Exploration of Zoo felids in North-East China for the prevalence and molecular identification of Cryptosporidium spp.
- Published
- Accepted
- Received
- Academic Editor
- Sharif Aly
- Subject Areas
- Genomics, Molecular Biology, Veterinary Medicine, Zoology
- Keywords
- Cryptosporidium, Zoo felids, 18S rRNA gene, Prevalence, Phylogenetic
- Copyright
- © 2021 Hussain et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Exploration of Zoo felids in North-East China for the prevalence and molecular identification of Cryptosporidium spp. PeerJ 9:e11819 https://doi.org/10.7717/peerj.11819
Abstract
Cryptosporidium spp. is a protozoan having the potential to cause zoonosis in humans and animals. Despite the zoonotic importance of this protozoan parasite, limited data are available about its prevalence in zoo felids in North-Eastern China. Hence, the current study was designed to determine the occurrence and molecular characterization of Cryptosporidium spp. from the fecal samples of captive zoo felids. Fecal samples (N = 244) were collected from different felids from five different zoos of North-Eastern China. 18S rRNA gene was amplified from the genomic DNA using species specific primers in nested polymerase chain reaction (nPCR) and Cryptosporidium parvum and Cryptosporidium spp. was found. The overall prevalence of Cryptosporidium was 9.43% (23/244). The 18S rRNA gene similarity analysis showed that 6 Cryptosporidium isolates were Cryptosporidium parvum and the remaining 17 Cryptosporidium isolates were resembling to a Cryptosporidium spp., which is similar to Cryptosporidium NEV10. Phylogenetic tree was constructed based on 18S rRNA of Cryptosporidium spp. The similarity of Cryptosporidium parvum was with its other isolates in China, India, Iran, Iraq, Turkey, Czech Republic, Spain and USA while Cryptosporidium NEV10 alike had a close relationship with Turkish isolates. In conclusion, Cryptosporidium was prevailing in feline animals of China zoo and zoo officials are directed to consider their control policy as it can be a cause of zoonosis.
Introduction
Cryptosporidium is an important enteric parasite/protozoan in human beings, pets, domesticated and wild animals and is among the top four causes of moderate-to-severe diarrheal disease in young children in developing countries (Cui et al., 2014; Liu et al., 2014; Ryan & Hijjawi, 2015). Cryptosporidium spp. causes a diarrhea leading to fatal illness in the immunocompromised host. Fecal contaminated water and food is usually responsible of the transmission of Cryptosporidium (Zahedi et al., 2016).
Genus Cryptosporidium has 38 species and about 70 genotypes (Feng, Ryan & Xiao, 2018). Among these 38 species, 20 have been identified in humans but most of them are caused by C. hominis and C. parvum (Holubová et al., 2016; Kváč et al., 2016; Li et al., 2015a; Ryan, Fayer & Xiao, 2014; Ryan et al., 2015) which are the significant source of human cryptosporidiosis. Additionally, some other Cryptosporidium species and genotypes have also been described in certain reports. It includes C. andersoni, C. fayeri, C. cuniculus, C. ubiquitum, C. suis, C. muris, C. canis, C. felis, and C. meleagridis species and five genotypes named as pig genotype II, monkey genotype, horse genotype, chipmunk I genotype and skunk genotype (Cama et al., 2003; Sulaiman et al., 2005; Feng et al., 2007; Robinson, Elwin & Chalmers, 2008; Fayer, 2010; Waldron, Cheung-Kwok-Sang & Power, 2010; Xiao, 2010). These animals have the capacity to contaminate the water resources with different waterborne zoonotic species of organisms like Cryptosporidium (Zahedi et al., 2016). Alike world countries, China is focusing on the epidemiological survey of Cryptosporidium infection in humans, environmental water, and domesticated and wild animals (Yin et al., 2013).
Cryptosporidium felis is the most common protozoan in the domestic cats first reported by Iseki (1979). Afterward, highly zoonotic Cryptosporidium parvum (Sargent et al., 1998) and Cryptosporidium muris (Pavlasek & Ryan, 2007) were also reported in the same host. Recently, molecular epidemiological investigations of cryptosporidiosis in captive wild felines have gained more attention.
Previously, the prevalence of Cryptosporidium has been studied in different captive wild felids (Yin et al., 2013; Gómez et al., 1996; Fagiolini et al., 2010; Li et al., 2015b; Lukášová et al., 2018; Alves et al., 2005; Mateo et al., 2017; Ziegler et al., 2007; Carver et al., 2012; Holsback et al., 2013). South China tiger (Panthera tigris tigris), Black Panther (Panthera pardus) and Manul (Felis manul) are the notable epidemiological studies on wild felids in China (Yin et al., 2013; Li et al., 2015b).
The present study was designed to find out the prevalence and molecular identification of Cryptosporidium spp. in zoos including Siberian tiger (Panthera tigris altaica), Bengal tiger (Panthera tigris tigris), white tiger (Panthera tigris), Jaguar (Panthera onca), white lion (Panthera leo), African lion (Panthera leo leo), cheetah (Acinonyx jubatus), caracal (Caracal caracal), African serval (Leptailurus serval) and bobcat (Lynx rufus) from the different zoos of Beijing, Dalian, Longyan and Harbin.
Materials & Methods
Sample collection
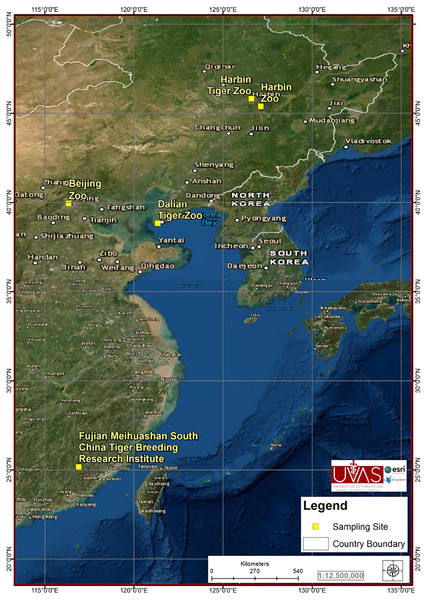
From May 2018 to September 2019, 244 samples (20–30 g each) were collected from Beijing zoo (38), Dalian tiger zoo (38), Fujian Meihuashan South China tiger Breeding Research Institute (10), Harbin zoo (56) and Harbin tiger zoo (102) of China (Fig. 1). The zoos selection criterion was the abundance of feline species in zoos, zoos with high number of feline species were included in the study. All the collected 244 samples were from the apparently healthy hosts of Siberian tiger (Panthera tigris altaica) (146), African lion (Panthera leo) (8), Bengal tiger (Panthera tigris tigris) (8), Caracal (Caracal caracal) (18), Cheetah (Acinonyx jubatus) (16), Jaguar (Panthera onca ) (11), Lynx (Lynx canadensis) (5), White lion (Panthera leo) (13), White tiger (Panthera tigris tigris) (8) and African serval (Leptailurus serval) (11). Estimated relative abundance of the sampled species was 60%. We observed maximum individual animals at different sites available within zoos for defection and collected freshly deposited fecal samples from the ground of the cages (fecal samples were collected within half hour after deposition). Fecal samples were mixed with 5% freshly prepared solution of potassium dichromate (Sigma–Aldrich, St. Louis, MO, USA) in 1:3 mass to volume ratio and stored at 4 °C.
Figure 1: City location in which the samples were collected.
Oocyst isolation
Sheather’s solution was used to isolate oocysts through the discontinuous sucrose gradients. This solution contained 320 ml water, 500 g sucrose, 9 ml phenol and diluted with distilled water and supplemented with Tween-80 (1%). The 1:2 solution and 1:4 solution have different specific gravity. In 50 ml centrifuge tubes, 15 ml of the 1:4 solutions were layered over 15 ml of the 1:2 solutions. A 10 ml aliquot of the sieved feces was layered at the top and centrifuged at 1,000×g for 25 min. The layer between the 1:2 solutions and 1:4 solutions was aspirated very gently and put into a new 50 ml centrifuge tube. Distilled water was supplemented up to 45 ml and centrifuged at 3,000×g rpm for 10 min. Discarded the supernatant and dissolved the sediment (oocysts) with 500 µL distilled water (Arrowood & Sterling, 1987).
DNA extraction
Genomic DNA was extracted from oocysts using QIAamp DNA Mini Stool Kit (Qiagen, Hilden, Germany) with slight modification in manufacturer instruction. The modification adjusted were lysis temperature at 80 °C and elution buffer to 80 ml to increase the DNA concentration. The genomic DNA was stored at −20 °C until further usage.
Nested PCR analysis of the 18S rRNA
A two-step nested PCR was optimized for diagnosis of Cryptosporidium spp. Forward primer (F1) 5′-TTCTAGAGCTAATACATGCG-3′ and reverse primer (R1) 5′-CCCTAATCCTTCGAAACAGGA-3′ was used in the first step of PCR to amplify a product of 1,325 bp. A product of 830 bp was amplified in the second PCR step using forward primer (F2) 5′-GGAAGGGTTGTATTTATTAGATAAAG-3′ and reverse primer (R2) 5′-AAGGAGTAAGGAACAACCTCCA-3′ (Xiao et al., 1999). Totals volume 25 μl containing 2.5 μl PCR buffer, 2 μl Magnesium Chloride, 2 μl dNTP, 1 μM of each primer, 1 μl of Taq DNA polymerase and 3 μl of DNA template. The optimized thermocyclic conditions were initial denaturation at 94 °C for 5 min followed by 30 cycles (94 °C for 30 s, 54 °C for 30 s and 72 °C for 90 s) and final extension at 72 °C for 7 min. The PCR products were run on 1% gel and purified with Gel Extraction Kit-200 (OMEGA, China). In second step, PCR products of 1st step were used as templates. In it, initial denaturation and final extension remained the same during but the 30 cycles consisted of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 60 s.
DNA sequencing and species/genotypes identification
The nPCR products were run on 1% gel and purified by Gel Extraction Kit-200 (OMEGA, China) and sent for sequencing to Comate Biosciences Co., Ltd. (Changchun, China). Successfully sequenced data was run on NCBI for similarity analysis using BLAST.
Phylogenetic analysis
MegAlign (Version 4.0 DNA Star, Madison, USA) was used to align the obtained sequences for genotyping. These sequences were submitted to GenBank (accession numbers can be seen in data availability). The phylogenetic tree was constructed using the Maximum-Likelihood method on MEGA version 6.0.
Statistical analysis
Descriptive frequencies of Cryptosporidium were shown in percentages by dividing the positive Cryptosporidium sample to the total number of samples. Chi square test of independence at significance level α = 0.05 was used to determine the association of Cryptosporidium prevalence with the sampling site. Sampling sites (zoos) and prevalence (positive and negative sample) were handled as nominal variable and Chi square test of independence was applied using IBM SPSS Statistics 20.
Results
Cryptotosporidium prevalence in Feline
A total of 244 samples were used for nested-PCR and 23 (9.43%) specimens were found positive for Cryptosporidium (Table 1). Among these 23 positives, 22 samples were from Harbin zoo and one from Beijing zoo while Dalian tiger zoo, Fujian Meihuashan South China tiger Breeding Research Institute and Harbin tiger zoo were negative for Cryptosporidium. The prevalence varied from 0.0 to 39.3% in different zoos. Prevalence of Cryptosporidium spp. was 8.22% (12/146) in Siberian tiger, 12.5% (1/8) in Bengal tiger, 37.5% (3/8) in white tiger, 15.38% (2/13) in white lion, 40% (2/5) in lynx and 25% (2/8) in African lion while it was not prevalent in caracal (18), cheetah (16), jaguar (11) and Serval (11) (Table 2).
| Sr. no. | Sampling site | No. of samples | Cryptosporidium parvum | Cryptosporidium sp. NEV10 Alike | Total positive tested | Chi square test of independence |
|---|---|---|---|---|---|---|
| 1 | Beijing Zoo | 38 | 01 | -- | 01 |
X2 (4) = 76.15 p < 0.01 φc = 0.56 n = 244 |
| 2 | Harbin Zoo | 56 | 05 | 17 | 22 | |
| 3 | Fujian Meihuashan South China tiger Breeding Research Institute | 10 | -- | -- | -- | |
| 4 | Dalian tiger zoo | 38 | -- | -- | -- | |
| 5 | Harbin tiger zoo | 102 | -- | -- | -- | |
| Total | 244 | 06 | 17 | 23 |
Note:
“φc” denoting Cramér’s V.
| Sr. no. | Host | No. of samples | Cryptosporidium Parvum | Cryptosporidium sp. NEV10 alike | Total positive tested | Negative samples | Percentage |
|---|---|---|---|---|---|---|---|
| 1 | Siberian Tiger | 146 | 03 | 09 | 12 | 134 | 8.22 |
| 2 | Bengal Tiger | 08 | 01 | – | 01 | 07 | 12.5 |
| 3 | White Tiger | 08 | 01 | 02 | 03 | 05 | 37.5 |
| 4 | White Lion | 13 | 01 | 02 | 03 | 10 | 23.1 |
| 5 | Caracal | 18 | – | – | – | 18 | – |
| 6 | Cheetah | 16 | – | – | – | 16 | – |
| 7 | Lynx | 05 | – | 02 | 02 | 03 | 40 |
| 8 | Jaguar | 11 | – | – | – | 11 | – |
| 9 | African Lion | 08 | – | 02 | 02 | 06 | 25 |
| 10 | Sarval | 11 | – | – | – | 11 | – |
| Total | 244 | 06 | 17 | 23 | 221 | 9.43 |
A chi-square test of independence was performed to examine the relationship between zoo and Cryptosporidium spp. prevalence. The relationship between these two was significant X2 (4, N = 244) = 76.15, p = 0.00. The effect size for these finding, Cramer’s V, was very strong, 0.56. As can be seen in Table 1, from the total positive sample tested in this study 95.65% of the positive sample were from the Harbin Zoo, it means feline species of Harbin zoo has significantly higher prevalence than the other zoo.
Cryptotosporidium spp./genotypes
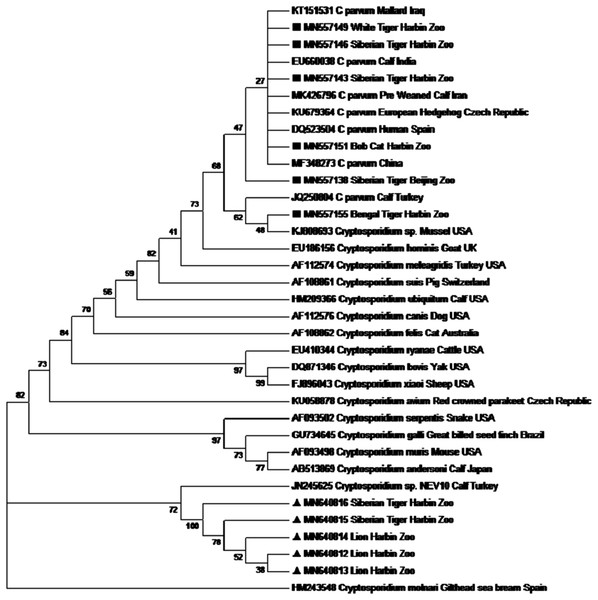
Similarity analysis of the 18S rRNA gene showed that 6 out of 23 were Cryptosporidium parvum and the remaining 17 isolates resembled to Cryptosporidium spp. NEV10 (JN245625). For explaining the phylogenetic relationship, a tree was constructed based on 18S rRNA using the maximum likelihood method (Fig. 2). Cryptosporidium with accession numbers MN640812, MN640813, MN640814, MN640815 and MN640816 was identical to the reference sequence (accession no. JN245625, CryptosporidiumNEV10) that was isolated from a diarrheic calf in Turkey. Six Isolates with accession numbers MN557138, MN557143, MN557146, MN557149, MN557151 and MN557155 were identical to that of Cryptosporidium parvum.
Figure 2: Phylogenetic tree showing the sequences of the present study, ▪ for Cryptosporidium parvum and ▴ for Cryptosporidium sp. NEV10 alike, remaining sequences are from NCBI GenBank data base.
Discussion
In present study, Cryptosporidium was prevalent in 9.43% (23/244) in five captive felids of Chinese Zoos. Lynx was having the highest prevalence rate for Cryptosporidium 40% (2/5) and lowest prevalence was observed in Siberian Tiger 8.22% (12/146). While, in term of locations the prevalence ranges from 0% to 39.3% in five different zoos. Harbin zoo was having the highest prevalence rate of Cryptosporidium.
Wildlife plays a crucial role in spreading various pathogens to animals (domestic, Pet, free-living) and humans (Thompson, Kutz & Smith, 2009). Various studies on epidemiological surveys have been conducted around the world considering their importance to public and veterinary health (Ryan, Fayer & Xiao, 2014; Feng et al., 2007). Although few studies have been conducted for the detection of Cryptosporidium in different wild and captive animals. In the case of zoo felids, only manul (Felis manul) was found infected with Cryptosporidium felis in Zhengzhou zoo of China (Li et al., 2015b) and Bobcat (Lynx rufus) with Cryptosporidium parvum in the USA (Carver et al., 2012). The present study will benefit our understanding on the prevalence of Cryptosporidium in zoo Felids.
The Cryptosporidium prevalence in the present study is 9.43% (23/244) in five captive felids of Chinese Zoos it coincides with the results reported previously in wild red fox in Spain 8% (7/87) and in animal sources (caged dogs, kennel cats, captive animals) in China 7% (6/84) (Yin et al., 2013; Mateo et al., 2017). Higher proportion of Cryptosporidium prevalence has been reported previously in wild and domestic mammals in South Africa (14%, 8/56) and in free-living wild felids in Brasil (15%, 2/13) (Lukášová et al., 2018; Holsback et al., 2013). Fewer studies also revealed lower rates of Cryptosporidium in wild and captive reptiles in USA 3% (15/528) and in captive Asiatic black bears (Ursus thibatenus) in China 2.4% (4/218) (Upton et al., 1989; Wang et al., 2020). In Italy, parasitic investigation in mammals of two zoological gardens reported the prevalence of Cryptosporidium in carnivores (10%), primates (66.7%) and artiodactyls (25%) (Fagiolini et al., 2010). In Lisbon Zoo, Cryptosporidium was found in one Indian star tortoise (Geochelone elegans), one Prairie bison (Bison bison bison) and one black wildebeest (Connochaetes gnou) (Alves et al., 2005). While in a study held in 1999, Cryptosporidium was found in 18 Artiodactyla, 14 Primate, 2 Perissodactyla, and 1 Proboscidea species of Barcelona zoo (Gomez et al., 2000). In wildlife populations within a watershed landscape, the prevalence rate of Cryptosporidium was 5% (312/6227) in southeastern New York State (Ziegler et al., 2007).
Current results of highest prevalence rate 39.3% in the Harbin zoo can be compared with the result of a previous study conducted in Zhengzhou zoo where prevalence rate was very low 1.5% (3/203) (Li et al., 2015b). Same in the Lisbon zoo, only three out of 274 fecal samples from mammals and reptiles were found positive of Cryptosporidium (Alves et al., 2005) while another study revealed 3.6% (14/388) in ruminants of the same zoo (Delgado et al., 2003). In 1996, Only six out of 51 mammal species were found to be infected from Cryptosporidium in Barcelona zoo (Gómez et al., 1996).
In our study, Cryptosporidium parvum and Cryptosporidium spp. NEV10 alike were identified in zoo felids. Cryptosporidium parvum has a zoonotic potential and previously found in preweaned calves and humans in China (Qi et al., 2015; Wang et al., 2013). Only from Chile, a sequence of Cryptosporidium parvum (accession no. GQ865534) in Bengal tiger has been submitted to NCBI GenBank before the findings of current study. To the best of our knowledge, manul (Felis manul) was infected from Cryptosporidium felis in Zhengzhou zoo which was the only published report of any zoo felid having cryptosporidiosis in China (Li et al., 2015b).
Another Cryptosporidium spp. was detected in the present study which showed up to 100% identity with the published sequence under the accession number JN245625 (Cryptosporidium spp. NEV10). Previously, Cryptosporidium spp. NEV10 was only found in diarrheic calf in Turkey. Because of the limited knowledge of this species, its significance about the public-health and whether zoo felids from China are the new host of this species is unclear. The prevalence of zoonotic Cryptosporidium parvum and Cryptosporidium spp. NEV10 alike might have link with the contaminated feed and water as zoo felids were fed with chicken and beef. Findings of this study demonstrated that, felid species were the suitable hosts for very common zoonotic Cryptosporidium sp. (C. parvum) and Cryptosporidium spp. NEV10 alike.
However, during this study all the members of a felid species available at zoo were not accessed, as animal freely roam in the garden of the zoo.
Conclusions
The study under taken showed that first evidence of molecular identification of Cryptosporidium parvum in the zoo felids of China and those hosts may act as carrier of zoonoses by shedding the oocysts of Cryptosporidium parvum. The identification of an unknown Cryptosporidium spp. which has a close relationship with Cryptosporidium NEV10 from Turkey directs for further investigation as it was detected from diarrheic calf but now it was found from zoo felids. This study directs the attention of animal welfare zoo officials to take control measures such as proper housing and management to provide environment that allow animal to grow, mature, reproduce and minimizes the effect of disease due to parasites.