A novel bicyclic 2,4-diaminopyrimidine inhibitor of Streptococcus suis dihydrofolate reductase
- Published
- Accepted
- Received
- Academic Editor
- Pedro Silva
- Subject Areas
- Microbiology, Molecular Biology, Infectious Diseases
- Keywords
- Streptococcus suis, Antibiotics, Antifolate, Dihydrofolate reductase, Pathogen Box, Diaminopyrimidine, Drug resistant, Drug discovery, Screening, Growth inhibition
- Copyright
- © 2021 Songsungthong et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. A novel bicyclic 2,4-diaminopyrimidine inhibitor of Streptococcus suis dihydrofolate reductase. PeerJ 9:e10743 https://doi.org/10.7717/peerj.10743
Abstract
Streptococcus suis is a Gram-positive bacterial pathogen of pigs and an emerging zoonotic pathogen. It has become increasingly resistant to multiple classes of antibiotics. New drug candidates and knowledge of their targets are needed to combat antibiotic-resistant S. suis. In this study, the open-source Pathogen Box compound library was screened. Thirty hits that effectively inhibited S. suis growth at 10 µM were identified. Among the most potent hits, MMV675968 (a diaminoquinazoline analog) was shown to target S. suis dihydrofolate reductase (SsDHFR) via (1) growth inhibition of an E. coli surrogate whose growth is dependent on exogenously expressed SsDHFR and (2) inhibition of in vitro SsDHFR activity. Thymidine supplement is able to reverse growth inhibition by MMV675968 in both E. coli surrogate and S. suis, indicating that a thymidine-related pathway is a major target of MMV675968. Comparison of MMV675968 with seven DHFR inhibitors representing different core structures revealed that bicyclic 2,4-diaminopyrimidines with long and flexible side chains are highly effective in inhibiting SsDHFR and S. suis growth. MMV675968 and related compounds thus may serve as starting points for developing antibiotics against drug resistant S. suis.
Introduction
It is estimated that antibiotic resistance will lead to 10 million deaths per year and economic loss of US$100 trillion by the year 2050 (O’Neill, 2014). Alarmingly, bacteria resistant to last resort antibiotics and to all commercially available antibiotics have already emerged and spread, signaling the start of a global public health crisis (McGann et al., 2016; Ruzauskas & Vaskeviciute, 2016; McCarthy, 2017). New antibiotics, preferably with new chemical scaffolds to bypass existing resistance mechanisms, are urgently needed. Pathogen Box, an open-source library of 400 compounds with chemical structures distinct from currently available antibiotics (https://www.mmv.org/mmv-open/pathogen-box/about-pathogen-box), demonstrated activity against selected neglected disease pathogens and low toxicity against human cells is a promising source of compounds for antibiotic discovery.
Streptococcus suis is a Gram-positive bacterium that can cause severe symptoms such as meningitis, septicemia, and arthritis in pigs and thus pose a high economic burden for the global pig industry (Wertheim et al., 2009a). Zoonotic transmission to humans occurs from eating uncooked contaminated pork or from occupation-related infection such as when farmers or abattoir workers come into contact with infected pigs or pig carcasses (Wertheim et al., 2009a; Ho et al., 2011; Goyette-Desjardins et al., 2014; Huong et al., 2014). S. suis isolates found in pigs and in human cases in Vietnam and China are of the same serotype and multi locus sequence typing (MLST), suggesting direct transmission (Hoa et al., 2011). In human infection, S. suis causes meningitis, sepsis, arthritis, vestibular dysfunction, and permanent hearing loss with fatality rate of approximately 13% (Huong et al., 2014). S. suis infection is a major cause of bacterial meningitis in Southeast Asia, and was responsible for a Streptococcal Toxic Shock Syndrome outbreak in China (Suankratay et al., 2004; Hui et al., 2005; Tang et al., 2006; Thi Hoang Mai et al., 2008; Wertheim et al., 2009b). The number of human S. suis cases has increased recently, especially in areas with high density pig farming, making S. suis infection an important emerging disease (Wertheim et al., 2009a; Gajdács et al., 2020).
Owing to widespread use of antibiotics for prophylaxis and as growth promoter in the pig farming industry, antibiotic resistance in S. suis is rising (Wertheim et al., 2009a; Varela et al., 2013; Hernandez-Garcia et al., 2017; Yongkiettrakul et al., 2019). S. suis isolates are recently reported to be resistant to multiple classes of antibiotics such as β-lactams (e.g., ampicillin and penicillin), macrolides (e.g., erythromycin and clarithromycin), lincosamide (e.g., clindamycin), tetracycline, fluoroquinolone (e.g., moxifloxacin), aminocyclitol (e.g., spectinomycin), aminoglycoside (e.g., gentamycin), and trimethoprim-sulfonamide (Yongkiettrakul et al., 2019; Tan et al., 2020; Riley et al., 2020; Gajdács et al., 2020). Multidrug resistance in S. suis thus prompts the need for new drugs. In this study, we screened the Pathogen Box library against two strains of S. suis, namely P1/7 and HE06 (Jacobs, Van den Berg & Loeffen, 1996; Maneerat et al., 2013) and identified 30 compounds with high growth inhibitory activity at 10 µM. The potential target of MMV675968, a diaminoquinazoline derivative, was identified as dihydrofolate reductase, which is a target of multiple classes of compounds including antibacterial, antimalarial, and anticancer compounds (Gao et al., 2019).
Materials and Methods
The experiments using S. suis were approved by BIOTEC Institutional Review Board on biosafety and biosecurity with approval number BT-IBC-59-028.
Bacteria strains and growth conditions
S. suis strains P1/7 and HE06 are serotype 2 isolates obtained from a pig with meningitis and an infected human, respectively (Jacobs, Van den Berg & Loeffen, 1996; Holden et al., 2009; Maneerat et al., 2013). Serotype 2 strains were chosen because of their association with pathogenicity (Lun et al., 2007; Wertheim et al., 2009a; Maneerat et al., 2013). S. suis was grown on brain heart infusion (BHI) plates (Beckton Dickinson, Franklin Lakes, NJ, USA) at 37 °C with 5% CO2. Escherichia coli PA414 lacking folA and thyA genes, encoding dihydrofolate reductase (DHFR) and thymidylate synthase (TS), respectively (Ahrweiler & Frieden, 1988) was used as a surrogate for expressing S. suis DHFR. The deletion of thyA is required for survival of DHFR deficient mutant (Ahrweiler & Frieden, 1988). E. coli PA414 transformed with various expression plasmids was grown in Luria Bertani (LB) broth supplemented as necessary with 50 µg/mL kanamycin, 100 µg/mL ampicillin, 10 µg/mL chloramphenicol, 0.2% (w/v) arabinose, and/or 50 µg/mL thymidine.
Bacterial growth inhibition assay
The Clinical Laboratory Standard Institute (CLSI) broth microdilution method was used to test antibacterial activity of Pathogen Box compounds and selected DHFR inhibitors (The Clinical and Laboratory Standards Institute, 2018). Briefly, 5 × 104 colony forming units (CFUs) of S. suis or E. coli PA414 were incubated with compounds at 10 µM (or other concentrations as indicated) in cation-adjusted Mueller Hinton broth (Beckton Dickinson, Franklin Lakes, NJ, USA) in 96-well plates at 37 °C with or without 5% CO2, respectively. Growth was monitored by measuring optical density at 600 nm (OD600) using a SpectraMax M5 spectrophotometer (Molecular Devices, San Jose, CA, USA). Percent growth of each compound-treated bacteria strain was calculated using the following formula:
Construction of expression plasmids
The dhfr gene encoding S. suis dihydrofolate reductase (SsDHFR) was amplified from clarified lysate of S. suis P1/7 and S. suis HE06 using high-fidelity Phusion DNA polymerase (New England Biolabs, Ipswich, MA, USA) and primers 5′ctggtgccgcgcggcagccatATGACTAAAAAGATTGTTGC3′ and 5′tcgggctttgttagcagccggatc CTAACCATCTCTTCTTTCATAG3′ (lower case denotes sequence homologous to pET15b while upper case denotes sequence homologous to the dhfr gene). The PCR products were cloned into NdeI and BamHI digested pET15b plasmid using a Gibson Assembly kit (New England Biolabs, Ipswich, MA, USA), according to manufacturer’s protocol. The resulting plasmids were subjected to Sanger sequencing (1st BASE, Singapore). Nucleotide sequences of the dhfr gene from S. suis P1/7 and S. suis HE06 were deposited in NCBI Genbank with accession number MH388486 and MH388487, respectively.
Complementation assay of E. coli surrogate
E. coli PA414 lacking functional DHFR and TS enzymes (Ahrweiler & Frieden, 1988) was used as a surrogate host to study the function of exogenously expressed SsDHFR. E. coli PA414 was co-transformed with (1) pBAD33 or pBAD33-EcTS and (2) pET15b or pET15b-SsDHFR. To test whether SsDHFR can complement for the loss of E. coli DHFR in the E. coli surrogate, overnight cultures of E. coli PA414 carrying various combination of plasmids were pelleted, washed, cell density adjusted to OD600 of 4, serially-diluted, and spotted onto LB plates supplemented with appropriate antibiotics and either 50 µg/mL thymidine or 0.2% (w/v) arabinose. Plates were incubated at 37 °C overnight. Growth on agar plate was observed.
Overexpression of SsDHFR in E. coli BL21(DE3)
Cultures of E. coli BL21(DE3) carrying pET15b or pET15b-SsDHFR were grown with shaking at 37 °C in LB supplemented with 100 µg/mL ampicillin. When OD600 reached 1, IPTG was added to 40 µM, temperature shifted to 16 °C, and incubated with shaking overnight. Bacterial pellets were collected by centrifugation, resuspended in lysis buffer (20 mM potassium phosphate buffer, 0.1 mM EDTA, 10 mM DTT, 50 mM KCl, 20% glycerol), and sonicated. Protein concentration of clarified bacteria lysate was determined by Bradford assay (Biorad, Hercules, CA, USA) according to the manufacturer’s protocol.
In vitro DHFR activity assay
DHFR uses NADPH as a cofactor for the conversion of dihydrofolate (DHF) to tetrahydrofolate (THF). Since the amount of NADPH consumed is directly proportional to the amount of THF product, monitoring NADPH consumed using absorbance at 340 nm (A340; ε340 of 12,300 M−1 cm−1) is indicative of DHFR enzymatic activity. The DHFR activity assay was performed as previously described (Tirakarn et al., 2012; Songsungthong et al., 2019). Briefly, using kinetics mode of a UV-Vis spectrophotometer (Agilent Technologies, Santa Clara, CA, USA), A340 value of the reaction mixture (50 mM TES, pH 7.0, 75 mM β-mercaptoethanol, 1 mM EDTA, 1 mg/mL BSA, 0.1 mM NADPH, 0.1 mM dihydrofolate) was set to blank (A340 = 0). Inhibitors were added to the reaction mixture as necessary. 1 µg of total protein from lysate of E. coli BL21(DE3) containing pET15b or overexpressing SsDHFR was added to reaction buffer to initiate the enzymatic reaction. The amount of SsDHFR containing lysate added was predetermined to give linear reaction kinetics. NADPH reduction was monitored by measuring A340 reduction for 100 s. A340 reduction per minute values were recorded.
Percent SsDHFR activity was calculated using the following formula:
(A340 reduction per minute of compound-treated lysate × 100)/(A340 reduction per minute of DMSO-treated lysate of BL21 expressing SsDHFR).
Statistical analysis
Data are shown as mean ± standard error of the mean (SEM). At least three independent experiments were performed. Statistical analysis using one-way ANOVA with Tukey’s post test was performed. Statistical significance is noted on the graph by asterisks. * denotes P < 0.05. ** denotes P < 0.01. ** denotes P < 0.001. *** denotes P < 0.0001. ns denotes not statistically significant.
Results
Identifying compounds with S. suis growth inhibitory activity from Pathogen Box
We screened the Pathogen Box library for compounds active against S. suis P1/7 and S. suis HE06 isolated from an infected pig and human, respectively (Jacobs, Van den Berg & Loeffen, 1996; Holden et al., 2009; Maneerat et al., 2013). Thirty compounds with high inhibitory activity (with average percent inhibition of 90–100%) at 10 µM were identified (Table S1). Seven of the hits including rifampicin, levofloxacin, and linezolid are reference compounds known to be broadly effective against Gram-positive bacteria (highlighted in blue in Table S1). Twenty three other hits are compounds with structures distinct from commercially available antibiotics (highlighted in green in Table S1). The mechanisms of action of most non-reference hits are unknown. MMV675968, one of the hits, has been shown to inhibit the growth of protozoan parasites and Gram-negative bacteria (Lau et al., 2001; Nelson & Rosowsky, 2001; Popov et al., 2006; Songsungthong et al., 2019) via inhibition of dihydrofolate reductase (DHFR), an enzyme in the thymidylate cycle.
MMV675968 is an inhibitor of S. suis DHFR
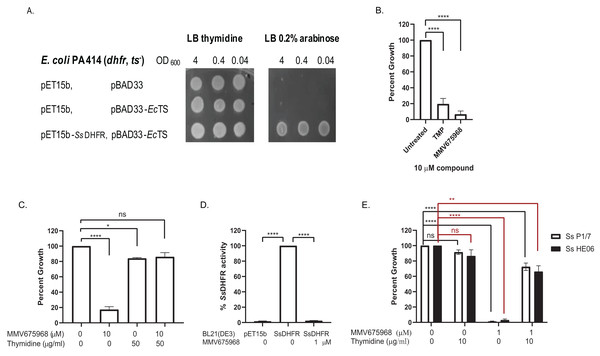
MMV675968 inhibits DHFR of protozoan parasites and of Gram-negative bacteria (Lau et al., 2001; Nelson & Rosowsky, 2001; Popov et al., 2006; Songsungthong et al., 2019). We therefore hypothesized that MMV675968 inhibited S. suis growth through inhibition of S. suis DHFR (SsDHFR). An E. coli surrogate assay and an in vitro DHFR activity assay were used to test this hypothesis. For the E. coli surrogate assay, E. coli PA414 deficient in E. coli DHFR (EcDHFR) and E. coli thymidylate synthase (EcTS) of the thymidylate cycle making the strain a thymidine auxotroph (Ahrweiler & Frieden, 1988), was used as a host for exogenously expressing SsDHFR. E. coli PA414 transformant controls carrying two empty vectors (pET15b and pBAD33), or expressing EcTS alone did not grow in the absence of thymidine supplement as expected (Fig. 1A). E. coli PA414 expressing EcTS together with SsDHFR was able to grow without thymidine supplement (Fig. 1A), indicating that SsDHFR was functional and complemented for the loss of EcDHFR. Growth of E. coli PA414 in the absence of thymidine was therefore dependent on the function of exogenously expressed SsDHFR.
Figure 1: MMV675968 inhibits Streptococcus suis. dihydrofolate reductase (Ss DHFR).
(A) Ss DHFR functions in E. coli PA414. Overnight cultures of E. coli PA414 carrying empty plasmids or expressing EcTS and SsDHFR were serially diluted and spotted onto LB plates supplemented with either 50 µg/mL thymidine or 0.2% (w/v) arabinose. Overnight growth was recorded. A representative picture from three independent experiment is shown. (B) MMV675968 inhibits SsDHFR in E. coli surrogate. E. coli PA414 expressing EcTS and SsDHFR was incubated with DMSO, trimethoprim (TMP), or MMV675968 (10 µM). Growth of compound-treated E. coli surrogate was calculated as percent of untreated (100%). (C) Growth inhibition of E. coli surrogate expressing SsDHFR by MMV675968 can be rescued by thymidine supplement. E. coli PA414 expressing EcTS and SsDHFR was incubated with or without 10 µM MMV675968 and with or without 50 µg/mL thymidine. Growth was monitored and compared with that of untreated control (100%). (D) MMV675968 inhibits SsDHFR activity. In vitro SsDHFR activity from clarified lysate of E. coli BL21(DE3) overexpressing SsDHFR was compared with that of E. coli BL21(DE3) harboring empty plasmid or in the presence of 1 µM MMV675968. (E) S. suis growth inhibition by MMV675968 can be rescued by thymidine supplement. S. suis P1/7 and S. suis HE06 was incubated with or without 1 µM MMV675968 and with or without 10 µg/mL thymidine. Growth was monitored and compared with that of untreated control (100%). The graphs (B–E) show mean ± standard error of the mean (SEM) from three independent experiments. One-way ANOVA with Tukey’s post test was performed to determine statistical significance compared with untreated control. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not statistically significant.Trimethoprim (TMP), a bacterial DHFR inhibitor (Baker et al., 1981), inhibited the growth of E. coli PA414 expressing SsDHFR to 19% of untreated (Fig. 1B), confirming that trimethoprim inhibits SsDHFR. MMV675968 was more effective than TMP as seen by its ability to inhibit the growth of E. coli PA414 expressing SsDHFR to 7% (Fig. 1B). E. coli PA414 growth inhibition by MMV675968 was reversed in the presence of thymidine supplement (Fig. 1C), suggesting that the thymidine-related pathway is the principal target of MMV675968 in E. coli surrogate consistent with DHFR being a target.
Next, we tested whether MMV675968 could inhibit SsDHFR activity directly by an in vitro DHFR activity assay. DHFR activity present in lysate of E. coli BL21(DE3) carrying an empty pET15b plasmid accounted for only 2% of DHFR activity of lysate of E. coli BL21(DE3) overexpressing SsDHFR (Fig. 1D), indicating that most of the DHFR activity observed was from overexpressed SsDHFR rather than endogenous EcDHFR. In the presence of 1 µM MMV675968, DHFR activity decreased to 2% of that of untreated lysate (Fig. 1D), indicating that MMV675968 inhibits SsDHFR directly. Results from both E. coli surrogate assay and DHFR activity assay thus indicate that MMV675968 inhibits SsDHFR.
We also tested whether thymidine-related pathway is the main target of MMV675968 in S. suis by testing whether growth inhibition was rescued by thymidine supplement. Growth of both S. suis P1/7 and S. suis HE06 were inhibited in the presence of 1 µM MMV675968 (Fig. 1E). Growth inhibition of both S. suis strains was reversed with thymidine supplement (Fig. 1E), indicating that the main target of MMV675968 within S. suis is a thymidine-related pathway.
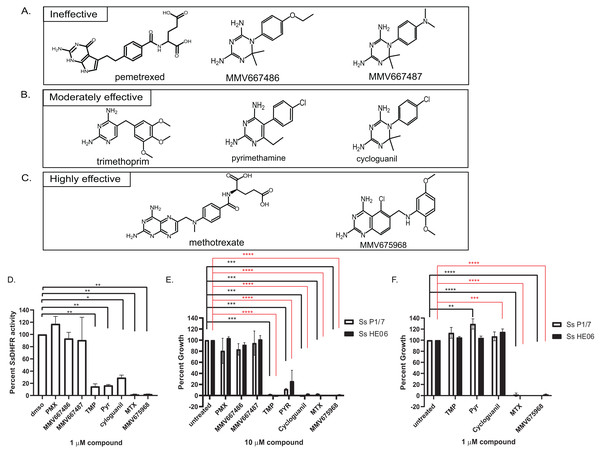
Figure 2: Efficacy of various dihydrofolate reductase (DHFR) inhibitors of Streptococcus suis DHFR (SsDHFR) and S. suis growth.
Structures of eight DHFR inhibitors tested in this study and grouped according to their inhibitory activity: (A) ineffective inhibitors, (B) moderately effective inhibitors, and (C) highly effective inhibitors. (D) SsDHFR activity in the presence of 1 µM DHFR inhibitors. (E) Percent growth of S. suis P1/7 (white bar) and S. suis HE06 (black bar) after 18 h incubation with 10 µM of DHFR inhibitors. (F) Percent growth of S. suis P1/7 (white bar) and S. suis HE06 (black bar) after 18 h incubation with 1 µM inhibitors. PMX, pemetrexed; TMP, trimethoprim; PYR, pyrimethamine; MTX, methotrexate. The graphs show mean ± standard error of the mean (SEM) from at least three independent experiments. One-way ANOVA with Tukey’s post test was performed to determine statistical significance compared with untreated control. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not statistically significant. For E and F, black asterisks show statistical significance between untreated and compound-treated S. suis P1/7 whereas red asterisks show statistical significance between untreated and compound-treated S. suis HE06.Bicyclic 2,4-diaminopyrimidines with long and flexible side chain associate with higher inhibitory activity against SsDHFR and S. suis
To investigate which chemical scaffolds are better at inhibiting SsDHFR and S. suis growth, eight DHFR inhibitors of various chemical structures (Figs. 2A–2C), which include commercially available compounds (pemetrexed, trimethoprim, pyrimethamine, cycloguanil, and methotrexate), compounds from Medicines for Malaria Venture (MMV)’s Malaria Box (MMV667486 and MMV667487), and a compound from Pathogen Box (MMV675968), were tested for SsDHFR inhibitory activity and S. suis growth inhibition. Pemetrexed, MMV667486, and MMV667487 did not show significant SsDHFR or S. suis growth inhibitory activity (Figs. 2A, 2D and 2E), indicating that these structures do not inhibit SsDHFR and consequently are unable to inhibit S. suis growth. Trimethoprim, pyrimethamine, and cycloguanil were moderately effective at inhibiting SsDHFR and S. suis growth (defined as being effective at 10 µM but not at 1 µM) (Figs. 2B, 2D–2F). Methotrexate and MMV675968 inhibited SsDHFR more effectively that trimethoprim, and completely inhibited S. suis growth, even at 1 µM (Figs. 2C–2F). Methotrexate and MMV675968, both of which are bicyclic 2,4-diaminopyrimidines with long and flexible side chains, are highly effective inhibitors of both SsDHFR and S. suis growth. Both S. suis P1/7 and S. suis HE06 strains shared a similar sensitivity pattern towards various DHFR inhibitors (Figs. 2E and 2F) consistent with the identical DHFR sequences between the two strains (Figs. S1A–S1B).
Discussion
Thirty compounds with high S. suis growth inhibitory activity were identified from the Pathogen Box library (Table S1). Among the thirty hits, seven are reference antibiotics known to be effective against Gram-positive bacteria, including levofloxacin, rifampicin, and linezolid (highlighted in blue in Table S1). The chemical structures of twenty three other hits are distinct from commercially available antibiotics (highlighted in green in Table S1). Approximately 60% of these hits (14 out of 23) also have activity against Mycobacterium tuberculosis (https://www.mmv.org/mmv-open/pathogen-box/about-pathogen-box), suggesting that they have broad activity against Gram-positive organisms and are attractive as starting points for developing antibiotics.
Among the 23 hits with novel structures, compounds MMV688508 and MMV687813 are oxazolidinone analogs. These compounds share the same core structure with radezolid, sutezolid, and linezolid, which are known to have broad spectrum activity against Gram-positive bacteria (Bozdogan & Appelbaum, 2004). Oxazolidinones inhibit bacterial protein synthesis by binding to the 50S ribosomal subunit and preventing the formation of a functional ribosome (Bozdogan & Appelbaum, 2004). It is therefore likely that MMV688508 and MMV687813 target S. suis protein synthesis in a similar fashion. Besides inhibiting the growth of S. suis, MMV688508 also inhibits the growth of other Gram-positive pathogens such as Mycobacterium tuberculosis, Mycobacterium abscessus, and Staphylococcus aureus (Bhandari et al., 2018; Jeong et al., 2018), showing broad-spectrum activity of MMV688508 against various Gram-positive pathogens. Interestingly, MMV687813 is not reported to have anti-Staphylococcus activity or anti-M. abscessus activity (Bhandari et al., 2018; Jeong et al., 2018) despite having activity against M. tuberculosis and S. suis (https://www.mmv.org/mmv-open/pathogen-box/about-pathogen-box, Table S1), pointing to structural differences among ribosomal targets and/or differences in compound permeability in various Gram-positive species.
DHFRs are present in various organisms including protozoan parasites, human, and bacteria. Even though DHFRs of different organisms perform the same function, i.e., the conversion of dihydrofolate (DHF) to tetrahydrofolate (THF) using NADPH as a cofactor, structures of DHFRs from different organisms are different and can be clustered into at least nine distinct clades, resulting in different druggable space and different structures of effective inhibitors (Bhosle & Chandra, 2016). DHFRs from various bacteria species do not necessarily cluster to the same clade. Specifically, SsDHFR does not belong in the same clade as DHFRs of Gram-negative bacteria previously shown to be inhibited by MMV675968 (Nelson & Rosowsky, 2001; Songsungthong et al., 2019). Consequently, it cannot be assumed a priori that MMV675968 will effectively inhibit DHFR from S. suis. Experiment testing inhibition of SsDHFR by MMV675968 was therefore needed.
This study is the first to show that MMV675968 has growth inhibitory activity against a Gram-positive bacterium, namely S. suis, by inhibiting SsDHFR (Table S1, Figs. 1B and 1D), raising a possibility that MMV675968 may serve as a broad-spectrum antibiotic candidate. Growth inhibition by MMV675968 is reversed by thymidine supplement (Figs. 1C and 1E), confirming that the principal target of MMV675968 is a thymidine-related pathway.
Eight DHFR inhibitors of various chemical structures were tested for their efficacy in inhibiting SsDHFR and S. suis growth, revealing structures associated with ineffective, moderately effective, and highly effective inhibitors (Fig. 2). Pemetrexed (a human DHFR inhibitor), MMV667486, and MMV667487 (Plasmodium DHFR inhibitors (Aroonsri et al., 2016)) were ineffective as inhibitors against SsDHFR and S. suis (Figs. 2A, 2D and 2E). Trimethoprim, pyrimethamine, and cycloguanil, which are known inhibitors of bacterial and parasite DHFRs (Gleckman, Blagg & Joubert, 1981; Rollo, 1955; Tonelli et al., 2017), were moderately effective at inhibiting SsDHFR and S. suis growth (Figs. 2B, 2D–2F). These moderately-effective inhibitors contain either diaminopyrimidine (trimethoprim and pyrimethamine) or diaminodimethyltriazine (cycloguanil) cores with a rigid or flexible side chain. Interestingly, even though MMV667486, MMV667487, and cycloguanil share the same diaminodimethyltriazine core and a phenyl ring at the same position, MMV667486 and MMV667487 were ineffective at inhibiting SsDHFR and S. suis growth whereas cycloguanil was effective (Figs. 2A, 2B, 2D and 2E). The data suggest that the SsDHFR active site can accommodate the shorter side chain of cycloguanil, whereas the longer/bulkier side chains of MMV667486 and MMV667487 may interfere with compound binding to the SsDHFR active site, rendering the compounds ineffective as inhibitors. A correlation between SsDHFR inhibitory activity and S. suis growth inhibition of various compounds was observed (Figs. 2D–2F), implying that the two S. suis strains are permeable to all DHFR inhibitors tested allowing access to the SsDHFR target, leading to growth inhibition. It is possible that other S. suis strains may have different compound permeability patterns.
Among the inhibitors tested, MMV675968 and methotrexate were highly effective SsDHFR and S. suis inhibitors (Figs. 2C–2F), both of which contain a bicyclic 2,4-diaminopyrimidine core with a long and flexible side chain. Such structures can serve as starting points for further rational drug design against SsDHFR and S. suis. Streptococcus pneumoniae DHFR is approximately 53% identical to SsDHFR and share high sequence homology at the folate and NADPH binding sites (Fig. S1C), raising a possibility that methotrexate and MMV675968 may also be effective against S. pneumoniae or other Streptococci. Other bicyclic 2,4-diaminopyrimidines were able to inhibit Streptococcus mutans growth (Zhang et al., 2015), confirming this hypothesis. Since methotrexate is a known inhibitor of human DHFR, whereas MMV675968 shows 59-83 fold selectivity for bacterial DHFRs over human DHFR (Nelson & Rosowsky, 2001; Songsungthong et al., 2019), and is not cytotoxic against human HL-60 and HepG2 cell lines (https://www.mmv.org/mmv-open/pathogen-box/about-pathogen-box), MMV675968 might be a better starting compound for antibiotic discovery.
Conclusions
A screen of the Pathogen Box compound library leads to identification of 23 new bioactive compounds against S. suis (Table S1). MMV688508 and MMV687813, oxazolidinone analogs, are strong inhibitors of S. suis growth (Table S1). Since MMV688508 is shown to inhibit the growth of multiple Gram-positive species (Bhandari et al., 2018; Jeong et al., 2018), MMV688508 may be able to serve as an antibiotic candidate against Gram-positive pathogens. MMV675968, a bicyclic 2,-4 diaminopyrimidine with a long and flexible side chain, was identified as a potent inhibitor of SsDHFR and of S. suis growth. MMV675968 may be able to serve as a broad-spectrum antibiotic candidate since it is shown to inhibit the growth of both Gram-positive and Gram-negative bacteria (Nelson & Rosowsky, 2001; Songsungthong et al., 2019).
Supplemental Information
Percent growth of two S. suis strains after 18 hour incubation with 10 µM compounds compared with growth of untreated control (100%)
Hit compounds are highlighted in green. Reference compounds are highlighted in blue. Data are shown as mean ± SD from three independent experiments. Ss P1/7, S. suis P1/7 strain; Ss HE06, S. suis HE06 strain.
Alignment of dhfr gene from S. suis P1/7 and HE06
(A) Nucleotide sequence of Ssdhfr gene (Genbank accession number MH388486 and MH388487) and (B) amino acid sequence of Ss DHFR from S. suis P1/7 and S. suis HE06, and (C) amino acid sequence of Ss DHFR and S. pneumoniae DHFR (Sp DHFR) were aligned using Clustal Omega. (Sievers & Higgins, 2014). Amino acids labeled in red are predicted to be folate binding site whereas amino acids labeled in blue are predicted to be NADPH binding site by NCBI conserved domain database.