PeerJ Section
Bioinformatics and Genomics
Welcome to your community’s home at PeerJ. Sections are community led and exemplify a research community’s shared values, norms and interests.
The citation average is 6.2 (view impact metrics).
65,493 Followers
Section Highlights
View all Bioinformatics and Genomics articles 
3 September 2025
BMP2 alterations in mucinous cystadenocarcinoma of the breast: insights from whole-exome sequencing
"The article presents a rare and understudied breast cancer subtype, and the integrative approach using whole exome sequencing (WES) and immunohistochemistry (IHC) provides valuable initial insights into its possible molecular pathogenesis. However, limited sample size (n=3) and lack of downstream functional analyses are limiting factors."
 Mehmet Burak Ateş, Handling Editor
Mehmet Burak Ateş, Handling Editor
 Mehmet Burak Ateş, Handling Editor
Mehmet Burak Ateş, Handling Editor
28 August 2025
Uncovering tissue-specific endophytic microbiota composition and activity in Rhizophora mangle L.: a metagenomic and metatranscriptomic approach
"Understanding the structure and functional activity of mangrove tree-associated microbiomes paves the way for mangrove ecosystem conservation and future biotechnological applications"
 Paripok Phitsuwan, Handling Editor
Paripok Phitsuwan, Handling Editor
 Paripok Phitsuwan, Handling Editor
Paripok Phitsuwan, Handling Editor
18 August 2025
Genomewide analysis of the Class III peroxidase gene family in apple (Malus domestica)
"The manuscript presents a robust genome-wide analysis of Class III peroxidases in Malus domestica, uncovering their structural diversity, evolution, and expression patterns. It highlights antagonistic roles of PRX subgroups in rootstock vigor and bud differentiation, offering valuable insights into ROS-mediated regulation and apple breeding strategies."
 Ganesh Nikalje, Handling Editor
Ganesh Nikalje, Handling Editor
 Ganesh Nikalje, Handling Editor
Ganesh Nikalje, Handling Editor
11 July 2025
An R package for survival-based gene set enrichment analysis
"This article presents SGSEA, a novel adaptation of traditional GSEA that incorporates survival data into pathway enrichment analysis. The development of both an R package and Shiny App makes this method accessible to researchers across disciplines. While standard GSEA has been effective for comparing differential expression between experimental conditions, SGSEA addresses the need to link transcriptomic variations with clinical outcomes like patient survival. The authors demonstrate the method's utility through a case study of kidney renal clear cell carcinoma, where pathways identified by SGSEA align with previously documented survival associations. This alignment with existing knowledge helps validate the approach. The method enables researchers to identify pathways relevant to disease progression and mortality risk in a systematic way, complementing insights from traditional GSEA. The provision of standardized tools addresses a gap in the current analytical landscape. While the approach shows promise, further validation across different cancer types and diseases would help establish its broader applicability. The development of SGSEA represents a practical contribution to the field of functional genomics and survival analysis, potentially aiding in the identification of clinically relevant pathways."
 Fanglin Guan, Section Editor
Fanglin Guan, Section Editor
 Fanglin Guan, Section Editor
Fanglin Guan, Section Editor
10 July 2025
Expression of the IL-18-related gene PTX3 correlates with clinicopathological features and prognosis in glioma patients
"This study provides a comprehensive analysis of PTX3's role in glioma progression through multi-dimensional research approaches. The investigation combines bioinformatics analysis of TCGA data with experimental validation, offering insights into both molecular mechanisms and clinical implications. The study's methodology is systematic, employing various analytical tools including LASSO Cox regression, functional enrichment analysis, and immune infiltration assessment. The identification of a nine-gene prognostic signature and the correlation between PTX3 expression and immune cell infiltration advances our understanding of glioma's molecular landscape. The research presents evidence linking PTX3 expression to clinical features and survival outcomes, supported by immunohistochemistry validation in patient specimens. While the findings are consistent with existing literature on tumor-associated molecular markers, this work adds specific insights about PTX3's potential as a prognostic indicator in glioma. The study's results may contribute to the development of targeted therapies, though additional validation studies would be beneficial. This research provides a foundation for future investigations into PTX3-based therapeutic strategies, while maintaining appropriate consideration of the complexities involved in translating molecular findings to clinical applications."
 Fanglin Guan, Section Editor
Fanglin Guan, Section Editor
 Fanglin Guan, Section Editor
Fanglin Guan, Section Editor
29 April 2025
A prognostic glycolysis-related gene signature in osteosarcoma: implications for metabolic programming, immune microenvironment, and drug response
"This is a comprehensive manuscript that has implications for understanding the biological mechanism, pathophysiology and response to treatment of osteosarcoma with the potential for the development of therapeutics. It is relevant to basic science researchers and clinicians."
 Katherine Mitsouras, Handling Editor
Katherine Mitsouras, Handling Editor
 Katherine Mitsouras, Handling Editor
Katherine Mitsouras, Handling Editor
19 March 2025
A comprehensive prognostic and immunological analysis of hexokinase domain containing protein-1 (HKDC1) in pan-cancer
"This paper demonstrates that the expression level of hexokinase domain-containing protein 1 (HKDC1) is involved in cancer prognosis, providing important insights into HKDC1-targeted cancer therapy."
 Tokuko Haraguchi, Handling Editor
Tokuko Haraguchi, Handling Editor
 Tokuko Haraguchi, Handling Editor
Tokuko Haraguchi, Handling Editor
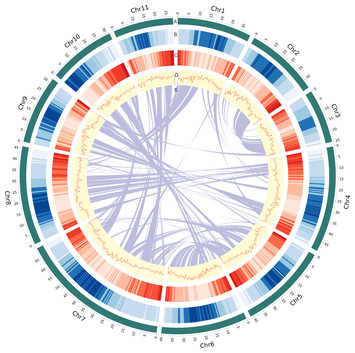
23 December 2024
A chromosome-scale genome assembly of mungbean (Vigna radiata)
"Together with the earlier two genome assemblies, this study would provide a triple reference, or would lead to deep genome insights."
 Tong Geon Lee, Handling Editor
Tong Geon Lee, Handling Editor
 Tong Geon Lee, Handling Editor
Tong Geon Lee, Handling Editor
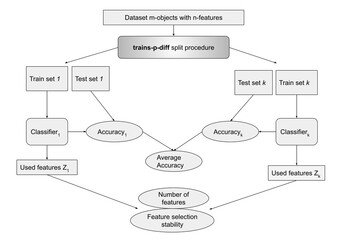
12 November 2024
Importance of feature selection stability in the classifier evaluation on high-dimensional genetic data
"This article makes an important contribution to its field by tackling a clearly identified research gap and presenting fresh insights with practical significance on feature selection stability on high-dimensional data in biology, delivering strong data and a well-established methodological approach that can be reproduced and expanded upon in future studies."
 Carlos Fernandez-Lozano, Handling Editor
Carlos Fernandez-Lozano, Handling Editor
 Carlos Fernandez-Lozano, Handling Editor
Carlos Fernandez-Lozano, Handling Editor

30 October 2024
Identification of common and specific cold resistance pathways from cold tolerant and non-cold tolerant mango varieties
"The manuscript is important as it cover the important aspect of cold stress in Mango"
 Kashmir Singh, Handling Editor
Kashmir Singh, Handling Editor
 Kashmir Singh, Handling Editor
Kashmir Singh, Handling Editor
Collections
View all 
PeerJ Expert Curations: Molecular Ecology
RiTA 2023: Robotics and AI for Digital Futures

International Conference on Machine Learning and Intelligent Systems (MLIS)

Revolutionizing Healthcare: The Role of AI and Machine Learning

Artificial Intelligence for Mental Health: Advancements, Challenges, and Ethical Implications

65,493 Followers


