PeerJ Section
Bioinformatics and Genomics
Welcome to your community’s home at PeerJ. Sections are community led and exemplify a research community’s shared values, norms and interests.
The citation average is 6 (view impact metrics).
67,096 Followers
Section Highlights
View all Bioinformatics and Genomics articles

5 December 2023
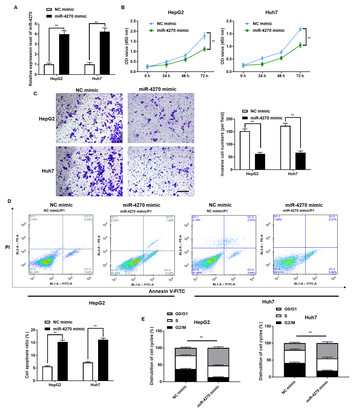
miR-4270 suppresses hepatocellular carcinoma progression by inhibiting DNMT3A-mediated methylation of HGFAC promoter
"The present study significantly contributes to the existing body of research pertaining to hepatocellular carcinoma (HCC). This study elucidates the functional significance of a particular gene, miR-4270, in its capacity as a tumor suppressor. The aforementioned discoveries not only contribute to the advancement of our understanding about the molecular mechanisms behind HCC, but also propose that miR-4270 has promise as a possible therapeutic target for HCC treatment. The aforementioned progress has significant significance within the discipline, perhaps establishing a foundation for more efficacious methodologies in the management of this particular kind of cancer."
 Peixin Dong, Handling Editor
Peixin Dong, Handling Editor
 Peixin Dong, Handling Editor
Peixin Dong, Handling Editor
4 December 2023
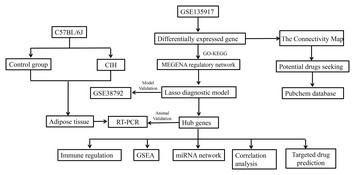
Identification of novel biomarkers in obstructive sleep apnea via integrated bioinformatics analysis and experimental validation
 Antonella Loperfido, Handling Editor
Antonella Loperfido, Handling Editor
28 November 2023
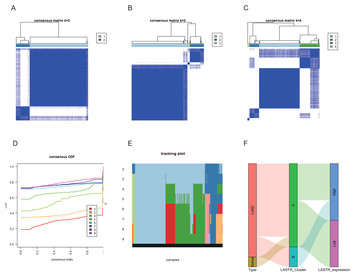
A novel long noncoding RNA (lncRNA), LINC02657(LASTR), is a prognostic biomarker associated with immune infiltrates of lung adenocarcinoma based on unsupervised cluster analysis
 Zhijie Xu, Handling Editor
Zhijie Xu, Handling Editor
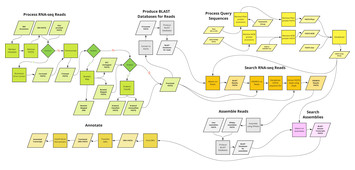
27 November 2023
kakapo: easy extraction and annotation of genes from raw RNA-seq reads
"Annotation of sequences from RNA-Seq is sorely needed for the many datasets being submitted to NCBI; this will be a great addition to bioinformatics tools"
 Brenda Oppert, Section Editor
Brenda Oppert, Section Editor
 Brenda Oppert, Section Editor
Brenda Oppert, Section Editor
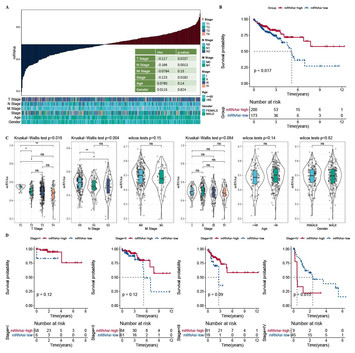
24 November 2023
Development of a 5-mRNAsi-related gene signature to predict the prognosis of colon adenocarcinoma
"This study advances the understanding of colon adenocarcinoma (COAD) by developing a prognosis model based on mRNA stemness index (mRNAsi). The researchers identified specific genes correlated with survival rates, clinical stages, and responses to treatments like immunotherapy. By highlighting the connections between these markers and various aspects of the disease, the findings could pave the way for more targeted and effective therapies for COAD, enhancing treatment outcomes."
 Peixin Dong, Handling Editor
Peixin Dong, Handling Editor
 Peixin Dong, Handling Editor
Peixin Dong, Handling Editor
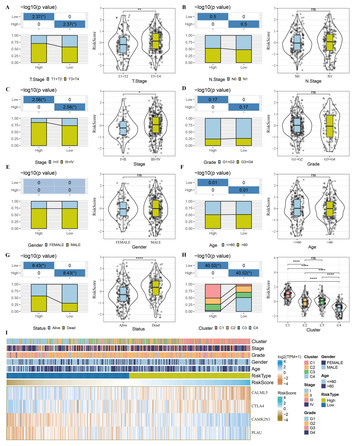
21 November 2023
Head and neck squamous cell carcinoma-specific prognostic signature and drug sensitive subtypes based on programmed cell death-related genes
"This article innovatively reports a prognostic model involved in the development and treatment of head and neck squamous cell carcinoma. It is useful for clinical management."
 Zhijie Xu, Handling Editor
Zhijie Xu, Handling Editor
 Zhijie Xu, Handling Editor
Zhijie Xu, Handling Editor
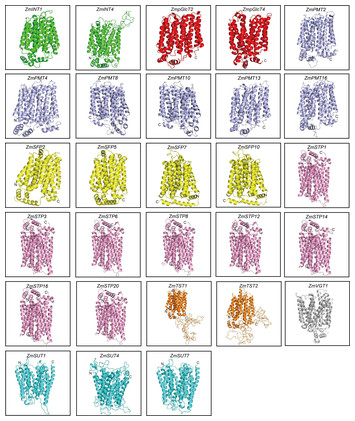
17 November 2023
Genome-wide analysis of sugar transporter genes in maize (Zea mays L.): identification, characterization and their expression profiles during kernel development
"The authors in this study performed a comprehensive search in the maize genome and identified 68 ZmST genes. The authors applied phylogenetic relationship, chromosome location, collinearity analysis, conservative structures, and expression patterns analyses. The study detected that ZmSTs have a substantial role in sugar transportation and seed development. These findings provide fairly robust information for further research on ZmSTs and enhance maize breeding."
 Elsayed Mansour, Handling Editor
Elsayed Mansour, Handling Editor
 Elsayed Mansour, Handling Editor
Elsayed Mansour, Handling Editor
15 November 2023
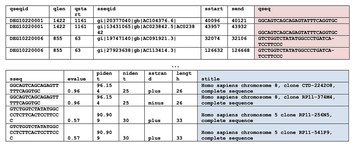
In-silico study of antisense oligonucleotide antibiotics
"This study underscores a significant advancement in the ongoing battle against antibiotic-resistant bacteria, a paramount concern in global health due to the dwindling development of new drugs. By pivoting from the traditional focus on small-molecule antibiotics to RNA-based therapeutics, particularly antisense oligonucleotides (ASOs), researchers have paved the way for a novel paradigm in antibiotic design. This research presents a bioinformatics pipeline tailored to identify potent ASO targets within essential bacterial genes, offering an efficient toolset to expedite ASO antibiotic research. By testing the approach on three pertinent bacterial species, the study not only confirms the efficacy of the pipeline but also provides a public database of ASO targets. This is invaluable for future research and holds the potential to significantly impact the development of next-generation antibiotics."
 Peixin Dong, Handling Editor
Peixin Dong, Handling Editor
 Peixin Dong, Handling Editor
Peixin Dong, Handling Editor
15 November 2023
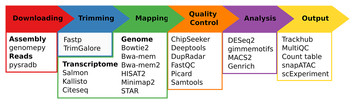
Seq2science: an end-to-end workflow for functional genomics analysis
"van der Sande and colleagues propose an easier and more reproducible way of conducting computational genomics analyses (like data download capabilities of the download-fastq sub-workflow), which can be expanded by adding workflow components for other genome-wide assays."
 Joseph Gillespie, Handling Editor
Joseph Gillespie, Handling Editor
 Joseph Gillespie, Handling Editor
Joseph Gillespie, Handling Editor

18 October 2023
Predicting protein and pathway associations for understudied dark kinases using pattern-constrained knowledge graph embedding
"The algorithm provides information on dark kinases that may be involved in disease processes, providing data that may be used to promote new treatments."
 Brenda Oppert, Section Editor
Brenda Oppert, Section Editor
 Brenda Oppert, Section Editor
Brenda Oppert, Section Editor
Collections
View all 
Open Data and Analysis: from morphology to evolutionary patterns Workshop at the Annual Meeting of the Paläontologische Gesellschaft 2019
Open Source Geospatial Research & Education Symposium (OGRS2018)
Thirteenth annual "Bioinformatics and Computational Biology in Campania" Collection
4th World Conference on Marine Biodiversity
German Conference on Bioinformatics 2017 Collection
67,096 Followers