Comparison between cultured and wild Pacific white shrimp (Penaeus vannamei) vitellogenesis: next-generation sequencing and relative expression of genes directly and indirectly related to reproduction
- Published
- Accepted
- Received
- Academic Editor
- Isabella Bordon
- Subject Areas
- Aquaculture, Fisheries and Fish Science, Marine Biology, Molecular Biology, Zoology
- Keywords
- Circadian rhythm, Transcriptome, Cultured shrimp, Relative expression, Vitellogenesis, Wild shrimp
- Copyright
- © 2021 Montes-Dominguez et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Comparison between cultured and wild Pacific white shrimp (Penaeus vannamei) vitellogenesis: next-generation sequencing and relative expression of genes directly and indirectly related to reproduction. PeerJ 9:e10694 https://doi.org/10.7717/peerj.10694
Abstract
Shrimp fisheries are among the most important fisheries worldwide, and shrimp culture has increased considerably in recent years. Most current studies on reproduction-related genes have been conducted on cultured shrimp. However, gene expression is intimately linked to physiological and environmental conditions, and therefore an organism’s growth environment has a great influence on reproduction. Thus, gene expression profiling, should be applied in fisheries studies. Here, we identified the expression patterns of 76 reproduction-related genes in P. vannamei via the analysis of pooled transcriptomes from a time-series experiment encompassing a full circadian cycle. The expression patterns of genes associated both directly (Vtg, ODP, and ProR) and indirectly (FAMet, CruA1, and CruC1) with reproduction were evaluated, as these genes could be used as molecular markers of previtellogenic and vitellogenic maturation stages. The evaluated genes were prominently upregulated during vitellogenic stages, with specific expression patterns depending on the organism’s environment, diet, and season. Vtg, ProR, ODP, and FaMet could serve as molecular markers for both wild and cultured organisms.
Introduction
Industrial shrimp fisheries are among the most important fisheries worldwide (FAO, 2020; Tirado-Ibarra et al., 2020). However, most biological processes have been studied in cultured organisms, due to the growing importance of the shrimp culture industry (CONAPESCA, 2020). As a result, reproduction-associated genes have been well characterized in cultured white shrimp Penaeus vannamei (Uengwetwanit et al., 2018; Wang et al., 2019) but not in wild organisms. Therefore, standardizing molecular approaches for gene expression analysis in wild organisms is crucial.
Reproduction studies in wild shrimp are based on morpho-colorimetric and histological primary analyses of the maturity stages, which characterize changes in gonadal and the oocyte development, respectively. These evaluated responses depend on the molecular mechanisms of vitellogenesis, which modulate the process of yolk synthesis and its accumulation in the oocyte and therefore also govern growth (Chen et al., 2018).
Molecular studies on crustacean reproduction primarily focus on gene characterization, as well as mRNA expression and transcriptome analyses of different vitellogenesis stages; which involves vitelline protein production via endoproteolysis of vitellogenin (Vtg; i.e., the precursor of vitelline protein). Therefore, Vtg is the most studied reproduction-related gene due to its role as the most important nutrient source for embryo development (Thongda et al., 2015; Boulangé-Lecomte et al., 2017; Jimenez-Gutierrez et al., 2019).
However, other ovary expressed genes are known to participate in reproduction regulation in crustaceans either directly or indirectly, including genes associated with gonadal maturation, physiological processes, among others, in addition to some genes in the hepatopancreas that regulate extraovarian Vtg sources and other nutrients (Shen et al., 2014; Jimenez-Gutierrez et al., 2019). Additionally, the X organ/sinus gland complex located in the eyestalk also regulates vitellogenesis and molting through hormone secretion (Bai et al., 2015). Previously reported transcriptomes feature between 25 and 33 reproduction-related genes (Gao et al., 2014; Jimenez-Gutierrez et al., 2019), some of which have been used as molecular markers of ovarian maturation, particularly Vtg, Vtg receptor (VtgR), gonadotropin-releasing hormone receptor (GnRHR), Vigillin, and Torso-like, among others (Shen et al., 2014; Tarrant et al., 2014; Uengwetwanit et al., 2018).
Additionally, many other P. vannamei genes have been previously characterized via next-generation sequencing; however, several of these genes have never been used as molecular markers (Jimenez-Gutierrez et al., 2019). In addition to Vtg, the most important genes directly involve in reproduction regulation include the ovary developing protein (ODP; which modulates oocyte maturation) and progesterone receptor (ProR). Moreover, other protein encoding genes indirectly involved in reproduction by regulating nutrient sources (this parameter constitutes the main difference between aquaculture and wild environments), are farnesoic acid O-methyltransferase (FAMet; which could have a role in reproduction and growth), and crustacyanins A1 and C1 (CruA1 and CruC1, respectively), which bind astaxanthin carotenoids (Gamiz-Hernandez et al., 2015; Meléndez, 2017). Therefore, our study sought to identify novel genes involved in reproduction regulation through next-generation sequencing, as well as their contribution to P. vannamei reproduction control and the expression of new molecular markers in cultured and wild organisms.
Materials and Methods
Experimental organisms
All animals were handled according to ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. Organisms were collected from two sources. Cultured P. vannamei specimens were obtained in November, 2018 from Fitmar farm (El Walamo, Mazatlan, Mexico). The organisms were kept under controlled conditions at 28 °C, a pH of 7.6 and 35 PSU (practical salinity units). Wild organisms, on the other hand, were donated by the Regional Center for Aquaculture and Fisheries Research of Mazatlan (Instituto Nacional de Pesca y Acuacultura; field study approval number PPF/DGOPA-002/18). These organisms were collected from the East Pacific (23°20′N 106°30′W) during open season (September, 2018 to March, 2019; INAPESCA, 2019). Two samplings were performed on November, 2018 (25.76 ± 0.89 °C and 35 ± 0.1 PSU) and March, 2019 (20.32 ± 1.33 °C and 34.9 ± 0.2 PSU). On both occasions, the shrimps were identified to the species level (Perez-Farfante & Kensley, 1997), and immediately frozen while still in the vessel.
Only mature females (56.48 ± 9.6 g and 18.92 ± 1.68 cm total length) from both organism sources were examined. The females were euthanized on ice and immediately measured and dissected thereafter. The ovary stages were primarily identified via morpho-colorimetric methods (Pérez-Ferro & Paramo-Granados, 2014). Hepatopancreas and eyestalk samples dissected from cultured organisms were kept at −80 °C until required for molecular analyses, whereas the ovaries were extracted and preserved separately for other procedures. A fragment of the ovarian tissue was preserved in Davidson’s solution (Bell & Lightner, 1988), for ovarian stage confirmation via histological methods (Bell & Lightner, 1988; Alfaro-Montoya, 2013) with hematoxylin and eosin staining (Humason, 1979). The rest of the ovary tissues were kept at −80 °C until used for molecular analyses. Complete ovaries dissected from wild organisms, were kept at −80 °C until required.
RNA isolation and illumina sequencing
To obtain mRNA transcription profiles throughout an entire circadian cycle under controlled conditions, only cultured organisms were used. For this purpose, eight samplings were conducted every three hours and five cultured females were collected per sampling for a total of 40 organisms. Total RNA from each tissue (ovaries, hepatopancreas and eyestalk) was obtained according to Jimenez-Gutierrez et al. (2019), and a pool from the 40 organisms for each tissue was submitted to Genoma Mayor, Universidad Mayor in Chile (Santiago de Chile) for next-generation sequencing. The library was constructed via the TruSeq Stranded mRNA (Illumina, San Diego, CA) protocol and sequenced in an Illumina MiSeq instrument according to the manufacturer’s instructions. De novo assembly and bioinformatic analyses were also conducted as previously described (Jimenez-Gutierrez et al., 2019).
mRNA expression of P. vannamei reproduction regulation genes
Based on our histologic results, the samples from wild and cultured organisms were classified as previtellogenic and vitellogenic ovaries. Stage III and post-spawning stages were omitted, as they were considered intermediate stages. A 3 × 2 factorial experimental design was used; with three different collection conditions and two vitellogenic stages. Moreover, given that cultured shrimp and Vtg represent the most well characterized organism source and reproduction-associated gene, respectively, these two were used as sample source and molecular marker controls.
Total RNA was extracted from at least four organisms from each evaluated condition in duplicate, and cDNA was synthesized with the RevertAid First Strand cDNA Synthesis Kit (Thermo scientific), according to the manufacturer’s instructions. Vtg, ODP, and ProR were deemed direct reproduction modulators, whereas FAMet, CruA1, and CruC1 were considered to indirectly regulate P. vannamei reproduction. Both L8 and β-actin were used as housekeeping genes for relative gene expression analysis.
cDNAs were amplified using the TopTaq Master Mix Kit (QIAGEN) according to the manufacturer’s instructions using the specific Tm for each gene (Supplemental Information 1). For gen expression analyses, 600 ng of total RNA template were employed for all target genes, whereas, 300 and 400 ng were used for L8 and β-actin, respectively. PCR products for each gene were first purified with the QIAquick PCR Purification Kit (Nucleospin), according to the manufacturer’s instructions and quantified with a spectrophotometer. All PCR products were electrophoretically separated (including the purified gene products), digitalized in the Gel Doc EZ System (Biorad), and analyzed with the ImageLab software.
Absolute expressions were quantified based on previously purified PCR products. The relative expression of each target gene was calculated via the 2−ΔΔCt method using the housekeeping genes and previtellogenic stage as first and second delta, respectively (Schmittgen & Livak, 2008), with modifications. Unlike Ct values, where lower values indicate higher mRNA concentrations and vice versa, absolute expression provides a direct gene expression quantification value. The following modification allows for the use of the absolute expression in the 2−ΔΔCtm formula: where Ctm is the modified Ct; 30 is a constant that represents the number of PCR cycles; AE is the absolute expression of each PCR product.
Statistical analysis
Relative expressions for each condition were analyzed for normality and variance homogeneity. Statistical significance was evaluated via one-way ANOVA (P ≤ 0.05) and differences between treatments were determined via Tukey-Kramer multiple-comparison tests. Finally, correlation coefficients from multivariate Principal Component Analysis (PCA) were used to verify the relationships between the evaluated genes using the InfoStat 2018 statistical software (Di Rienzo et al., 2018).
Results
Histological results from ovary development
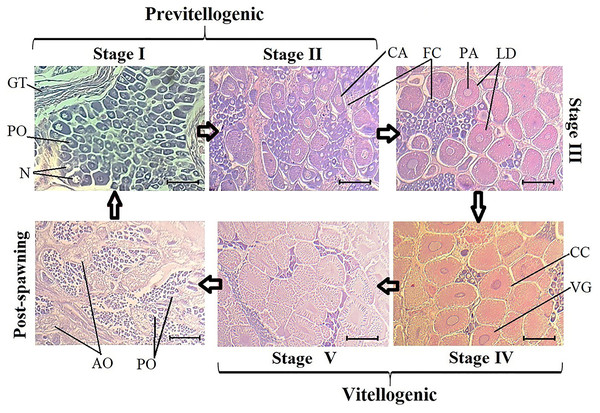
Upon analyzing all samples to characterize ovary development, five development stages and one post-spawning stage were identified. In previtellogenic stages I and II, germinal tissue, follicular cells, perinuclear oocytes, and cortical alveolus were observed. Stage III is a transition between previtellogenic and vitellogenic oocytes, and lipid droplets and perinuclear alveoli were observed in the first vitellogenic oocytes.
Vitellogenic stages IV and V exhibited larger oocytes, with elongated nuclei, vitellogenic grains, and cortical crypts. Finally, in the post-spawning stage, flaccid tissue, atretic oocytes, and a group of stage I oocytes began to appear (Fig. 1). The V-Red semi-stage, where stage V ovaries turn red, was only observed in wild organisms from samples acquired in March (Supplemental Information 2).
Figure 1: Histological sections of oocyte differentiation in ovaries from Penaeus vannamei.
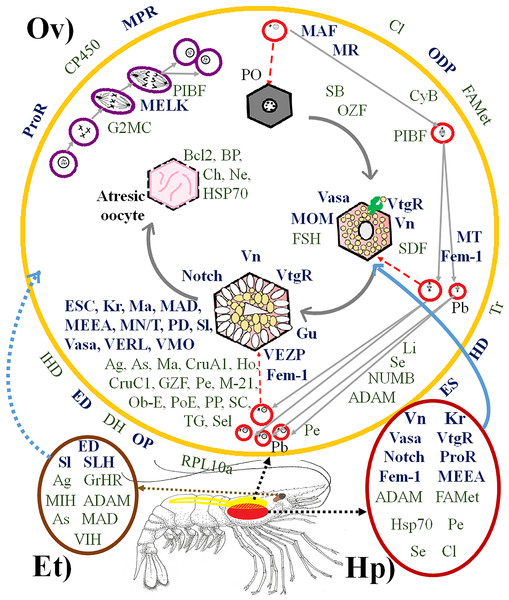
GT, germinal tissue. PO, perinuclear oocytes. N, Nucleus. CA, cortical alveolus. FC, Follicular cells. PA, perinuclear alveolus. LD, lipid drops. CC, cortical crypts. VG, vitellic grains. AO, atresic oocytes. Scaled bar represent 20 µm.Figure 2: Proteins directly and indirectly related to Penaeus vannamei reproduction regulation.
(Ov) Ovary, represented in yellow. (Hp) Hepatopancreas, represented in dark red. (Et) Eyestalk, represented in brown. Violet cells represent mitotic process. Red cells represent meiotic process. Hexagonal cells represent oocyte development. Proteins in bold dark blue are directly reproduction-related. Proteins in green are indirectly reproduction-related. Proteins outside the ovary are involved in several places of the ovary at the same time. PO, Primary oocyte. Pb, Polar bodies. Abreviated Genes: Ag, Agrin. As, Asterix. BP, Blastula protease. Ch, Chitinase. Cl, Clathrin. CruA1, Crustacyanin A1. CruC1, Crustacyanin C1. CP450, Cytochrome P450. CyB, Cyclin B. DH, Diuretic Hormone. ED, Estradiol 7 beta Dehydrogenase. ES, Estrogen Sulfotransferase. ESC, Extra sex comb. FAMet, Farnesoic Acid O-Methyltransferase. Fem-1, Sex-determining protein Fem-1. FSH, Female Sterile Homeotic. G2MC, G2 mitotic specific cyclin. GrHR, Gonadotropin-releasing hormone receptor. Gu, Gustavus. GZF, Gastrula zinc finger. HD, Hydroxysteroid Dehydrogenase. Ho, Homeobox. Hsp70, Heat Shock Protein 70. IHD, Inactive Hydroxyesteroid Dehydrogenase. JH, Juvenile hormone epoxide hydrolase. Kr, Krueppel. Li, Lin-9. MN/T, Complex Mago Nashi/TsunagiY14. M-21, Mab-21. Ma, Masquerade. MAD, Mothers Against Decapentaplegic. MAF, Meiosis arrest female. MEEA, Maternal effect embryo arrest. MELK, Maternal Embryonic Leucine zipper Kinase. MIH, Molt-inhibiting hormone. MOM, Missing oocyte meiosis. MPR, Membrane progestin receptor. MR, Meiosis regulator. MT, Maternal protein Tudor. Ne, Neuroparsin. Notch, Notch proteins (Notch, Notchless protein and Strawberry Notch). Ob-E, Obstructor-E. ODP, Ovary Development Protein. OP, Ovaric Peritrophin. OZF, Oocyte zinc finger. PD, Partitioning Defective. Pe, Pelota protein. PIBF, Progesterone-Induced Blocking Factor. PoE, Purity of essence. PP, Peter Pan. ProR, Progesterone Receptor. RPL10a, Ribosomal Protein 10a. SB, Singles Bar. SC, Shuttle craft. SDF, Se: Sel-1. Sel, protein Seele. Sex Determination Fruitless. Sl, Slowmo. SLH, Sex-lethal homolog. TG, Twisted Gastrulation. Tr, Trithorax. Vasa, Vasa protein. VERL, Vitelline envelope receptor of lysin. VIH, Vitellogenesis-inhibitinghormone. VMO, Vitelline Membrane Outer layer. Vn, Vitellin. VtgR, Vitellogenin Receptor. (Artist: Rafael Serrano-Quiñonez).P. vannamei ovary, hepatopancreas, and eyestalk transcriptomes
The transcriptomes from the ovary, hepatopancreas and eyestalk circadian rhythm pools exhibited 76 genes related to P. vannamei reproduction regulation, 32 and 44 of which were directly and indirectly related to reproduction, respectively. As expected, most of them were only found in the ovary transcriptome. Additionally, 14 of them were expressed in both the ovary and hepatopancreas, of which six were expressed in both the ovary and eyestalk, and five were expressed exclusively in the eyestalk (Fig. 2). Among all identified genes, some had never been reported in the GenBank database. The codified proteins were found to be related to one or more steps of the mitotic or meiotic processes, and some of them were specifically involved in oocyte development, such as Vitelline (for which the precursor is Vtg), VtgR, Vasa, Missing oocyte meiosis, TsunagiY14, Fem-1, Crustacyanin A1 and C1, among others.
Some other encoded proteins were simultaneously involved in several parts of the ovary, such as ODP, FAMet, ProR, Membrane progestin receptor, diuretic hormone, ovarian peritrophin, trithorax, among others. The function of each gene is summarized in Supplemental Information 3.
Relative expression of Vtg, ODP, ProR, FAMet, CruA1, and CruC1
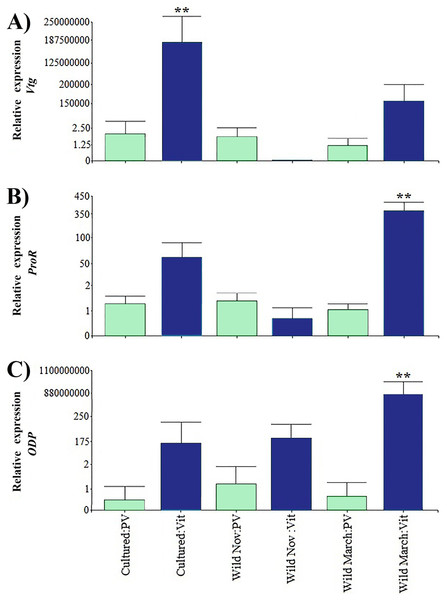
Relative expression of directly related reproduction regulation genes was greater than that of indirectly related genes; Vtg displayed the highest expression levels among all the evaluated genes (Fig. 3). As expected, Vtg expression was greater in vitellogenic stages than in previtellogenic stages; this was more evident in cultured organisms with highly significant differences (i.e., three orders of magnitude greater than the vitellogenic stages of wild-caught shrimp captured in March and eight orders of magnitude greater than the rest of the treatments).
Figure 3: Relative expression of directly shrimp reproduction-related genes.
Bars indicates average ± standard error. n = 4. Light green bars indicate previtellogenic states. Dark blue bars indicate vitellogenic states. Asterisks (**) indicate highly statistically significant differences at P ≤ 0.01. (A) Vtg. (B) ProR. (C) ODP.A different pattern was observed in ODP and ProR, where the highest expression was seen in wild shrimp caught in March during vitellogenic stages, exhibiting two and six orders of magnitude upregulation relative to the rest of the treatments, respectively. However, gene expression was generally lower in previtellogenic stages.
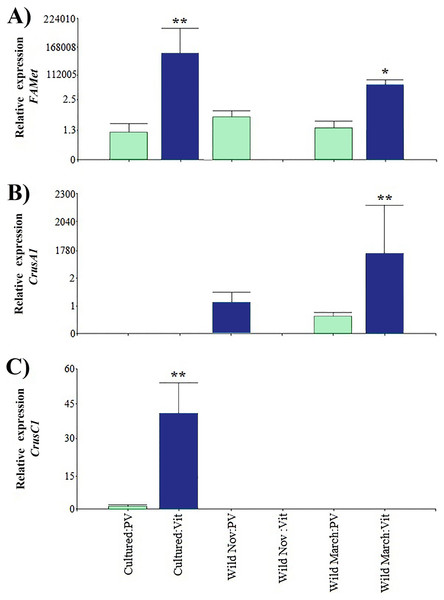
Regarding the genes that were indirectly associated with reproduction, the relative expression of CrusC1 was the lowest. FAMet showed the same pattern as Vtg expression in cultured vitellogenic stages; however, it did not present significant differences in the vitellogenic stages of wild shrimp caught in March, and both treatments had expression values five orders of magnitude higher than those of previtellogenic stages (Fig. 4).
Figure 4: Relative expression of indirectly shrimp reproduction-related genes.
Bars indicates average ± standard error. n = 4. Light green bars indicate previtellogenic states. Dark blue bars indicate vitellogenic states. Two asterisks (**) indicate highly statistically significant differences at P ≤ 0.01. One asterisk (*) indicates statistically significant differences at P ≤ 0.05. (A) FAMet. (B) CrusA1. (C) CrusC1.Furthermore, both crustacyanin genes exhibited a particular expression pattern. Specifically, CrusA1 was only found in wild organisms, whereas CrusC1 was only found in cultured organisms. As in the previously discussed genes, the previtellogenic stages exhibited highly significant minimum expression values, with three and one orders of magnitude of difference, respectively.
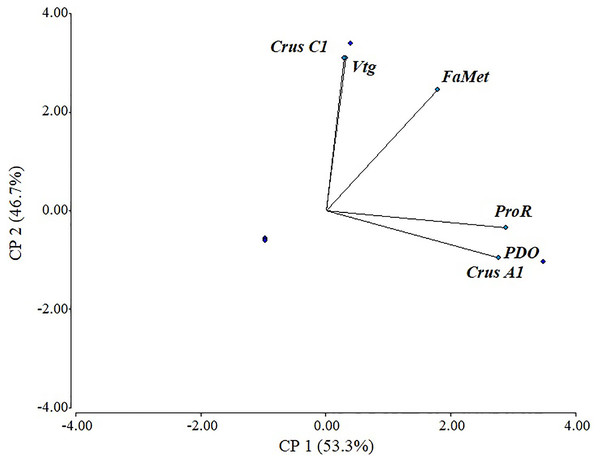
There was a positive correlation in most evaluated genes, with a cophenetic correlation of 1 (Fig. 5). Correlations between variables were particularly strong between Vtg and CrusC1 (1.0), followed by ProR, CrusA1, and ODP (0.98), and finally FAMet and Vtg (0.85). Although some variables exhibited a negative correlation (e.g., CrusC1 and CrusA1), none were statistically significant.
Figure 5: Scaled principal component of relative expression of directly and indirectly shrimp reproduction-related genes.
Discussion
Depending on the authors, ovarian development in shrimp is often classified into three to five stages (Perdichizzi et al., 2012; Pérez-Ferro & Paramo-Granados, 2014; Nguyen et al., 2018) plus one post-spawning stage. However, wild shrimp presented a V-Red semi-stage in organisms sampled in March, which was likely due to the rich carotenoid diet of shrimp in spring (Yanar, çelik & Yanar, 2004). Red ovaries had not been reported in cultured organisms. Here, reproduction-related differences between cultured and wild organisms were observed at the anatomical and molecular level.
Transcriptome analysis throughout an entire circadian cycle in cultured organisms allowed for the discovery of a greater number of reproduction-related genes than in previous reports (Gao et al., 2014; Jimenez-Gutierrez et al., 2019). However, infradian rhythms and different collection areas must also be considered to obtain a better estimation of the number of these genes, without considering the large number of genes whose functions remain unknown.
Moreover, a close anatomical and molecular relationship were observed between the ovaries and hepatopancreas, as evidenced by the great number of genes involved in P. vannamei reproduction expressed in the hepatopancreas, as well as the presence of ProR and VtgR in both tissues, thereby demonstrating communication between membranes. The genes examined in this work were divided into directly related reproduction genes, whose encoded proteins have a direct function in oocyte maturation or meiosis, and indirectly related genes, whose encoded proteins bind to directly related proteins, thereby inhibiting or activating them.
Most differential expression reports are based on directly related genes, which are used as molecular markers (Bae et al., 2017; Boulangé-Lecomte et al., 2017; Chen et al., 2017). The differences in the relative expression of the genes in both groups were of several orders of magnitude, with Vtg being the most highly expressed, as expected.
Vtg expression is known to be species- and tissue-specific, in addition to the effects of external and physiological conditions (Jimenez-Gutierrez et al., 2019). This was confirmed by the differential expression between wild and cultured organisms. Furthermore, the remaining evaluated genes in this work had never been examined in the conditions evaluated herein.
Only a few sequences for ODP are available in the GenBank database, of which only one corresponds to crustaceans (Shanghai crab Eriocheir sinensis; Zhu et al., 2016). Despite its direct relationship with ovary maturation, its expression patterns had never been documented in crustacean species. In this study, ODP was confirmed to be linked to both ovary development and seasonal maturity.
Additionally, the seasonal variation of ProR in wild organisms was more noticeable than that of ODP. ProR is not only well known as a membrane surface receptor that is mainly present in follicular cells (Ye et al., 2010; Wu et al., 2014), but also as a transcriptional activator or repressor, involving cofactors or coactivators that regulate many aspects of the female reproductive system (Klotzbücher et al., 1997; Chen et al., 2008). Its role as a reproduction regulator in the estrogen pathway in vertebrates and invertebrates is well documented in the ovary, as well as its synthesis in the hepatopancreas (Ye et al., 2010; Wu et al., 2014; Thongbuakaew et al., 2016).
This receptor belongs to the steroidogenic-related proteins including estradiol 17 beta dehydrogenase, estrogen sulfotransferase, and hydroxysteroid dehydrogenase, all of which directly modulate estrogen enzyme biosynthesis, and cytochrome P450, which modulates these mechanisms indirectly (James & Boyle, 1998; Thongbuakaew et al., 2016; Subramoniam, 2017). All genes listed above were also found in this study. The relationship between Vtg during the ovarian cycle with progesterone levels and the aforementioned enzymes have been previously reported (Subramoniam, 2017).
Progesterone translocates from follicular cells through the oocyte plasma membrane and binds to ProR to form a progesterone-ProR complex to reach the nuclei and regulate transcriptional activity. Many studies have characterized progesterone activity and several of them have confirmed a considerable increase in progesterone levels during vitellogenesis (Ye et al., 2010; Wu et al., 2014; Thongbuakaew et al., 2016). ProR has been studied via immunohistological approaches (Wu et al., 2014); however, the analysis of its expression had not been implemented for the evaluation of progesterone’s role in ovary maturation or as a molecular marker in crustaceans until now.
Only three indirectly related reproduction genes were evaluated herein due to their key role in nutrition to account for the differences in dietary quality and availability between cultured and wild shrimp. FAMet has been reported to participate in P. vannamei reproduction (Hui, Tobe & Chan, 2008). Furthermore, its substrate (i.e., methyl farnesoate) is also related to neurohormones and ecdysteroids (Ye et al., 2010). FAMet was evaluated in several tissues, molt conditions, between sexes, and between larvae and juvenile organisms (Hui, Tobe & Chan, 2008); however, its expression in ovaries and its role in gonad maturation had not been characterized. In this work, FAMet exhibited the same expression pattern as Vtg, and both shared a significant correlation according to our PCA results.
Finally, crustacyanin had not previously been studied as a reproduction-related protein. However, the presence of the V-Red semi-stage warranted the assessment of the aforementioned gene in this work. Crustacyanins are specific crustacean proteins that bind to astaxanthin (Cianci et al., 2002; Budd et al., 2017) and belong to the lipocalin family, which also includes retinol-binding protein (RBP) and apolipoproteins (Ma et al., 2020). There are five precursor subtypes of crustacyanins reported in the GenBank databases: A1, A2, A3, C1, and C2. Nevertheless, only CruA1 and CruC1 were found in P. vannamei transcriptomes.
These results indicate an evident relationship between the red ovaries from wild organisms caught in March and CruA1, suggesting a more intimate link with carotenoids, unlike CruC1, which is only found in cultured organisms. The above-mentioned relationships were confirmed by our PCA results. Adult cultured organisms are fed with the same formulated diet throughout most of the year, which is specifically formulated to promote maturation; however, differences between natural diets limit the availability of bioactive metabolites for natural growth and development (Liñán Cabello, Paniagua-Michel & Zenteno-Savín, 2003). Wild shrimp are mostly scavengers that feed on smaller crustaceans, fish, mollusks, plants, and organic detritus (INAPESCA, 2019). Food availability and quality are affected by different population dynamics, regions, and seasons (Yanar, çelik & Yanar, 2004; FAO, 2016).
In this sense, differences in the carotenoid content between cultured and wild shrimp were previously studied, and their possible relationship with gonadal maturation was reported, with extensive focus on wild organisms. This suggests that these carotenoids are able to bind to Vtg to form a lipo-carotene-glyco-protein (Liñán Cabello, Paniagua-Michel & Zenteno-Savín, 2003), which translates into seasonal differences in gonad maturation (Thongda et al., 2015; Jimenez-Gutierrez et al., 2019).
There are several genes involved in shrimp reproduction regulation that possess great potential as molecular markers but are yet to be evaluated and validated. Moreover, despite the wealth of studies that have reported on the physiology and gene expression patterns of cultured shrimp, additional studies should be conducted in wild shrimp to account for the environmental variability that wild organisms face.
Conclusions
This study identified more than 76 genes related to reproduction regulation in P. vannamei. Vtg exhibited the highest expression among all evaluated genes. The genes directly- and indirectly-associated with shrimp reproduction studied herein act as molecular markers between previtellogenic and vitellogenic maturation stages. Variations in anatomical and specific gene expression patterns were observed between wild and cultured shrimps. Vtg, ProR, ODP, and FaMet could serve as molecular markers for both wild and cultured organisms, whereas CrusA1 and CrusC1 were exclusively suitable to characterize wild and cultured organisms, respectively.
Supplemental Information
Colorimetric maturity scale from Penaeus vannamei ovaries
(A) colorimetric scale. (B) complete gonad from stage V-Red.
Function of reproduction-related proteins
Directly related genes.
Indirectly related genes.
Accession numbers of reported genes.