PeerJ Section
Bioinformatics and Genomics
Welcome to your community’s home at PeerJ. Sections are community led and exemplify a research community’s shared values, norms and interests.
The citation average is 6.4 (view impact metrics).
65,102 Followers
Section Highlights
View all Bioinformatics and Genomics articles 
12 November 2024
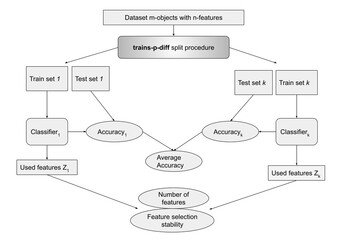
Importance of feature selection stability in the classifier evaluation on high-dimensional genetic data
"This article makes an important contribution to its field by tackling a clearly identified research gap and presenting fresh insights with practical significance on feature selection stability on high-dimensional data in biology, delivering strong data and a well-established methodological approach that can be reproduced and expanded upon in future studies."
 Carlos Fernandez-Lozano, Handling Editor
Carlos Fernandez-Lozano, Handling Editor
 Carlos Fernandez-Lozano, Handling Editor
Carlos Fernandez-Lozano, Handling Editor

30 October 2024
Identification of common and specific cold resistance pathways from cold tolerant and non-cold tolerant mango varieties
"The manuscript is important as it cover the important aspect of cold stress in Mango"
 Kashmir Singh, Handling Editor
Kashmir Singh, Handling Editor
 Kashmir Singh, Handling Editor
Kashmir Singh, Handling Editor
16 October 2024
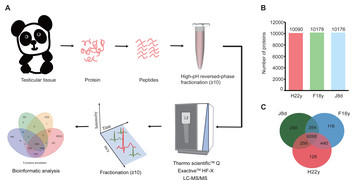
Proteomic analysis of giant panda testicular tissue of different age groups
"Due to the giant panda's status as an endangered species and conservation symbol, and the difficulty in accessing samples for research, I believe this paper is an important piece of work."
 Matthew Barnett, Handling Editor
Matthew Barnett, Handling Editor
 Matthew Barnett, Handling Editor
Matthew Barnett, Handling Editor
14 October 2024
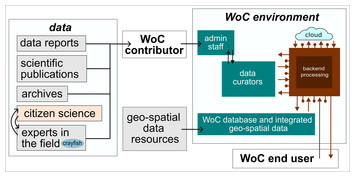
World of Crayfish™: a web platform towards real-time global mapping of freshwater crayfish and their pathogens
"An interactive biodiversity tool for crayfish - we need more of these kinds of tools for endangered species."
 Brenda Oppert, Section Editor
Brenda Oppert, Section Editor
 Brenda Oppert, Section Editor
Brenda Oppert, Section Editor

27 September 2024
SpLitteR: diploid genome assembly using TELL-Seq linked-reads and assembly graphs
"This assembly algorithm to address gaps in current reference assemblies is needed to promote more complete genome assemblies in functional and evolutionary genomics research."
 Brenda Oppert, Handling Editor
Brenda Oppert, Handling Editor
 Brenda Oppert, Handling Editor
Brenda Oppert, Handling Editor
19 September 2024
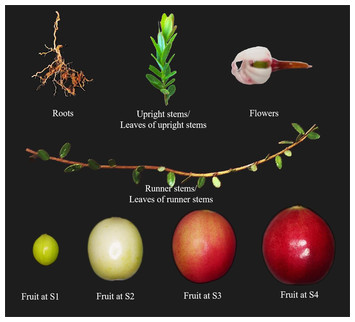
Genome-wide identification and expression analyses of SWEET gene family reveal potential roles in plant development, fruit ripening and abiotic stress responses in cranberry (Vaccinium macrocarpon Ait)
"This manuscript outlines the identification of members of the SWEET family of sugar transporters in cranberry.
Post identification, the authors have used expression analyses to assign putative function for individual family members in cranberry development and in the response of cranberry to various abiotic stresses."
 Andrew Eamens, Handling Editor
Andrew Eamens, Handling Editor
 Andrew Eamens, Handling Editor
Andrew Eamens, Handling Editor
16 September 2024
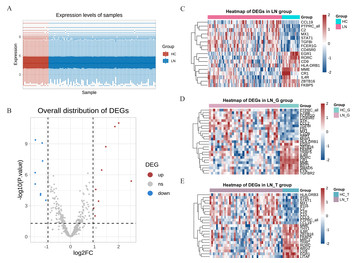
MME and PTPRC: key renal biomarkers in lupus nephritis
"This study identified seven key renal biomarkers through bioinformatics analysis using the GEO and NephroSeq databases. It was identified that MME and PTPRC may have a high predictive value as renal biomarkers in the pathogenesis of LN, as confirmed by animal validation."
 Rohit Upadhyay, Handling Editor
Rohit Upadhyay, Handling Editor
 Rohit Upadhyay, Handling Editor
Rohit Upadhyay, Handling Editor
13 September 2024
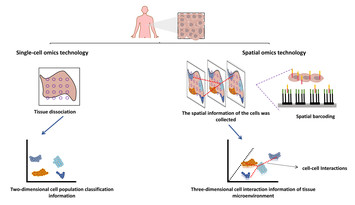
The burgeoning spatial multi-omics in human gastrointestinal cancers
"Spatial multi-omics technology holds promising applications in gastrointestinal tumors by unraveling the spatial and cellular heterogeneity, enhancing biomarker discovery, and facilitating precision medicine. Its integration of multi-omics data with spatial information enables detailed insights into tumor progression and immune microenvironment, leading to targeted therapies and improved patient outcomes."
 Ning Li, Handling Editor
Ning Li, Handling Editor
 Ning Li, Handling Editor
Ning Li, Handling Editor
13 September 2024
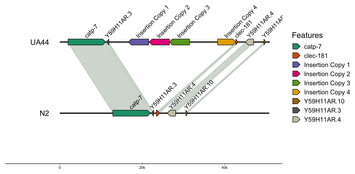
Identifying transgene insertions in Caenorhabditis elegans genomes with Oxford Nanopore sequencing
"The ability to use Oxford Nanopore kits to improve genomes is important in the field. As such the paper will appeal beyond the community interested in C. elegans to the wider community doing new or revised genome sequencing and using experimental transgene manipulations."
 Clement Kent, Handling Editor
Clement Kent, Handling Editor
 Clement Kent, Handling Editor
Clement Kent, Handling Editor
5 August 2024
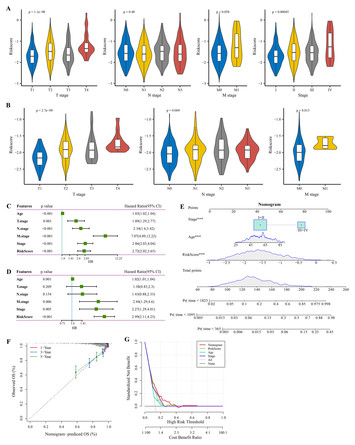
A glycolysis-related signature to improve the current treatment and prognostic evaluation for breast cancer
"This manuscript presents a study on breast cancer, focusing on the development of a glycolysis-related gene signature for prognostic evaluation. The research utilizes glycolysis-related genes to create a prognostic model, offering an alternative approach to breast cancer risk stratification. The authors employ various analytical methods, including GSVA, LASSO regression, and immune infiltration analysis, to develop their model. The proposed Riskscore and nomogram are designed to predict breast cancer patient outcomes, though their clinical utility remains to be fully established. The study examines potential relationships between glycolysis, tumor immune microenvironment, and breast cancer progression. The findings are supported by both computational analysis and some laboratory experiments. This work introduces a potential tool for assessing breast cancer prognosis and explores the role of metabolic factors in cancer progression. While the research contributes to the current understanding of glycolysis in cancer biology, further validation and investigation may be needed to determine its full impact on breast cancer patient care and prognostic evaluation. The study provides a foundation for future research in this area, but additional work will be required to confirm its clinical significance."
 Fanglin Guan, Handling Editor
Fanglin Guan, Handling Editor
 Fanglin Guan, Handling Editor
Fanglin Guan, Handling Editor
Collections
View all 
Pulsed Resources Impacts on Biological Processes: From Genes to Ecosystems

Advancing the Environmental DNA and RNA Toolkit for Aquatic Ecosystem Monitoring and Management
Analysis and Mining of Social Media Data

Equity in Diabetes: World Diabetes Day
Emergence Phenomena in Biology
65,102 Followers


