance estimation confirmed species identification, having revealed 99.6% similarity to the sequence of Z. auratis (KC849425) available from gallbladder infections in specimens of S. aurata also reared in Southern Portuguese fish farms (Rocha et al., 2013), and 99.0% to a second sequence of Z. auratis (MF978273) obtained from the brain of farmed striped snakehead Channa striata (Bloch, 1793) in India (Paul et al., 2020). All other sequences retrieved less than 94.5% similarity.
Discussion
The present study reports Ceratomyxa sargus n. sp. and Zschokkella auratis Rocha et al., 2013 infecting the gallbladder of reared white seabream D. sargus from two Southern Portuguese fish farms. While C. sargus n. sp. was present at both sampling locations, Z. auratis was never observed in specimens obtained from the EPPO. We suspect this to be correlated with differences in the annelid communities established in the earth ponds of the two fish farms and nearby wild locations in the affluents used for water supply. In order to evaluate this hypothesis and understand if infections of reared specimens are dependent or not on the earth pond annelids, we are presently surveying the annelid communities present at these locations for the detection of actinospore development.
The species identifications performed in this study were based on combined microscopic and molecular data. Ultrastructure was further performed for C. sargus n. sp. and revealed developmental features congruent with previous studies of Ceratomyxa, i.e., asynchronous development, with pseudoplasmodia displaying different shapes and sizes, and lacking the peripheral projections typical of other myxosporean coelozoic plasmodia (Rocha et al., 2015, 2016). The coiling of the polar tubule in two rows, although uncommon among Myxosporea, has been reported in ultrastructural studies of other Ceratomyxa spp., such as C. diplodae (Rocha et al., 2016) and Ceratomyxa tenuispora (Casal, Costa & Azevedo, 2007).
Molecular comparison to myxozoan sequences available in GenBank confirmed the identity of the Zschokkella samples here studied to Z. auratis (KC849425) with 99.6% of similarity, and revealed C. sargus n. sp. presenting 99.4% similarity to the sequence of an undescribed Ceratomyxa sp. (DQ333430). Though genetic differences higher than 1% are usually accepted for establishing myxozoan differentiation, there is no specified benchmark for Ceratomyxa interspecific variation, making it necessary for species identifications to be corroborated by differences in other taxonomic characters such as myxospore morphometry, host species, and tissue specificity (Gunter, Whipps & Adlard, 2009; Heiniger & Adlard, 2013; Atkinson et al., 2015; Bartošová-Sojková et al., 2018). According to the GenBank record, Ceratomyxa sp. (DQ333430) was sequenced from the gall bladder of marbled spinefoot Siganus rivulatus Forsskål & Niebuhr, 1775 (Acanthuriformes, Siganidae) from Israel. No data are available for myxospore morphology and dimensions. Thus, undoubtable conspecificity between these two isolates cannot be solely based on the small genetic difference found between their sequences (0.6%). Future analyses targeting the re-sequencing and morphological study of the Israeli isolate are necessary to either confirm or deny conspecificity to C. sargus n. sp., and acknowledge host specificity of this species.
Although ceratomyxids are usually accepted to be host specific (Gunter & Adlard, 2009; Gunter, Whipps & Adlard, 2009; Heiniger & Adlard, 2013; Alama-Bermejo, Raga & Holzer, 2011), there are molecularly confirmed exceptions to this rule (see Gunter & Adlard, 2008; Heiniger & Adlard, 2013; Fiala et al., 2015; Thabet et al., 2016; Bartošová-Sojková et al., 2018). Among sparid-infecting Ceratomyxa spp., Ceratomyxa diplodae Lubat et al., 1989 and C. pallida have been reported from multiple hosts but were characterized based on myxospore morphology alone. Ceratomyxa diplodae was originally described from the gall bladder of the annular seabream Diplodus annularis, and later reported from D. labrax, D. dentex and D. puntazzo (Lubat et al., 1989; Alvarez-Pellitero & Sitjà-Bobadilla, 1993; Rigos et al., 1997; Katharios et al., 2007), while C. pallida has been reported to infect both S. salpa and B. boops (Thélohan, 1895; Thabet et al., 2019). Considering that morphology alone is also a weak character for determining conspecificity among Myxosporea (Atkinson et al., 2015), molecular data are needed from these species to better support hypotheses and speculation about host specificity and potential host shifting/switching of sparid-infecting Ceratomyxa.
The phylogenetic analysis performed here is congruent with previous studies through supporting a close phylogenetic relationship between Ceratomyxa spp. The SSU rRNA gene sequences belonging to this genus have been shown to cluster together, distributed among five well-defined subclades that further encompass representatives of the genera Palliatus, Pseudoalataspora, Myxodavisia and Unicapsulocaudum (Fiala et al., 2015; Rocha et al., 2015; Yang et al., 2017). This topology was retrieved in our phylogenetic trees, which showed subclade E as the most divergent and taxon-rich subclade displaying unresolved deeper nodes, in accordance with previous studies (Gunter, Whipps & Adlard, 2009; Fiala et al., 2015; Bartošová-Sojková et al., 2018).
Overall, the sequences available for sparid-infecting Ceratomyxa, including C. sargus n. sp., were retrieved positioned within subclades A, E1, E2, and E3, forming several independent lineages of sparid-infecting Ceratomyxa spp. Their radiation within the most divergent subclades (E1–E3) further supports the occurrence of more recent evolutionary events driving rapid speciation of these parasites in sparid fish. This is suggested by the soft inner polytomies in subclades E1 and E3, which demonstrate rapid species evolution, with the data currently available being unable to resolve inner nodes and ascertain exact interspecific relationships within these groups.
The sequence obtained for C. sargus n. sp. appeared positioned separately from other sparid-infecting Ceratomyxa spp. reported from Southern European countries, demonstrating that this species does not share a more immediate common ancestor with its closest relatives based on host affinity and geography. This reinforces the occurrence of multiple evolutionary entries of Ceratomyxa into sparid fish in the Southern European region alone. Our study further strengthens the contention that geographic location is not a main driver of Ceratomyxa radiation (Gunter & Adlard, 2009; Alama-Bermejo, Raga & Holzer, 2011; Rocha et al., 2015), as it shows C. sargus n. sp. clustering together with Ceratomyxa spp. from the Persian Gulf and Australia, rather than with other Southern European species.
Molecular systematics have shown a surprising radiation of Ceratomyxa within certain host families (Heiniger, Gunter & Adlard, 2008; Gunter & Adlard, 2009; Alama-Bermejo, Raga & Holzer, 2011; Heiniger & Adlard, 2013; Bartošová-Sojková et al., 2018). Ceratomyxa sargus n. sp. is the 12th species of the genus described from sparid fishes in the Mediterranean and North Atlantic off Southern European regions. This supports the existence of a high species richness in sparids inhabiting this geographic region, demonstrating the need for integrative taxonomic studies to shed insight into this host-parasite association with potential economic impact for aquaculture and fishery industries. Pathological changes induced by Ceratomyxa are mostly mild and may include swelling, vacuolization, sloughing, damage, and necrosis of the gallbladder’ epithelial cells (Sitjà-Bobadilla & Alvarez-Pellitero, 1993; Palenzuela, Sitjà-Bobadilla & Alvarez-Pellitero, 1997), but high mortality rates have also been associated with these myxosporeans (Katharios et al., 2007). Despite clinical signs of infection and disease having not been observed in D. sargus specimens infected with C. sargus n. sp. and Z. auratis, it is our intention to proceed with pathological studies trying to assess the real pathogenicity and impact that these parasites might have on cultured stocks.
Our study reports for the first time the occurrence of Zschokkella auratis in D. sargus. This myxosporean was originally described from the gall bladder of another sparid fish, the gilthead seabream S. aurata, also reared in a Southern Portuguese fish farm (Rocha et al., 2013). A second report was recently performed from infections in the brain of farmed striped snakehead Channa striata (Anabantiformes, Channidae) in India (Paul et al., 2020). However, species identification was based in a short 618 bp-long SSU rRNA fragment (MF978273) at a highly conservative region close to the 3′ end of the gene and cannot provide undoubtable evidence of conspecificity.
The occurrence of Z. auratis in more than a single host species agrees with the broad host specificity that has been reported for Zschokkella. According to a recent synopsis, 24 Zschokkella spp. have been reported from multiple hosts; half from various fishes belonging to the same taxonomic order, even if from distinct families, and the other half from a broader range of taxonomically distant hosts (Matsche et al., 2021). For instance, Z. leptatherinae Su & White, 1995 and Z. macrocapsula Su & White, 1995 have been reported from multiple Atheriniformes; Z. hildae Auerbach, 1910, Z. meglitschi Moser & Noble, 1977 and Z. microcapsula Moser & Noble, 1977 from multiple Gadiformes; and Z. carassii Nie & Li, 1973, Z. oviformis Ma et al., 1982 and Z. striata Shulman, 1962 from multiple Cypriniformes. In turn, Z. nova Klokačewa, 1914 has been reported from an array of fish species belonging to the orders Cypriniformes, Salmoniformes, Perciformes, Mugiliformes and Anguiliformes; Z. candia Kalatzis et al., 2015 from fishes belonging to Perciformes, Cichliformes and Eupercaria incertae sedis; with several others reported from fish species belonging to at least two taxonomic orders (Matsche et al., 2021). Most of these reports were performed based solely on myxospore morphology, for which this broad host specificity requires corroboration through sequencing of species isolates from supposed hosts.
The molecular data available for the genus Zschokkella is rather limited, representing ca. 15% of the number of species thus far described, and including information for different host isolates of only Z. candia, Z. nova, and now also Z. auratis. Identical SSU rRNA gene sequences are available in GenBank for Z. candia infections in Sparisoma cretense (Eupercaria incertae sedis), Scorpaena porcus (Perciformes), and Oreochromis niloticus (Cichliformes). Zschokkella nova accounts for a total of 10 sequences from the SSU and LSU genetic markers ascertaining identification of the parasite in Carassius auratus gibelio, Ctenopharyngodon idella (Cypriniformes), and Sander lucioperca (Perciformes). The present study further reveals molecular evidence for the development of Z. auratis in at least two sparid fish, showcasing a potential host shift phenomenon of this myxozoan among sparids. This highlights a necessity for myxozoans surveys to target the molecular description of known Zschokkella spp., to determine their true host range and capacity of this morphotype to undertake host shift/witch.
Conclusions
The present study sheds some insight into the diversity of myxosporean parasites potentially affecting the sustainable production of white seabream Diplodus sargus in Southern European aquacultures. Two gall bladder dwelling myxosporeans are reported from specimens reared in Southern Portuguese fish farms. The description of Ceratomyxa sargus n. sp. reinforces the previously reported high species richness of Ceratomyxa infecting sparids in the Southern European region, with phylogenetic analyses demonstrating the lack of a more immediate common ancestor for these species and highlighting multiple evolutionary entries into this host family in Southern Europe. The possibility of conspecificity between C. sargus n. sp. and the unnamed Ceratomyxa sp. reported from the acanthuriform Siganus rivulatus in Israel requires investigation through the morphological and molecular analysis of novel material from the Israeli isolate, as confirmation of conspecificity would provide evidence of host switch potentially allowing geographical spreading of C. sargus n. sp. from co-habitation sites in the eastern Mediterranean.
Contextualization of evidence of host shift of Zschokkella auratis among sparids with the available literature suggest a high capability of the Zschokkella morphotype to undergo host shift/switch that requires confirmation through the molecular description of the numerous Zschokkella spp. reported from multiple hosts and geographical locations.