Improving robustness in X-ray image classification through attention mechanisms in convolutional neural networks
- Published
- Accepted
- Received
- Academic Editor
- Consolato Sergi
- Subject Areas
- Artificial Intelligence, Computer Vision
- Keywords
- Deep learning, Self-supervised learning, Attention mechanism, Feature fusion, Feature selection, Convolutional network, X-ray, Robustness, Majority voting
- Copyright
- © 2025 Alammar et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ Computer Science) and either DOI or URL of the article must be cited.
- Cite this article
- 2026. Improving robustness in X-ray image classification through attention mechanisms in convolutional neural networks. PeerJ Computer Science 12:e3447 https://doi.org/10.7717/peerj-cs.3447
Abstract
Aim
Musculoskeletal radiographs (X-rays) are commonly used to diagnose bone and joint abnormalities because they are affordable and accessible. Convolutional neural networks (CNN) have demonstrated strong performance in medical image classification. However, interpreting these images reliably is challenging due to a lack of labelled data, inherent image noise, and the lack of explainable artificial intelligence (AI). This research aims to improve the robustness against noise, accuracy, and interpretability of musculoskeletal radiograph classification by addressing these key challenges.
Method
This study proposes an attention-enhanced deep learning framework that combines in-domain self-supervised pretraining, region-specific attention-based convolutional neural networks, and hierarchical feature fusion. To reduce redundancy and improve generalisation, descriptor-based feature selection is applied before classification. The final classification is performed through an ensemble of classifiers, and visual explanation techniques (such as Grad-CAM) are used to enhance model transparency.
Finding
The proposed model demonstrated high classification accuracy, achieving 95.48% on the MURA dataset and 99.80% on the Chest X-ray dataset. The model worked reliably even when images were noisy and was able to detect essential signs in unclear scans. The visualisations showed that the attention layers focused on meaningful areas in the X-rays, making the results easier to understand.
Implication
Integrating attention mechanisms, feature selection, and ensemble learning significantly mitigated overfitting and improved generalisation and explainability. The model demonstrated consistent performance across datasets and provided interpretable outputs, supporting its potential for clinical deployment.
Conclusion
This study presents a robust and interpretable deep learning framework for musculoskeletal X-ray analysis. By addressing core limitations in existing models, namely, data scarcity, noise sensitivity, and a lack of explainability, the proposed method enhances the reliability of AI-assisted diagnosis in medical imaging.
Introduction
Convolutional neural networks (CNNs) have demonstrated strong performance in medical-image classification and many other domains (Alzubaidi et al., 2024c). However, medical-image classification typically faces challenges such as data scarcity and noise (including overlaid text, irrelevant anatomy, or low-quality scans), all of which can compromise the robustness and interpretability of deep-learning (DL) models (Datta et al., 2021; Alzubaidi et al., 2024b). The first challenge in medical-image classification is data scarcity. Medical imaging datasets often face limitations due to privacy concerns, a lack of expert annotations, and the high costs associated with data curation and annotation (Alzubaidi et al., 2025; Colussi, 2024). This scarcity hampers the training of DL models, resulting in overfitting and decreased generalisability in real-world clinical settings. To address the limited availability of annotated data, recent research has explored transfer learning and self-supervised learning (SSL) strategies. These methods pretrain models on large, unlabelled datasets to enhance downstream performance (Alzubaidi et al., 2020; Alammar et al., 2023). In particular, in-domain SSL has emerged as a powerful approach for learning domain-specific representations from grayscale radiographs without relying on colour-natural image datasets such as ImageNet (Alzubaidi et al., 2024a, 2019).
The second challenge is domain adoption, which involves training DL models to perform multiple tasks without the need for complete retraining from scratch (Nareshkumar & Nimala, 2025; Alzubaidi et al., 2024d). This process requires the models to generalise across different imaging domains while retaining their performance. To address this issue, it is essential to develop strategies that enable models to transfer knowledge effectively while maintaining stability and consistency. Recent research demonstrates the effectiveness of combining in-domain self-supervised learning with multi-task learning techniques to facilitate domain adaptation (Alammar et al., 2024; Liu et al., 2025; Alammar et al., 2022). Their framework allows models to retain previously learned representations while adapting to new tasks. This approach helps mitigate catastrophic forgetting and enhances model robustness across diverse datasets. Therefore, ensuring resistance to catastrophic forgetting is crucial for developing reliable medical-image classification.
Another critical challenge in medical image analysis is ensuring the robustness of DL models against various forms of noise, including overlaid text, irrelevant anatomical structures, and low-resolution or artefact-laden scans (Hussain & Hyeon Gu, 2024; Alzubaidi et al., 2024b). Such noise can mislead the model by drawing attention to non-diagnostic regions, thereby compromising classification accuracy and clinical reliability. Traditional CNNs often lack the spatial awareness needed to focus on important regions in the presence of distractions (Natha et al., 2025).
To overcome this limitation, attention mechanisms have been widely adopted to enhance spatial focus within DL architectures (Niu, Zhong & Yu, 2021). These mechanisms enable models to assign dynamic weights to different regions of an image, effectively suppressing irrelevant information while amplifying areas that are diagnostically significant. As a result, attention-based networks have demonstrated improved resilience to noise, better localisation of pathological features, and enhanced interpretability, making them especially well-suited for complex and heterogeneous medical imaging scenarios.
In this context, attention mechanisms have proven effective in enabling DL models to selectively prioritise salient regions of an image (Soydaner, 2022). When integrated into CNNs, attention modules enhance both robustness and interpretability by promoting spatial discrimination and reducing susceptibility to background artifacts (Bello et al., 2019; Alzubaidi et al., 2024d). These mechanisms are particularly valuable in clinical applications where subtle radiographic cues must be consistently identified across heterogeneous imaging conditions.
However, despite the growing adoption of attention-based models in medical imaging, several studies continue to exhibit key limitations. One notable study examines the integration of self-attention mechanisms within a recurrent CNN-LSTM architecture, specifically designed for structured healthcare data. While this approach shows promise in processing complex information, it falls short in terms of multi-task learning capabilities. It lacks the necessary features for clinical interpretability, which are crucial for practical applications in healthcare settings (Usama et al., 2020). In addition, a cross-modal attention mechanism has been implemented within a supervised multimodal framework to identify paediatric pneumonia. However, this approach lacks the integration of feature selection and ensemble learning techniques, which ultimately restricts its robustness and effectiveness in clinical applications (Li, Chen & Hu, 2024). A semi-supervised framework utilising adaptive aggregated attention has been developed for the classification of wireless capsule endoscopy images. However, this approach is limited to single-task learning and does not provide adequate explainability regarding its decision-making processes, as noted in Guo & Yuan (2020). Hierarchical attention mechanisms have been designed to enhance the accuracy of fracture localisation in medical imaging. However, these approaches fall short in addressing two critical areas: continual learning and anatomical generalisation. While they effectively identify fractures within specific contexts, they do not adequately adapt to new data or evolve in their understanding of various anatomical structures over time (Shaw, Uszkoreit & Vaswani, 2018; Bello et al., 2019).
In this study, we propose a unified framework to overcome the limitations as mentioned earlier. Our key contributions are:
We introduce a novel framework for multi-task classification of musculoskeletal abnormalities. This framework combines in-domain self-supervised learning, attention-based multi-region feature extraction, and descriptor-guided feature selection, all of which are explicitly designed for interpretable diagnosis across multiple anatomical tasks.
We design a dual-region attention mechanism that processes humerus and wrist radiographs in parallel. This enables spatially adaptive feature extraction that is both anatomically aware and task-specific, thereby supporting the concurrent learning of multiple classification tasks within a unified pipeline.
We apply self-supervised pretraining optimised for grayscale medical images to initialise the feature extractor. This approach eliminates dependence on natural image datasets, such as ImageNet, and improves domain adaptation across tasks.
Attention-augmented CNN models are trained using hierarchically fused features, which are further enhanced through Scale-Invariant Feature Transform (SIFT)-based feature selection. This selective refinement improves robustness and task-level discrimination.
We integrate an ensemble classification pipeline that combines decision tree, K-Nearest Neighbours (KNN), and Support Vector Machine (SVM) classifiers. This leverages the strengths of multiple classifiers to enhance diagnostic reliability. Majority voting across these models ensures reliable predictions for each task and anatomical site.
We incorporate explainability tools, such as Grad-CAM, Occlusion Sensitivity, and t-SNE, to evaluate the model’s interpretability at both spatial and latent feature levels.
We validate the framework extensively on the MURA and chest X-ray datasets, demonstrating consistent performance gains across multiple tasks and superior interpretability compared with baseline and single-task approaches.
The article is structured as follows: ‘Related Work’ reviews the limitations of existing studies; ‘Research Methodology’ presents the proposed methodology; ‘Results and Discussion’ discusses and interprets the experimental results; ‘Validation of the Proposed Method with Chest X-ray Classification’ validates the approach using the chest X-ray dataset; and ‘Discussion’ provides concluding remarks.
Related work
Recent advances in computer vision have produced increasingly sophisticated image feature extractors, with many architectures now incorporating multi-scale convolutional operations and skip connections to improve representational capacity (Khan et al., 2020; Alwzwazy et al., 2025). Automated design methods based on convolutional building blocks have also delivered state-of-the-art accuracy in large-scale classification challenges, and these design principles have been successfully adapted to medical imaging applications (Ming, Yin & Li, 2022).
In parallel, attention mechanisms have become integral to deep-learning architectures due to their ability to capture long-range dependencies and emphasise the most relevant features within complex data (Ding & Jia, 2022). Initially introduced in recurrent neural networks for sequence alignment in machine translation (Bhadauria et al., 2023), attention methods have since been combined with convolutional operations across a wide range of vision tasks (Galassi, Lippi & Torroni, 2020). Their adoption in medical imaging has been particularly impactful, enabling models to focus on subtle yet clinically significant patterns that might be overlooked by conventional convolutional approaches (Li et al., 2023).
In medical imaging, attention mechanisms have been applied to diverse modalities and clinical objectives, from lesion localisation to disease progression tracking (Zhao et al., 2022). A summary of representative state-of-the-art attention-based approaches, together with their key contributions and limitations, is provided in Table 1. These methods include innovations in architecture design, integration strategies, and interpretability tools, each aiming to address specific challenges in diagnostic accuracy, generalisability, and clinical trust.
| Source | Application | Method | Limitation |
|---|---|---|---|
| Zou & Arshad (2024) | X-ray | Attention mechanism | Missed or incorrect detections in the test set. |
| Sanzida Ferdous Ruhi, Nahar & Ferdous Ashrafi (2024) | X-ray | Attention mechanism | Requires improved reliability. |
| Chang et al. (2024) | Musculoskeletal ultrasound | Self-Attention U-Net | Limited in dataset, interpretability, and generalisability concerns. |
| Hu et al. (2022) | Lesion localisation | Squeeze-and-Excitation attention | Focused only on inter-channel feature relationships. |
| Ilse, Tomczak & Welling (2018) | Histopathology | Attention mechanism | Small sample size. |
| Han et al. (2020) | COVID-19 classification | AD3D-MIL | Small sample size. |
| Al-Shabi, Shak & Tan (2022) | Lung cancer | Non-local attention | Applied only to a simplified example. |
| Datta et al. (2021) | Skin cancer | Soft attention mechanism | Performs poorly with noisy images. |
| Guo & Yuan (2020) | WCE images | Adaptive aggregated attention | Less effective with varied symptoms. |
| Valanarasu et al. (2021) | Brain segmentation (Ultrasound) | Multi-head attention | Small dataset. |
| Zhao et al. (2022) | Medical images | Attention-based GAN | Poor repeatability and interpretability. |
| Dai, Gao & Liu (2021) | Dermatoscopic images | Soft attention mechanism | Dataset imbalance. |
For example, Sanzida Ferdous Ruhi, Nahar & Ferdous Ashrafi (2024) proposed a bottleneck attention module (BAM) within a transfer learning framework for X-ray image classification, achieving 90.48% accuracy and establishing a strong benchmark for the dataset in use. Chang et al. (2024) introduced SEAT-UNet, which combines U-Net with self-attention for improved lesion classification and localisation in wrist joint radiographs. However, its reliance on annotated datasets raises concerns about bias and generalisability. Zou & Arshad (2024) integrated attention into YOLOv7 for complex bone fracture detection, improving localisation and bounding box precision.
Beyond radiography, Ilse, Tomczak & Welling (2018) developed a gated attention mechanism within a multiple-instance learning framework for colon cancer classification on histopathology slides, while Hu, Shen & Sun (2018) introduced squeeze-and-excitation (SE) networks to model inter-channel relationships with low computational overhead. Shuffle Attention (SA) (Zhang & Yang, 2021) and CABNet (He et al., 2020) have further advanced small-lesion detection by combining spatial–channel attention and category-aware modules. Other applications include glioma subtype classification using sparse attention blocks (Lu et al., 2021), diabetic retinopathy diagnosis with gated attention (Bodapati, Shaik & Naralasetti, 2021), and hybrid CNN-Transformer approaches for ophthalmic image classification (Yang, Chen & Xu, 2021).
Attention mechanisms have also been explored in CT and MRI applications, such as non-local attention for lung cancer classification with curriculum-based training (Al-Shabi, Shak & Tan, 2022), interpretability-enhanced histopathology classification (Moranguinho et al., 2021), and CNN-Transformer models for head, neck, and knee MRI scans (Dai, Gao & Liu, 2021). Additional studies include soft attention for skin cancer classification (Datta et al., 2021), ScopeFormer for cerebral bleeding detection (Barhoumi & Rasool, 2021), and attention-augmented ResNet for Alzheimer’s diagnosis using brain MRI (Liang & Gu, 2020).
After reviewing the literature, several limitations recurrent in prior studies are outlined below, each of which is explicitly addressed in our proposed approach:
-
Effectiveness and limitations of attention mechanisms: Attention mechanisms have demonstrated strong performance across various tasks. However, their diagnostic accuracy varies when applied to different types of clinical imaging. For example, studies using the same dataset have reported classification accuracies ranging from 81% to 91%, depending on factors such as model architecture, label quality, and task complexity (Li et al., 2023; Zhao et al., 2022). The wide variation in performance highlights a key limitation: traditional attention mechanisms often struggle to generalise across imaging scenarios, particularly when detecting subtle or low-contrast abnormalities.
To address this limitation, our framework introduces a multi-level attention mechanism that adaptively fuses global and local feature representations. This design enhances the model’s sensitivity to fine-grained and low-contrast patterns, which is particularly critical for the accurate classification of pneumoconiosis, an area where traditional attention models often struggle to perform. The manuscript has been updated to provide a more precise explanation of this rationale, with a detailed discussion included in both the introduction and methods sections.
-
Limitations of Transfer Learning with ImageNet for grayscale medical images: Although transfer learning using ImageNet pre-trained models is common, it often performs poorly on grayscale medical images due to differences in colour channels and spatial textures. Previous studies have attempted to overcome this by freezing or fine-tuning layers (Matsoukas et al., 2022). However, these methods often overlook the distinct feature distributions of grayscale data.
To overcome this limitation, our study leverages self-supervised learning (SSL) tailored to domain-specific adaptation. Rather than relying on colour-rich external datasets like ImageNet, we perform in-domain pretraining using unlabelled grayscale medical images. This approach enables the model to learn meaningful representations directly from the target domain, capturing clinically relevant structures, contrast variations, and spatial dependencies inherent to grayscale modalities. By pretraining the backbone network on the same type of imaging data without manual labels, the model develops a stronger initialisation that improves downstream classification performance and generalisability, especially in data-scarce or label-limited scenarios. This domain-aligned pretraining strategy substantially reduces the risk of negative transfer and enhances robustness across diverse medical imaging tasks.
-
Need for explainability in clinical applications: Explainability is essential for the clinical adoption of DL models, as clinicians must be able to understand and trust the model’s predictions. However, many previous studies either excluded interpretability methods or relied only on basic saliency maps (Moranguinho et al., 2021; Ilse, Tomczak & Welling, 2018).
To address this limitation, the current study employs Grad-CAM and occlusion sensitivity to visually verify that the model consistently attends to clinically relevant areas within the MURA and chest X-ray datasets. These explainability techniques enhance the model’s transparency, improve clinical interpretability, and ensure that the model’s predictions align with established diagnostic standards.
The collective findings from previous studies indicate persistent gaps between existing attention-based deep learning methods and the requirements for clinically robust, generalisable, and interpretable diagnostic tools. Addressing these gaps requires a comprehensive framework that not only enhances accuracy but also adapts to various imaging modalities, employs domain-specific pretraining, and incorporates reliable interpretability mechanisms.
To address the limitations identified in the literature, the proposed methodology integrates several complementary strategies designed to enhance the robustness and reliability of the system’s performance across a range of diagnostic tasks.
Research methodology
This section outlines a proposed methodology designed to enhance the classification of musculoskeletal radiographs by systematically addressing several significant challenges. These challenges include the limited availability of labelled data, domain shift, catastrophic forgetting, visual noise, and the increasing demand for explainable artificial intelligence (XAI). The framework integrates complementary components to ensure robustness, generalisability, and clinical interpretability.
We implement self-supervised learning (SSL) specifically tailored for grayscale radiographs, allowing the model to extract domain-aligned representations from unlabelled datasets. This method addresses the scarcity of annotations and facilitates domain adaptation by aligning the internal features of the model with the distinct statistical properties of medical imagery.
To enhance spatial focus and interpretability, we incorporate attention mechanisms that direct the model’s attention toward diagnostically relevant regions, even in the presence of artefacts or unrelated structures. This integration enhances both the robustness and transparency of the decision-making process. Additionally, we introduce a feature fusion module that extracts hierarchical deep features from CNNs. This fusion enriches the feature space by capturing both global semantic patterns and local invariant structures, which increases the model’s discriminative power and resilience to noise and anatomical variability. Moreover, the framework supports multi-task learning, enabling the simultaneous classification of multiple anatomical regions while preserving previously acquired knowledge. This approach mitigates the risk of catastrophic forgetting during incremental learning or when applying the model to evolving datasets. Finally, we adopt an ensemble classification strategy that integrates the outputs of multiple independent classifiers through majority voting. This ensemble design reduces individual model biases and enhances predictive stability, particularly under uncertain or atypical imaging conditions.
Collectively, these components contribute to a robust, adaptable, and interpretable framework that is well-suited for real-world clinical deployment in the field of musculoskeletal radiology.
Self-supervised learning (In-domain pre-training)
In the first stage of our framework, we implement a self-supervised learning (SSL) strategy to pre-train the model using unlabelled X-ray images. Instead of using weights from models trained on natural RGB datasets, such as ImageNet, we adapt the network by selectively unfreezing only the final convolutional blocks. This approach enables the model to learn domain-specific features, such as bone density, anatomical structures, and grayscale contrast characteristics, which are unique to radiographs. There are two widely adopted strategies in SSL: freezing selected layers of the pre-trained model and end-to-end fine-tuning. In our framework, we adopt the layer-freezing approach. To retain useful low-level representations, we freeze the first 50% of the convolutional layers, which include early and mid-level filters responsible for capturing edges and textures. The remaining layers are fine-tuned to focus on high-level features relevant to musculoskeletal abnormalities, such as bone fractures. To facilitate domain adaptation, we apply a transform-based pretext task using targeted image augmentations. These include horizontal flipping, small rotations ( 10°), contrast normalisation, and random cropping, each designed to introduce variability without compromising anatomical structures. During training, different augmented views of the same image are assigned the same label, encouraging the model to learn invariant and domain-specific features. The network is optimised using categorical cross-entropy loss. This SSL process produces robust pretrained weights, which are then used to initialise the full model for subsequent supervised training focused on fracture and abnormality classification.
Self-attention mechanism
The attention mechanism allows DL models to prioritise the most informative regions within an image by modelling the relationships between spatial features. This is particularly beneficial in medical imaging tasks, such as fracture detection using the MURA dataset and chest X-ray classification, where accurate diagnosis often depends on subtle contextual cues. By comparing anatomical structures across different parts of an image, attention mechanisms help the model to detect fine-grained abnormalities that might otherwise be overlooked.
At the core of this mechanism is the concept of self-attention, which enables the model to assess and weigh the relevance of each input element relative to the others. This process involves the following steps:
-
For each input position in a sequence , the model derives three vectors:
-
–
Query ( ), Key ( ), Value ( )
These are computed via trainable linear projections: (1) where is the input vector at position , and , , and are learnable weight matrices used to project the input into the query, key, and value representations, respectively. These projections enable the model to transform the input into subspaces tailored for attention computation.
-
The similarity between the -th query and all keys is measured using the scaled dot-product: (2) where denotes the attention score and is the dimension of the key vectors used for scaling.
The attention scores are normalised using the softmax function: (3) yielding a weight matrix S that reflects the relative importance of other positions to the -th element.
Each output vector is computed as a weighted sum of the value vectors: (4) providing a context-aware representation that incorporates information from across the input sequence.
In our proposed framework, we enhance this mechanism using three parallel convolutional layers denoted as , , and —to extract the query, key, and value feature maps from the input (Vaswani et al., 2017). The attention map is calculated by applying a softmax operation to the product of the query and key feature maps. These attention scores are then used to weight the value features in a component referred to as softmax-mult. The result is added back to the query features to preserve the original structure, forming an updated representation:
(5)
To ensure dimensional consistency between the input and the output, we apply a convolutional adjustment. The resulting attention-enhanced features are merged with the original input via residual addition, enabling the model to retain low-level details while benefiting from global contextual awareness.
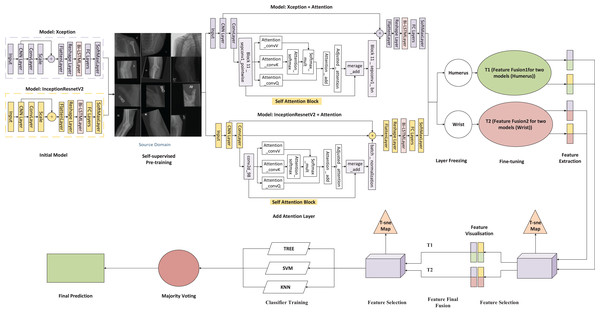
This attention block is integrated into both the InceptionResNetV2 and Xception architectures used in our study, as shown in Fig. 1. By positioning the self-attention module between the convolutional and batch normalisation layers, we enable the model to emphasise diagnostically significant regions more effectively. This architectural refinement improves the network’s ability to model long-range dependencies, thereby enhancing its interpretability and performance on complex medical imaging tasks.
Figure 1: Architecture of the proposed framework.
Feature fusion and feature selection
Feature fusion is designed to combine multiple complementary representations to enhance model performance, particularly in medical imaging tasks where multiscale and modality-specific information must be integrated. By incorporating attention mechanisms, the fusion process is optimised to prioritise clinically relevant regions while maintaining global contextual awareness. Complementing this, feature selection focuses on retaining the most informative components from high-dimensional data, improving computational efficiency and reducing overfitting, which ultimately leads to a more generalisable model.
In the proposed framework (Fig. 1), two convolutional neural networks, Xception and InceptionResNetV2, are independently applied to each anatomical region, resulting in four parallel pipelines: two for the humerus task and two for the wrist task. Each model is augmented with a self-attention mechanism to enhance the capture of both local and contextual features relevant to diagnostic interpretation.
Following feature extraction, the Scale-Invariant Feature Transform (SIFT) is employed to detect distinctive local patterns that remain stable under changes in scale, rotation, and illumination. For each task, deep features obtained from the two attention-based CNNs are horizontally concatenated, producing two task-specific fused representations: Fusion 1 (T1) for the humerus and Fusion 2 (T2) for the wrist. This fusion strategy preserves diverse spatial characteristics while aligning the dimensional structure of the original samples.
The fused representation integrates the high-level semantic information derived from both CNN architectures (3,584 feature dimensions) with the fine-grained local descriptors obtained via SIFT (128 dimensions), forming a comprehensive 3,712-dimensional feature vector. This combination allows the model to retain global contextual features while enriching them with structural details, such as bone edges, trabecular textures, and minor cortical discontinuity features, which are often attenuated in deep convolutional layers. In this study, incorporating SIFT proved particularly beneficial in capturing subtle musculoskeletal variations and improving cross-anatomical generalisation, where deep models alone showed limited sensitivity to fine-scale patterns. To improve computational efficiency and eliminate redundancy, Principal Component Analysis (PCA) is applied, retaining 95% of the total variance and reducing the feature space to 51 dimensions for subsequent classification.
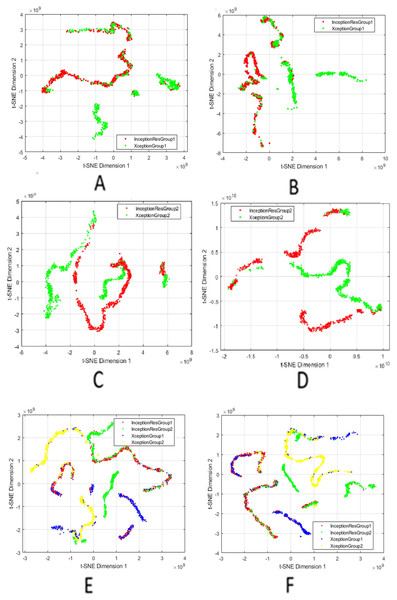
The compact and distinctive features are visualised using t-distributed Stochastic Neighbour Embedding (t-SNE) to evaluate clustering behaviour and class separability qualitatively. The t-SNE projections confirm that the SIFT-enhanced feature space offers more precise class boundaries and improved intra-class cohesion, demonstrating its effectiveness in enhancing discriminative capacity.
The refined feature sets from T1 and T2 are combined vertically to form a unified high-dimensional feature space that captures the differences across tasks. This multitask fusion strategy enhances the model’s flexibility and supports generalisation across various anatomical regions. The consolidated feature set is then used to train several classical machine-learning classifiers, including KNN, SVM, and decision tree models. A comparative evaluation of these classifiers identifies the most effective architecture for each task. Additionally, the ensemble approach leverages its complementary strengths to achieve stable, reliable performance.
This two-stage fusion and feature-selection framework provides a comprehensive, compact, and interpretable representation that balances global semantic understanding with local structural precision, enhancing robustness and clinical relevance for musculoskeletal classification tasks.
Ensemble prediction via majority voting
The final classification output is generated using a majority voting ensemble method that combines predictions from three different machine learning classifiers: decision tree, KNN, and SVM. As illustrated in Fig. 1, it is essential to note that t-SNE and Gradient-weighted Class Activation Mapping (Grad-CAM) are used solely for post-hoc visualisation and model interpretability; they do not influence the feature selection process and are not used as inputs for the classifiers.
Before classification, the integrated feature representations go through a systematic process of feature extraction and selection. Initially, SIFT is utilised to extract robust local features and descriptors from the fused feature space. This step significantly enhances the model’s ability to identify distinguishing patterns, such as bone fractures and relevant regions, across varying scales, orientations, and imaging conditions. After feature extraction, the extracted features are normalised using min-max scaling and evaluated for dimensional consistency across all samples, ensuring they are compatible with the classifiers used in the later stages.
Each of the three classifiers is trained independently on a cleaned and structured feature set. The final predictions are determined through a majority voting strategy, which identifies the class that receives the most votes as the definitive prediction. This ensemble method reduces the impact of individual classifier biases, strengthens robustness, and improves the model’s generalisation capabilities.
In summary, this study presents a robust and scalable framework for medical image analysis that integrates self-supervised learning, attention mechanisms, SIFT-based feature selection, and ensemble decision-making. By incorporating domain-specific pre-training and context-aware feature extraction, the proposed methodology is designed to improve interpretability and support robust diagnostic decision-making. Its modular design allows adaptation to a variety of medical imaging tasks, providing a scalable foundation for future AI-driven diagnostic systems.
Data collection
This study utilised two primary datasets: the MURA dataset and a curated collection of chest X-ray images. The MURA dataset, introduced by Rajpurkar et al. (2017), comprises musculoskeletal radiographs across seven anatomical regions: elbow, finger, forearm, hand, humerus, shoulder, and wrist. Each image is labelled as either normal (negative) or abnormal (positive), resulting in a total of 40,561 images divided into separate training and test sets. Additionally, a chest X-ray dataset was utilised, comprising images from four diagnostic categories: COVID-19, normal, pneumonia, and tuberculosis. The dataset was divided into two groups: Group 1 (COVID-19 and normal) and Group 2 (pneumonia and tuberculosis). Moreover, to simulate diverse imaging conditions and anatomical variations, we employ data augmentation techniques, including rotation, scaling, translation, and contrast variation, during the training process. This approach significantly improves the model’s ability to generalise across imperfect and heterogeneous clinical data.
Experimental scenario
To evaluate the proposed framework systematically, two distinct case studies are defined based on target datasets: musculoskeletal X-rays (humerus and wrist) and chest X-rays. Correspondingly, two independent self-supervised learning (SSL) sources were constructed to support robust domain-specific representation learning for each target.
Target datasets
-
1.
Case study 1-Bone X-ray Classification (Target and SSL source)
This experiment focuses on binary classification (positive or negative) for two specific anatomical regions (humerus and wrist) from the MURA dataset. These tasks were selected to evaluate the framework’s effectiveness in detecting abnormalities in localised musculoskeletal areas.
SSL source: To facilitate domain-specific representation learning, approximately 40,000 unlabelled bone X-rays were collected from the MURA dataset, excluding humerus and wrist images. These included scans of the shoulder, elbow, hand, finger, and forearm. Additionally, 30,000 external bone-related X-rays (e.g., hip, knee, ankle) were incorporated to construct a diverse and comprehensive SSL source.
-
2.
Case study 2-Chest X-ray Classification (Target and SSL source) This study assesses the generalisability of the proposed framework by applying it to chest X-ray images. The model was tasked with classifying scans into two distinct categories: COVID-19 versus normal, and pneumonia versus tuberculosis.
SSL source: A separate pretraining corpus of 50,000 unlabelled chest X-ray images was constructed from publicly available datasets, allowing the model to learn thoracic imaging features before fine-tuning.
All experiments used the Adam optimiser with an initial learning rate of 0.001 and a mini-batch size of 15, conducted over a maximum of 100 epochs. The training data was shuffled at the beginning of each epoch. All experiments were performed using MATLAB 2024a (The MathWorks, Natick, MA, USA) on a system equipped with an Intel Core i7 processor, 32 GB of RAM, a 1 TB SSD, and an NVIDIA RTX A3000 GPU with 12 GB of memory.
The average training duration for each deep learning model was approximately 148 min over 15 epochs for the Xception model, while it took about 296 min over 15 epochs for the InceptionResNetV2.
Base models
Two high-performing CNN backbones, Xception and InceptionResNetV2, were selected for their established efficacy in large-scale visual tasks. Both models underwent initial pretraining through self-supervised learning on their respective SSL sources (bone or chest X-ray) before being fine-tuned to classify specific targets (humerus, wrist, or chest X-ray).
To enhance spatial understanding and representation learning, we integrated multi-head self-attention layers into each model. A partial fine-tuning strategy was implemented, freezing approximately 50% of the layers to retain learned representations while effectively adapting to task-specific features. Each classification task was trained independently within a multi-task framework, thereby preserving prior knowledge and minimising the risk of catastrophic forgetting, while maintaining adaptability to new imaging tasks without requiring complete retraining.
In summary, we employed two CNN models as our foundation, as displayed in Table 2, which involved the following steps:
-
Model selection and justification: After evaluating the performance of several baseline CNN architectures, including ResNet, MobileNet, Xception, and InceptionNet on the ImageNet dataset, two pre-trained models were selected for the proposed framework: Xception (Chollet, 2017) and InceptionResNetV2 (Szegedy et al., 2017). These models are well-suited to address training challenges commonly encountered in DL, such as vanishing gradients, by employing residual learning. Rather than mapping inputs directly to outputs, they learn residual functions, which facilitates the training of deeper and more stable networks. Their architectures have significantly influenced the development of modern neural networks and are widely adopted in both academic research and practical applications. Additionally, both models exhibit strong feature representation capabilities and demonstrate resilience against overfitting.
Integration of Attention Mechanisms: To enhance model performance, a multi-head self-attention mechanism was integrated into both the Xception and InceptionResNetV2 architectures. This enhancement enables the models to focus selectively on the most informative regions of the input data, which is particularly beneficial in medical image classification tasks where subtle patterns, such as fine fractures or lesions, are crucial for accurate diagnosis. The attention mechanism dynamically adjusts the model’s focus based on contextual relevance, improving interpretability and classification accuracy.
In the first ablation study, attention layers were integrated into models pre-trained using SSL. SSL is a DL paradigm that enables models to learn meaningful representations from unlabelled data through in-domain pretraining, making it particularly effective in scenarios with limited annotated samples. This approach facilitates knowledge transfer from a related domain, thereby improving performance on downstream tasks.
| Model | Input size | Parameters (106) | Depth |
|---|---|---|---|
| Inceptionresnetv2 | 299 299 3 | 55.9 | 164 |
| Xception | 299 299 3 | 22.9 | 71 |
Results and discussion
Experimental evaluation
This section outlines the experimental design and evaluation of the proposed framework for classifying musculoskeletal abnormalities, with a specific focus on tasks derived from the MURA dataset, which includes X-ray images of the humerus, wrist, and chest. In addition to describing the model configurations, we emphasise the vital role that dataset characteristics play in assessing the framework’s generalisability. We report comprehensive performance metrics, including accuracy, precision, recall, F1-score, specificity, and Cohen’s Kappa, to provide a thorough assessment of the model’s effectiveness.
Evaluation metrics
To evaluate the effectiveness of the proposed framework, a confusion matrix (CM) was employed to summarise the classification outcomes. In the context of binary classification, the confusion matrix consists of four key components:
-
1.
True positives (TP): Correctly identified instances of the target class.
-
2.
True negatives (TN): Correctly identified instances of the non-target class.
-
3.
False positives (FP): Non-target instances incorrectly classified as the target class.
-
4.
False negatives (FN): Target instances incorrectly classified as non-target.
These components serve as the foundation for computing various performance metrics. The following equations define the key evaluation measures used in this study:
(6) (7) (8) (9) (10)
Cohen’s Kappa equation
(11) (12) (13)
(14)
Cohen’s Kappa score(Kappa.) =
(15)
Results of the proposed framework solution
The predictive performance of each classifier is shown along with the overall results obtained from the ensemble approach. The architecture of the base model includes transformer blocks that employ a self-attention mechanism. This allows the model to dynamically focus on different regions of the input feature space, enhancing its ability to capture complex relationships and dependencies within the data.
Feature fusion was conducted by combining the outputs of two DL models for each task: Fusion 1 (T1) for the humerus and Fusion 2 (T2) for the wrist. Afterwards, a feature selection technique was applied to each fusion output to retain the most informative attributes. The refined features from both tasks were then integrated into a single, unified fusion pool. This consolidated feature set served as the input for training multiple machine learning classifiers, which were evaluated based on their ability to classify the combined humerus and wrist datasets accurately. For further analysis, we selected three classifiers: KNN, decision tree, and SVM. The classification results for both the humerus and wrist tasks are illustrated in Table 3. In the majority voting ensemble approach, each classifier casts a vote for a predicted class label, and the label receiving the most votes is selected as the final output. This strategy enhances overall accuracy by leveraging the complementary strengths of individual models while mitigating the effect of their respective weaknesses.
| Evaluation metric (%) | Tree | KNN | SVM | Final decision |
|---|---|---|---|---|
| Humerus task | ||||
| Accuracy | 98.96 | 94.10 | 94.10 | 94.79 |
| Recall | 98.57 | 92.86 | 93.57 | 94.78 |
| Precision | 99.28 | 94.89 | 94.24 | 94.80 |
| Specificity | 99.32 | 95.20 | 94.57 | 94.78 |
| F1-score | 98.92 | 93.86 | 93.91 | 94.79 |
| Kappa | 97.90 | 88.00 | 88.20 | 89.60 |
| Wrist task | ||||
| Accuracy | 98.79 | 91.20 | 91.20 | 94.99 |
| Recall | 98.31 | 86.78 | 86.44 | 95.23 |
| Precision | 98.98 | 93.09 | 93.41 | 94.70 |
| Specificity | 99.17 | 94.78 | 95.05 | 94.70 |
| F1-score | 98.64 | 89.82 | 89.79 | 94.91 |
| Kappa | 97.50 | 82.00 | 82.30 | 90.21 |
By combining the predictions of the Tree, KNN, and SVM classifiers through majority voting, the ensemble achieved an overall classification accuracy of 95.48% across the combined humerus and wrist datasets. This result is summarised in Table 4.
| Evaluation metric | Majority voting of combination |
|---|---|
| Accuracy | 95.48 |
| Recall | 92.46 |
| Precision | 95.74 |
| Specificity | 96.57 |
| F1-score | 94.08 |
| Cohen’s kappa | 90.91 |
Ablation studies
This section examines three fundamental components of the proposed framework, exploring their significance and interconnections. Each aspect will be examined to highlight its unique contributions and how they collectively support the framework’s overarching objectives.
Experimental evaluation of attention layers in CNN models: ablation study 1
In Ablation Study 1, self-attention layers were integrated into two models of CNN architectures, specifically Xception and InceptionResNetV2, to enhance performance in classification tasks related to the humerus and wrist. The incorporation of attention mechanisms facilitated the models’ ability to prioritise diagnostically relevant regions within X-ray images, thereby improving the quality of the extracted features while simultaneously reducing sensitivity to extraneous information. Xception, which utilises depthwise separable convolutions for efficient spatial feature extraction, demonstrated an enhanced capacity for recognising fine-grained anatomical structures after implementing attention layers. Similarly, InceptionResNetV2, which combines residual connections with multi-scale inception modules, exhibited improved discriminative performance due to attention-guided refinement. The resultant models displayed enhanced robustness, diagnostic accuracy, and interpretability, as validated by confusion matrix evaluations across both classification tasks. The incorporation of attention layers has yielded notable enhancements in both interpretability and classification performance. In the case of the humerus task, the attention-augmented Xception and InceptionResNetV2 models recorded accuracies of 94.10% and 92.00%, respectively. For the wrist task, the Xception model achieved an accuracy of 89.20%, while the InceptionResNetV2 model attained 88.60%. These findings are summarised in Table 5.
| Evaluation metric | AttentionXception (Humerus) | AttentionInception (Humerus) |
|---|---|---|
| Accuracy | 94.10 | 92.00 |
| Recall | 93.60 | 88.60 |
| Precision | 94.20 | 94.60 |
| Specificity | 94.60 | 95.30 |
| F1-score | 93.90 | 91.50 |
| Cohen’s kappa | 88.00 | 84.00 |
| Evaluation metric | AttentionXception (Wrist) | AttentionXception (Wrist) |
| Accuracy | 89.20 | 88.60 |
| Recall | 85.10 | 84.00 |
| Precision | 90.60 | 89.90 |
| Specificity | 92.90 | 92.30 |
| F1-score | 87.80 | 86.80 |
| Cohen’s kappa | 78.00 | 76.00 |
Experimental evaluation of deep feature fusion: ablation study 2
This section presents the results of Ablation Study 2, which evaluated a task-specific hybrid fusion strategy combining attention-guided deep learning features with handcrafted local descriptors. For each anatomical region, humerus and wrist features were independently extracted using two self-attention-augmented CNNs: Xception and InceptionResNetV2. Additionally, SIFT descriptors were computed to capture local structural information. For each task, features from the two deep models and SIFT were concatenated horizontally, resulting in two task-specific fused representations: Fusion 1 for the humerus and Fusion 2 for the wrist. This approach preserved the dimensional structure of individual samples while integrating multiscale and contextual features. Following fusion, model-specific feature selection techniques were applied to refine the feature space and enhance its discriminative power. These refined features were used to train separate classifiers for each task. The classification outcomes are summarised in Table 6. The models achieved 94.40% accuracy for humerus and 91.00% for wrist, confirming that task-specific horizontal fusion of deep and handcrafted features enhances musculoskeletal image classification performance.
| Evaluation metric | Humerus + ML classifier | Wrist + ML classifier |
|---|---|---|
| Accuracy | 94.40 | 91.00 |
| Recall | 94.40 | 90.70 |
| Precision | 94.50 | 91.12 |
| Specificity | 94.40 | 90.70 |
| F1-score | 94.40 | 91.00 |
| Cohen’s kappa | 88.88 | 81.80 |
Ablation study 3: proposed framework for multitask feature fusion using attention models on two x-ray tasks
This section outlines the findings of Ablation Study 3, which assessed the generalisation capability of the proposed multi-task framework across related anatomical regions. In this evaluation, a single machine learning classifier was trained exclusively on the fused features derived from the humerus task and subsequently applied to classify wrist X-rays without any further training or fine-tuning. To facilitate this assessment, a consistent fusion strategy was employed for both tasks. Features for each anatomical region were extracted using attention-augmented Xception and InceptionResNetV2 models, which were then combined with SIFT descriptors. These features were concatenated vertically across samples, enabling the classifier, trained solely on humeral features, to generalise effectively to the wrist task utilising aligned feature dimensions.
The classification performance is summarised in Table 7, with the corresponding confusion matrix for the combined evaluation. The classifier demonstrated an accuracy of 93.80% on the humerus task and 91.40% on the wrist task, despite being trained exclusively on humerus data. This result underscores strong representational transfer and affirms the robustness of the proposed fusion strategy. Notably, there was no evidence of catastrophic forgetting, further validating the framework’s capacity to generalise across related musculoskeletal classification tasks.
| Evaluation metric (%) | Humerus + ML classifier | Wrist + ML classifier |
|---|---|---|
| Accuracy | 93.80 | 91.40 |
| Recall | 93.70 | 90.90 |
| Precision | 93.80 | 91.70 |
| Specificity | 93.70 | 90.90 |
| F1-score | 93.70 | 91.30 |
| Cohen’s kappa | 87.49 | 82.39 |
The results in Tables 7 and 8 provide a comparative analysis of our multi-task learning framework under two conditions: with and without the use of SIFT for feature selection before classification.
| Evaluation metric (%) | Humerus + ML classifier | Wrist + ML classifier |
|---|---|---|
| Accuracy | 93.00 | 89.40 |
| Recall | 92.90 | 86.10 |
| Precision | 93.00 | 89.80 |
| Specificity | 92.80 | 89.10 |
| F1-score | 92.80 | 87.90 |
| Cohen’s kappa | 86.00 | 78.44 |
Through the collected comparative experiment results, we have the following observations:
-
1.
As shown in Table 7, the model achieved 93.80% accuracy for humerus tasks and 91.40% for wrist tasks.
-
2.
High Cohen’s Kappa scores (87.49 and 82.39) indicate strong agreement beyond chance and reflect reliable generalisation.
-
3.
These results were obtained even though the classifier was only trained on humerus data, showcasing the effectiveness of the attention mechanism, feature fusion, and transferability between tasks.
Meanwhile, the other experiment that has been conducted without SIFT Feature Selection:
-
1.
In contrast, removing SIFT from the pipeline (Table 8) resulted in a notable drop in accuracy: 93.00% 89.40% for wrist, and 93.80% 93.00% for humerus.
-
2.
Cohen’s Kappa also declined (Humerus: from 87.49 to 86.00; Wrist: from 82.39 to 78.44), suggesting weaker generalisation and increased uncertainty in predictions.
These findings underscore the crucial role of SIFT in improving the model’s capacity to identify discriminative local features that are invariant to scale and orientation. The combination of self-supervised attention-based fusion and SIFT yields substantial improvements in performance and cross-task generalisation, while mitigating the risks of performance degradation and catastrophic forgetting.
Visualisation and validation
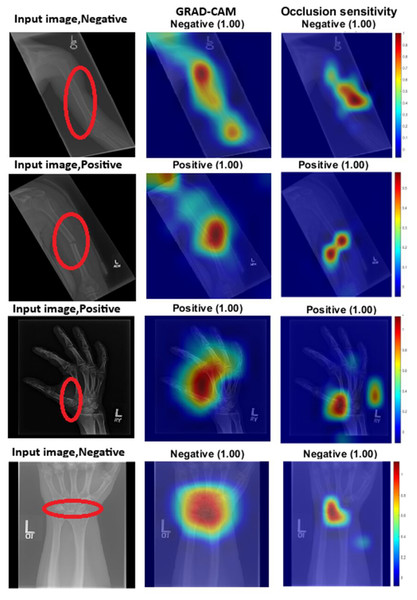
Medical image analysis is a domain where diagnostic decisions carry critical implications. Traditionally, such decisions have relied on the expertise of human specialists, whose reasoning processes are transparent and interpretable. In contrast, CNNs, due to their deep and complex architectures, often lack inherent interpretability, making it challenging to understand the rationale behind their predictions. To evaluate the robustness of our attention-enhanced CNN model and to gain insight into its decision-making processes, we employed several explainable artificial intelligence (XAI) techniques, gradient-weighted class activation mapping (Grad-CAM), occlusion sensitivity, and t-distributed stochastic neighbour embedding (t-SNE). These methods offer complementary perspectives. Grad-CAM and occlusion sensitivity highlight spatial regions that influence predictions, while t-SNE visualises the structure of the learned feature space.
Grad-CAM works by using the gradients of output classification scores to generate heat maps that highlight the areas most significant to the model’s predictions. Regions with high gradient activations are deemed crucial to the network’s decision-making process. On the other hand, occlusion sensitivity involves systematically masking portions of the input image and observing the changes in the output. This technique helps identify which areas are essential for accurate predictions. Figure 2 provides both visual and quantitative comparisons of these two methods in humerus and wrist classification tasks, confirming that the models consistently focus on anatomically and diagnostically relevant regions.
Figure 2: GRAD-CAM and occlusion sensitivity visualisation of fused features for both tasks with the baseline of two models with attention mechanism.
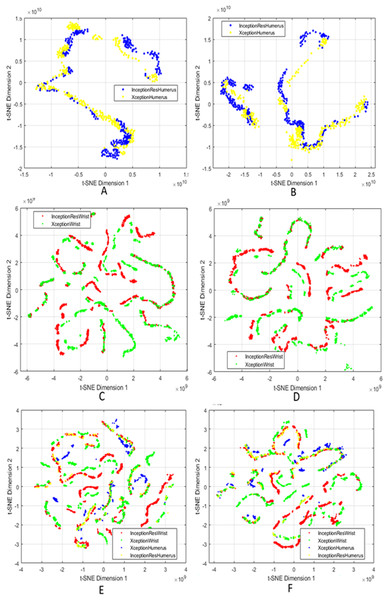
We used t-SNE to visualise the high-dimensional fused features generated after applying attention mechanisms, feature fusion, and SIFT-based selection. By projecting these features into two dimensions, t-SNE allows us to qualitatively assess the compactness within classes and the separability between classes. It is crucial to understand that t-SNE is used solely for visualisation and does not impact on model training, feature selection, or classification decisions.
Figure 3 presents a comprehensive series of t-SNE plots that depict how the fused features evolve with different processing stages. The progression across these subfigures highlights the effectiveness of combining attention, fusion, and feature selection in generating representations that are not only robust for classification but also interpretable in terms of latent structure. The integration of these XAI techniques underscores that the model’s predictions are informed by valid, domain-relevant features, both in terms of spatial saliency and latent feature structure. This enhances confidence in the reliability and clinical applicability of the proposed approach.
Figure 3: t-SNE visualisation of fused features after attention and SIFT extraction.
Points from the same class form tight clusters, showing separable representations. (A and B) Humerus task before and after feature selection; (C and D) Wrist task before and after selection; (E and F) Aggregated features from all models before and after selection.Comparison with the state-of-the-art
Table 9 presents the performance of the proposed method, which demonstrates superior efficacy compared with existing approaches by integrating attention mechanisms, optimised feature selection, and a majority voting ensemble strategy. The framework has achieved state-of-the-art results, reflecting a significant improvement in classification accuracy relative to previous studies. For example, Feng et al. (2021) reported an accuracy of 70.23%, followed by subsequent research indicating continuous advancements. Rath, Reddy & Singh (2022) attained an accuracy of 80.06%, while Zou & Arshad (2024) reached 86.20%. Additional improvements were documented by Alammar et al. (2023) at 87.85% and Alammar et al. (2024), who achieved 92.71%. More recently, Ounasser et al. (2023) and Sanzida Ferdous Ruhi, Nahar & Ferdous Ashrafi (2024) reported accuracies of 90.00% and 90.48%, respectively, highlighting a consistent trend towards increasingly robust and effective frameworks for X-ray image classification.
| Source | Accuracy | Recall | Precision | Specificity | F1-score | Kappa |
|---|---|---|---|---|---|---|
| Feng et al. (2021) | 70.23% | 68.23% | 70.25% | – | 67.55% | – |
| Rath, Reddy & Singh (2022) | 80.06% | – | – | – | – | – |
| Zou & Arshad (2024) | 86.20% | – | – | – | – | – |
| Alammar et al. (2023) | 87.85% | 88.57% | 86.71% | 87.16% | 87.63% | 75.69% |
| Oh, Hwang & Lee (2023) | 87.50% | – | – | – | – | – |
| Ounasser et al. (2023) | 90.00% | – | 90.50% | – | 90.70% | – |
| Sanzida Ferdous Ruhi, Nahar & Ferdous Ashrafi (2024) | 90.48% | – | – | – | – | – |
| Alammar et al. (2024) | 92.71% | 92.14% | 92.81% | 93.24% | 92.47% | 85.40% |
| Our proposal | 95.48% | 92.46% | 95.74% | 96.57% | 94.08% | 90.91% |
Although the proposed framework has demonstrated state-of-the-art performance, it is important to critically evaluate its limitations and potential generalisability in real-world clinical environments. While the model has shown robust results on the MURA dataset, its performance may be influenced by external datasets or clinical archives, which may present variations in factors such as image quality, resolution, and acquisition protocols. The dataset used in this study has a relatively balanced distribution of normal and abnormal cases. However, it is essential to recognise that real-world clinical data often exhibits significant class imbalances, especially for rare conditions such as uncommon fractures or degenerative changes, which are typically under-represented in these datasets. This discrepancy between the training data and actual prevalence may result in reduced model sensitivity for low-frequency diagnostic categories, ultimately limiting the framework’s overall effectiveness and reliability in routine clinical practice.
Validation of the proposed method with chest X-ray classification
This section outlines the validation process for the proposed framework in chest X-ray classification, utilising a newly curated dataset. The framework incorporates several advanced components designed to enhance both predictive accuracy and interpretability. For a structured evaluation, the dataset was partitioned into two distinct groups, each containing two diagnostic categories. This grouping strategy facilitates targeted feature learning and enables performance assessment across clinically relevant classification scenarios.
Chest X-ray dataset
The chest X-ray dataset used in this study was publicly available. To evaluate the performance and generalisability of the proposed classification framework, the dataset was organised into two distinct groups: COVID-19, normal, pneumonia, and tuberculosis. Data augmentation techniques were applied to increase dataset diversity and mitigate the risk of overfitting. For a thorough and impartial evaluation, each group was further divided into three subsets: training, validation, and test sets. The training set was used to optimise model parameters, while the validation set facilitated hyperparameter tuning and early stopping to prevent overfitting. The test set was reserved for the final performance evaluation. This partitioning strategy ensured a comprehensive assessment of the model’s ability to generalise across various respiratory conditions, while also maintaining robustness and mitigating the risk of catastrophic forgetting.
The two groups were defined as follows:
Group 1: COVID-19 and normal classes. This collection includes chest X-ray images classified into two categories: patients diagnosed with COVID-19 and individuals exhibiting no apparent lung pathology (normal). The dataset provides the model with an opportunity to identify and learn distinct features that differentiate COVID-19 abnormalities from healthy lung patterns. This supports binary classification within the context of viral infections.
Group 2: pneumonia and tuberculosis classes. This collection comprises chest X-ray images indicative of pneumonia, characterised by lung inflammation and fluid accumulation, as well as tuberculosis, which frequently manifests with nodules, infiltrates, or cavitary lesions. This combination facilitates an assessment of the model’s capability to differentiate between bacterial and mycobacterial pulmonary infections.
The diagnostic categories outlined in Table 10 were employed to assess the proposed framework’s capability to generalise across a variety of respiratory conditions. This evaluation focused on the model’s ability to accurately learn, retain, and transfer knowledge across multiple classification tasks. Additionally, the assessment sought to validate the model’s robustness and its resilience to catastrophic forgetting when confronted with diverse diagnostic categories.
| Training | Testing | |
|---|---|---|
| Group 1 | ||
| Covid19 | 460 | 106 |
| Normal | 1,341 | 234 |
| Group 2 | ||
| Pneumonia | 7,610 | 780 |
| Tuberculosis | 650 | 41 |
Results of the proposed framework solution with chest X-ray
This section outlines the experimental design and results of the proposed framework for chest X-ray classification. The dataset was divided into two groups, each comprising two diagnostic categories. The primary objective of these experiments was to assess the effectiveness of attention-based DL models, feature fusion strategies, and ensemble classification methods in accurately identifying respiratory conditions across both groups.
Initially, the Xception and InceptionResNetV2 models were trained on chest X-ray images using SSL. Both models were augmented with transformer blocks incorporating self-attention mechanisms, enabling them to focus on spatially and contextually significant regions within the images. This design enhanced the models’ ability to capture clinically relevant features.
Subsequently, feature fusion was applied to combine the learned representations from the two classes within each diagnostic group, resulting in Fusion 1 for Group 1 and Fusion 2 for Group 2. A feature selection technique was then employed to reduce dimensionality while preserving the most discriminative features from each fusion output. The selected features from both groups were merged into a unified combined fusion pool, representing a comprehensive set of patterns across the chest X-ray dataset.
This consolidated feature pool was used to train and evaluate multiple machine learning classifiers, including KNN, decision tree, and SVM. The classification performance of these classifiers is summarised in Table 11.
| Evaluation metric (%) | Tree | KNN | SVM | Final decision |
|---|---|---|---|---|
| Group 1 | ||||
| Accuracy | 99.40 | 99.70 | 100 | 99.40 |
| Recall | 99.60 | 99.50 | 100 | 99.60 |
| Precision | 99.10 | 99.80 | 100 | 99.10 |
| Specificity | 99.60 | 99.50 | 100 | 99.60 |
| F1-score | 99.30 | 99.70 | 100 | 99.30 |
| Cohen’s kappa | 98.64 | 99.31 | 100 | 98.64 |
| Group 2 | ||||
| Accuracy | 100 | 100 | 100 | 100 |
| Recall | 100 | 100 | 100 | 100 |
| Precision | 100 | 100 | 100 | 100 |
| Specificity | 100 | 100 | 100 | 100 |
| F1-score | 100 | 100 | 100 | 100 |
| Cohen’s kappa | 100 | 100 | 100 | 100 |
To improve robustness and generalisation, a majority voting strategy was implemented across the three classifiers, KNN, decision tree, and SVM. In this ensemble approach, each classifier independently predicts a class label, and the final prediction is determined by the class receiving the majority of votes. This method effectively mitigates the limitations of individual classifiers while capitalising on their complementary strengths.
The final ensemble decision, derived through majority voting, achieved an overall classification accuracy of 99.80% across the combined chest X-ray groups. This outstanding performance is summarised in Table 12. These results highlight the effectiveness of the proposed framework in reliably distinguishing between diagnostic categories, supported by a structured and interpretable learning pipeline.
| Evaluation metric | Majority voting of combination |
|---|---|
| Accuracy | 99.80 |
| Recall | 99.80 |
| Precision | 99.55 |
| Specificity | 99.80 |
| F1-score | 99.55 |
| Cohen’s kappa | 99.32 |
Experimental evaluation of attention layer in the CNN models for ablation study 1
This section provides an overview of integrating self-attention layers into two CNN models. In this study, we applied SSL on two distinct sets of chest X-ray images. The Test set results are presented in Table 13, the Training set results are in Table 14, and the Validation set results are shown in Table 15. Upon reviewing these three tables, it is evident that the models consistently exhibit strong performance across the training, validation, and test datasets. This observation suggests effective generalisation and indicates that there has been no significant overfitting.
| Evaluation metric | AttentionXception (Group 1) | AttentionInception (Group 1) |
|---|---|---|
| Accuracy | 97.40 | 98.50 |
| Recall | 98.10 | 98.20 |
| Precision | 96.10 | 98.40 |
| Specificity | 98.10 | 98.20 |
| F1-score | 97.10 | 98.30 |
| Cohen’s kappa | 93.97 | 96.56 |
| Evaluation metric | AttentionXception (Group 2) | AttentionInception (Group 2) |
| Accuracy | 99.80 | 100 |
| Recall | 98.80 | 100 |
| Precision | 99.90 | 100 |
| Specificity | 98.80 | 100 |
| F1-score | 99.30 | 100 |
| Cohen’s kappa | 98.64 | 100 |
| Evaluation metric | AttentionXception (Group 1) | AttentionInception (Group 1) |
|---|---|---|
| Accuracy | 100 | 99.20 |
| Recall | 100 | 98.50 |
| Precision | 100 | 99.50 |
| Specificity | 100 | 98.50 |
| F1-score | 100 | 99.00 |
| Cohen’s kappa | 100 | 97.97 |
| Evaluation metric | AttentionXception (Group 2) | AttentionInception (Group 2) |
| Accuracy | 100 | 100 |
| Recall | 100 | 100 |
| Precision | 100 | 100 |
| Specificity | 100 | 100 |
| F1-score | 100 | 100 |
| Cohen’s kappa | 100 | 100 |
| Evaluation metric | AttentionXception (Group 1) | AttentionInception (Group 1) |
|---|---|---|
| Accuracy | 99.70 | 99.20 |
| Recall | 99.50 | 98.40 |
| Precision | 96.80 | 99.40 |
| Specificity | 99.50 | 98.40 |
| F1-score | 99.60 | 98.90 |
| Cohen’s kappa | 99.27 | 97.79 |
| Evaluation metric | AttentionXception (Group 2) | AttentionInception (Group 2) |
| Accuracy | 99.97 | 99.90 |
| Recall | 100 | 99.60 |
| Precision | 99.97 | 100 |
| Specificity | 99.62 | 99.60 |
| F1-score | 99.98 | 99.80 |
| Cohen’s kappa | 99.80 | 99.58 |
Experimental evaluation of deep feature fusion for ablation study 2 (Chest X-ray)
This ablation study investigated the effect of deep feature fusion by combining attention-based DL models with descriptors within a task-oriented framework. Two attention-enhanced CNN architectures, Xception and InceptionResNetV2, were utilised for the classification of Group 1 and Group 2 chest X-ray images, with the attention layers activated to improve feature representation. To further augment the feature space, local descriptors were extracted using the SIFT method.
Each classification task (Group 1 and Group 2) was trained and evaluated independently using separate classifiers in a standard classification setup. The fused feature representations, combining deep-model outputs with SIFT descriptors, were used to train multiple machine learning classifiers, and their performance was systematically evaluated. The top-performing classifiers for each group were selected, with the results summarised in Table 16. The hybrid strategy, which combines attention-based deep features with SIFT descriptors, produced significant accuracy gains, achieving 98.50% for Group 1 and 100% for Group 2. These findings demonstrate that integrating feature extraction with DL substantially improves classification performance, validating the effectiveness of hybrid feature fusion in chest X-ray image analysis.
| Evaluation metric | Group 1 + ML classifier | Group 2 + ML classifier |
|---|---|---|
| Accuracy | 98.50 | 100 |
| Recall | 98.20 | 100 |
| Precision | 98.40 | 100 |
| Specificity | 98.20 | 100 |
| F1-score | 98.30 | 100 |
| Cohen’s kappa | 96.56 | 100 |
Experimental evaluation of deep feature fusion for ablation study 3 (Chest X-ray)
During this phase, we established two fusion pools designed to extract features from attention-enhanced models, specifically Xception and InceptionResNetV2, for Group 1 of the chest X-ray dataset. This methodology was similarly applied to Group 2, utilising the same models to maintain consistency across both diagnostic groups. To further enhance the feature representations, we implemented the SIFT, a technique recognised for its ability to detect and describe robust local features accurately. The integration of SIFT significantly improved the feature selection process by emphasising discriminative regions that are critical for classification.
The features derived from the four DL models, XceptionGroup1, InceptionResNetV2Group1, XceptionGroup2, and InceptionResNetV2Group2, were utilised as inputs for training multiple machine learning classifiers across both chest X-ray groups. As demonstrated in Table 17, there was a marked improvement in classification accuracy when applying the same classifier to both groups. Specifically, Group 1 achieved an accuracy of 98.53%, while Group 2 achieved 100% accuracy. These findings underscore the effectiveness of the proposed feature fusion and selection strategy, highlighting its capacity to enhance the performance of machine learning classifiers in chest X-ray classification tasks.
| Evaluation metric | Group 1 + ML classifier | Group 2 + ML classifier |
|---|---|---|
| Accuracy | 98.53 | 100 |
| Recall | 97.17 | 100 |
| Precision | 98.10 | 100 |
| Specificity | 99.15 | 100 |
| F1-score | 97.63 | 100 |
| Cohen’s kappa | 96.56 | 100 |
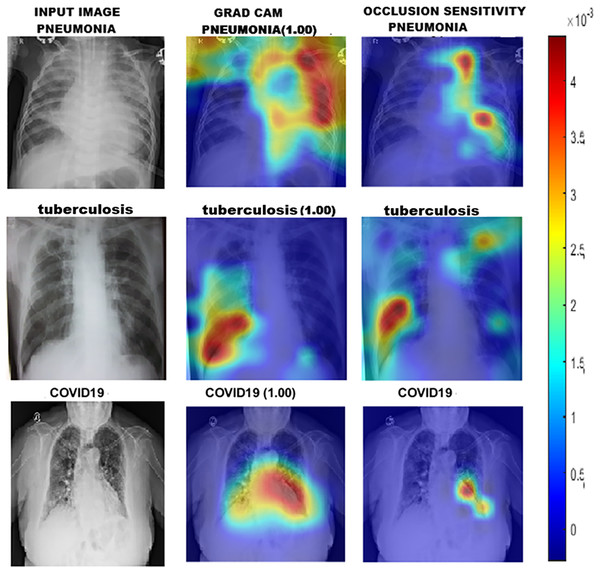
To gain a deeper understanding of how DL models classify chest X-ray images into Group 1 and Group 2, we utilised a range of interpretability techniques, including Grad-CAM, occlusion sensitivity, and t-SNE visualisation. Grad-CAM and Occlusion Sensitivity were employed to identify the specific regions within each image that significantly influenced the model’s predictions. These techniques highlighted the areas on which the models relied most for classification, confirming their focus on clinically relevant structures. The resulting visual explanations are presented in Fig. 4.
Figure 4: GRAD-CAM and occlusion sensitivity visualisation of fused features for both groups of chest X-ray.
Finally, we utilised t-SNE to project high-dimensional feature representations into a two-dimensional space as illustrated in Fig. 5. The resulting t-SNE plot revealed well-separated clusters for both chest X-ray groups, indicating that the model successfully learned distinct, discriminative features for each class. This combination of visualisation techniques enhances the model’s interpretability and fosters trust in its clinical reliability.
Figure 5: t-SNE visualisation of fused features.
(A and B) Group 1 before and after feature selection; (C and D) Group 2 before and after selection; (E and F) Combined models (Groups 1 and 2) before and after selection.Discussion
This study demonstrates the effectiveness of integrating attention mechanisms into CNNs for classifying musculoskeletal radiographs, including X-rays of the humerus, wrist, and two diagnostic groups of chest X-rays. The proposed end-to-end framework addresses challenges such as image noise and anatomical complexity by combining the local feature extraction capabilities of CNNs with the global contextual awareness offered by attention mechanisms. This architecture enables more precise identification of abnormalities and consistently achieves strong predictive performance across all evaluated tasks.
Implications
The proposed framework not only achieves high classification accuracy but also significantly enhances interpretability, a crucial factor for clinical adoption. By effectively identifying diagnostically relevant areas within noisy or ambiguous X-ray images, the model serves as a dependable decision-support tool for clinicians. Additionally, its demonstrated generalisability across various medical imaging domains indicates its broad applicability to a wide range of diagnostic tasks. With classification accuracies of 95.48% on MURA X-ray images and 99.80% on chest X-rays, the framework demonstrates considerable promise in supporting radiologists in high-stakes clinical environments, ultimately enhancing both diagnostic confidence and efficiency.
Limitations
The proposed framework, although demonstrating promising results, presents certain limitations. The integration of multiple attention layers within a CNN can introduce practical challenges, such as increased model complexity, heightened computational overhead, and extended training durations. This computational intensity could potentially impede deployment in environments with limited resources, especially where access to high-performance hardware is constrained. Moreover, the framework’s lack of integration with non-imaging data, such as patient history, laboratory results, and clinical notes, restricts its capacity to incorporate essential contextual information, which is vital for making accurate diagnoses and informed decisions in clinical practice.
Future work
Future efforts will focus on optimising the model for enhanced efficiency and smooth integration into clinical workflows. Additionally, validation within real-world settings will be prioritised to ensure the model’s robustness. Subsequent iterations will integrate multimodal data, including laboratory results and patient history, to facilitate more comprehensive clinical decision-making.
Conclusion
This study introduces a robust attention-enhanced CNN framework designed for the classification of X-ray images. The proposed framework demonstrates high accuracy and strong interpretability, even in the presence of noise and structural complexity. While there are notable limitations, such as the computational demands and the necessity for well-curated datasets, the model exhibits significant promise as a dependable tool for medical image analysis. Continued refinement, particularly in real-world clinical environments, is essential for maximising its potential.