Adaptive machine learning approaches utilizing soft decision-making via intuitionistic fuzzy parameterized intuitionistic fuzzy soft matrices
- Published
- Accepted
- Received
- Academic Editor
- Dragan Pamucar
- Subject Areas
- Algorithms and Analysis of Algorithms, Data Mining and Machine Learning, Optimization Theory and Computation
- Keywords
- Intuitionistic fuzzy sets, Soft sets, ifpifs-matrices, Soft decision-making, Machine learning
- Copyright
- © 2025 Memiş et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ Computer Science) and either DOI or URL of the article must be cited.
- Cite this article
- 2025. Adaptive machine learning approaches utilizing soft decision-making via intuitionistic fuzzy parameterized intuitionistic fuzzy soft matrices. PeerJ Computer Science 11:e2703 https://doi.org/10.7717/peerj-cs.2703
Abstract
The exponential data growth generated by technological advancements presents significant challenges in analysis and decision-making, necessitating innovative and robust methodologies. Machine learning has emerged as a transformative tool to address these challenges, especially in scenarios requiring precision and adaptability. This study introduces two novel adaptive machine learning approaches, i.e., AIFPIFSC1 and AIFPIFSC2. These methods leverage the modeling ability of intuitionistic fuzzy parameterized intuitionistic fuzzy soft matrices (ifpifs-matrices). This state-of-the-art framework enhances the classification task in machine learning by employing soft decision-making through ifpifs-matrices. The proposed approaches are rigorously evaluated against leading fuzzy/soft-based classifiers using 15 widely recognized University of California, Irvine datasets, including accuracy and robustness, across six performance metrics. Statistical analyses conducted using Friedman and Nemenyi tests further substantiate the reliability and superiority of the proposed approaches. The results consistently demonstrate that these approaches outperform their counterparts, highlighting their potential for solving complex classification problems. This study contributes to the field by offering adaptable and effective solutions for modern data analysis challenges, paving the way for future advancements in machine learning and decision-making systems.
Introduction
The amount of data made possible by technological advancement is constantly growing. This increasing amount of data may be analyzed and interpreted using machine learning, a technical improvement. Numerous industries frequently use this technology, including defense, finance, psychology, medicine, meteorology, astronomy, and space sciences. Such fields are encountered with many uncertainties. Several mathematical tools, such as fuzzy sets (Zadeh, 1965) and intuitionistic fuzzy sets (Atanassov, 1986), have been propounded to model these uncertainties. Furthermore, modeling such uncertainties is a crucial process to enhance the performance of machine learning algorithms. To this end, many classical machine learning algorithms, such as k-nearest neighbor (kNN) (Fix & Hodges, 1951; Keller, Gray & Givens, 1985), have been successfully modified as fuzzy kNN (Keller, Gray & Givens, 1985) using fuzzy sets.
Although fuzzy sets provide a mathematical framework for dealing with uncertainties where classical sets are inadequate, soft sets (Molodtsov, 1999) further this approach by offering additional flexibility in modeling uncertainties. In the last two decades, these concepts have evolved into various hybrid forms such as fuzzy soft sets (Maji, Biswas & Roy, 2001a), fuzzy parameterized soft sets (Çağman, Çıtak & Enginoğlu, 2011), and fuzzy parameterized fuzzy soft sets (fpfs-sets) (Çağman, Çıtak & Enginoğlu, 2010), which have manifested their utility in modeling scenarios where parameters or objects possess fuzzy values.
Developing fuzzy parameterized fuzzy soft matrices (fpfs-matrices) (Enginoğlu & Çağman, 2020) ensures a significant advancement in the field, particularly in computerizing decision-making scenarios involving a large amount of data. It has been applied to performance-based value assignment problems in image denoising by employing generalized fuzzy soft max-min decision-making method (Enginoğlu, Memiş & Çağman, 2019), and operability-configured soft decision-making methods (Enginoğlu et al., 2021; Enginoğlu & Öngel, 2020) and classification problems in machine learning (Memiş & Enginoğlu, 2019). Moreover, classification algorithms based on soft decision-making methods constructed by fpfs-matrices have been suggested, namely FPFS-CMC (Memiş, Enginoğlu & Erkan, 2022a) and FPFS-AC (Memiş, Enginoğlu & Erkan, 2022c). However, fpfs-matrices lack modeling ability for intuitionistic fuzzy uncertainties. Therefore, the concepts of intuitionistic fuzzy soft sets (ifs-sets) (Maji, Biswas & Roy, 2001b), intuitionistic fuzzy parameterized soft sets (ifps-sets) (Deli & Çağman, 2015), and intuitionistic fuzzy parameterized fuzzy soft sets (ifpfs-sets) (El-Yagubi & Salleh, 2013) have been proposed. In addition, intuitionistic fuzzy parameterized intuitionistic fuzzy soft sets (ifpifs-sets) (Karaaslan, 2016) and matrices (ifpifs-matrices) (Enginoğlu & Arslan, 2020) have enabled the modeling of problems with both parameters and objects containing intuitionistic fuzzy uncertainties. Besides, various mathematical tools, such as bipolar soft sets (Mahmood, 2020), intuitionistic fuzzy mean operators (Hussain et al., 2023), and picture fuzzy soft matrices (Memiş, 2023b), have been proposed to deal with uncertainty and are still being studied. However, when the related literature is investigated, it is noteworthy that there are almost no applications, especially in real-world problems/data (Bustince Sola et al., 2016; Karakoç, Memiş & Sennaroglu, 2024). Although several applications have recently been conducted to the classification problem in machine learning, which is a real problem modeled by fpfs-matrices, only a few studies employ ifpifs-matrices in machine learning (Memiş et al., 2021). Therefore, applying ifpifs-matrices, a pioneer mathematical tool to model uncertainty, to machine learning is a topic worthy of study.
Recently, Memiş et al. (2021) have defined metrics, quasi-metrics, semi-metrics, and pseudo-metrics, as well as similarities including quasi-, semi-, and pseudo-similarities over ifpifs-matrices. Besides, Memiş et al. (2021) has proposed an Intuitionistic Fuzzy Parameterized Intuitionistic Fuzzy Soft Classifier (IFPIFSC), which utilizes six pseudo-similarities of ifpifs-matrices. This classifier has been simulated using 20 datasets from the University of California, Irvine Machine Learning Repository (UCI-MLR) (Kelly, Longjohn & Nottingham, 2024). Its performance has been evaluated using six metrics: accuracy (Acc), precision (Pre), recall (Rec), specificity (Spe), macro F-score (MacF), and micro F-score (MicF). The results have manifested that IFPIFSC has outperformed fuzzy and non-fuzzy-based classifiers in these metrics. Although machine learning algorithms have been suggested via ifpifs-matrices for real problems, machine learning datasets in UCI-MLR and other databases usually consist of real-valued raw data. In these datasets, no serious uncertainties, such as intuitionistic uncertainty, can be modeled with mathematical tools. However, our aim in this study is to propose a new machine learning algorithm that can work with a dataset that contains intuitionistic fuzzy uncertainties by first converting the raw data to fuzzy values and then to intuitionistic fuzzy values. On the other hand, the most significant disadvantage of the two algorithms described above based on ifpifs-matrices, i.e., IFPIFSC and IFPIFS-HC, is that working by fixed and values. In this article, instead of fixed and values, we aim to develop an equation that allows the algorithm to determine these values based on the dataset and a machine learning algorithm that can work adaptively using this equation. Moreover, the soft decision-making methods constructed by ifpifs-matrices, one of the efficacious decision-making techniques, can be integrated into the machine learning process. As a result, in this study, we focus on proposing a machine learning algorithm based on pseudo-similarities of ifpifs-matrices, adaptive and values for intuitionistic fuzzification of the data, and soft decision-making methods constructed by ifpifs-matrices. The significant contributions of the article can be summed up as follows:
Improving two adaptive machine learning algorithms employing ifpifs-matrices,
Utilizing two soft decision-making methods constructed by ifpifs-matrices in machine learning.
The fact that this article is one of the pioneer studies combining soft sets, intuitionistic fuzzy sets, and machine learning.
Applying the similarity measures of ifpifs-matrices to classification problems in machine learning, in contrast to many soft set-based studies on hypothetical problems.
A key innovation in our approach is the adaptive modification of lambda values in the classification algorithms, which significantly enhances the adaptability and Acc of the classifiers. By integrating ifpifs-matrices with two classification algorithms and dynamically adjusting lambda values, our method represents a novel contribution to machine learning. This adaptive approach not only addresses the limitations of previous classifiers but also demonstrates superior performance in handling uncertainties and dynamic natures of real-world data, setting a new benchmark for future research in the domain.
The rest of the present study is organized as follows: The second section provides some fundamental definitions, notations, and algorithms to be needed for the following sections. The third section presents the basic definitions for the two proposed algorithms and their algorithmic designs. The fourth section details the utilized datasets and performance metrics. Secondly, it simulates the proposed algorithms with the well-known and state-of-the-art fuzzy and soft sets/matrices-based classification algorithms. Finally, it statistically analyzes the simulation results using the Friedman and Nemenyi tests. The final section discusses the proposed approaches, their performance results, and the need for further research.
Preliminaries
This section first introduces the notion of ifpifs-matrices (Enginoğlu & Arslan, 2020) and several of its fundamental characteristics. During this research, let E and U represent a parameter set and an alternative (object) set, respectively.
Definition 1 (Atanassov, 1986) Let and represent two functions from E to such that , for all . The set is referred to as an intuitionistic fuzzy set (if-set) over E.
Here, for all , and are called the membership and non-membership degrees, respectively. In addition, the indeterminacy degree of is defined by .
Across the study, the set of all the if-sets over E is denoted by IF(E) and . Briefly, the notation can be employed instead of . Thus, an if-set over E can be denoted by
Definition 2 (Karaaslan, 2016) Let and be a function from to IF(U). Then, the set is called an ifpifs-set parametrized via E over U (or over U for brevity) and denoted by .
In the present study, the set of all the ifpifs-sets over U is denoted by .
Definition 3 (Enginoğlu & Arslan, 2020) Let . Then, is called ifpifs-matrix of and is defined by
such that and , or briefly . Here, if and , then is an ifpifs-matrix.
Hereinafter, the set of all the ifpifs-matrices over U is denoted by .
Secondly, we provide iCCE10 (Arslan et al., 2021) and isMBR01 (Arslan et al., 2021) in Algorithms 1 and 2, respectively, by considering the notations used across this study.
| Input: ifpifs-matrix |
| Output: Score matrix , Optimum alternatives |
| 1: |
| 2: |
| 3: for i from 2 to m do |
| 4: for j from 1 to n do |
| 5: |
| 6: |
| 7: end for |
| 8: |
| 9: |
| 10: end for |
| 11: |
| 12: |
| 13: for i from 1 to do |
| 14: if AND then |
| 15: |
| 16: else |
| 17: |
| 18: end if |
| 19: end for |
| 20: |
| Input: ifpifs-matrix |
| Output: Score matrix , Optimum alternatives |
| 1: and |
| 2: for i from 1 to do |
| 3: for k from 1 to do |
| 4: for j from 1 to n do |
| 5: |
| 6: |
| 7: end for |
| 8: end for |
| 9: end for |
| 10: |
| 11: for i from 1 to do |
| 12: if AND then |
| 13: |
| 14: |
| 15: else |
| 16: |
| 17: |
| 18: end if |
| 19: end for |
| 20: |
| 21: for i from 1 to do |
| 22: |
| 23: end for |
| 24: |
| 25: for i from 1 to do |
| 26: |
| 27: end for |
| 28: |
Proposed adaptive machine learning approaches
This section overviews the fundamental mathematical notations essential for the proposed classifier based on ifpifs-matrices. In this article, we represent the data with a matrix , where represents the number of samples, is the number of parameters, and the last column of D contains the labels for the data. The training data matrix, denoted as , along with the corresponding class matrix , is used to generate a testing matrix derived from the original data matrix D, where . Additionally, we employ the matrix to stand for the unique class labels extracted from . Notably, and refer to the -th rows of and , respectively. Similarly, and denote the -th columns of and . Furthermore, represents the predicted class labels for the testing samples.
Definition 4 Let . Then, is a function defined by
is denoted as the Pearson correlation coefficient for the variables and .
Definition 5 Let and . The normalizing vector of is defined as , where
Definition 6 Let be a data matrix, , and . A column normalized matrix of D is defined by , where
Definition 7 Let be a training matrix obtained from , , and . A column normalized matrix of is defined by , where
Definition 8 Let be a testing matrix obtained from , , and . A column normalized matrix of is defined by , where
Definition 9 (Memiş et al., 2021) Let and be a training matrix and its class matrix obtained from a data matrix , respectively. Then, the matrix is called intuitionistic fuzzification weight (ifw) matrix based on Pearson correlation coefficient of and defined by
and
such that and .
Definition 10 (Memiş et al., 2021) Let be a column normalized matrix of a matrix . Then, the matrix is called intuitionistic fuzzification of and defined by
and
such that , , and .
Definition 11 (Memiş et al., 2021) Let be a column normalized matrix of a matrix . Then, the matrix is called intuitionistic fuzzification of and defined by
and
such that , , and .
Definition 12 (Memiş et al., 2021) Let be a column normalized matrix of a matrix and be the intuitionistic fuzzification of . Then, the ifpifs-matrix is called the training ifpifs-matrix obtained by -th row of and and defined by
such that and .
Definition 13 (Memiş et al., 2021) Let be a column normalized matrix of a matrix and be the intuitionistic fuzzification of . Then, the ifpifs-matrix is called the testing ifpifs-matrix obtained by -th row of and and defined by
such that and .
Secondly, it presents the concept of pseudo-similarities over and seven pseudo-similarities over .
Definition 14 (Memiş et al., 2021) Let be a mapping. Then, is a pseudo-similarity over if and only if satisfies the following properties for all , , :
(i) ,
(ii) ,
(iii) .
Thirdly, this part provides the Minkowski, Hamming, Euclidean, Hamming-Hausdorff, Chebyshev, Jaccard, and Cosine pseudo-similarities over using the normalized pseudo-metrics of ifpifs-matrices (For more details see (Memiş et al., 2023)).
Proposition 15 (Memiş et al., 2023) Let . Then, the mapping defined by
is a pseudo-similarity and referred to as Minkowski pseudo-similarity.
In this case, and are represented by and , respectively, and are referred to as Hamming pseudo-similarity (Memiş et al., 2021) and Euclidean pseudo-similarity.
Proposition 16 (Memiş et al., 2023) Let . Then, the mapping defined by
is a pseudo-similarity and referred to as Hamming-Hausdorff pseudo-similarity.
Proposition 17 (Memiş et al., 2023) Let . Then, the mapping defined by
is a pseudo-similarity and is referred to as Chebyshev pseudo-similarity.
Proposition 18 (Memiş et al., 2023) The mapping defined by
such that
and
is known as the Jaccard pseudo-similarity, and it is a pseudo-similarity. Here, , for example, .
Proposition 19 (Memiş et al., 2023) The mapping defined by
such that
and
is known as the Cosine pseudo-similarity, and it is a pseudo-similarity. Here, , for example, .
Fourthly, it propounds the classification algorithms AIFPIFSC1 and AIFPIFSC2 and provides the pseudocodes of normalize and intuitionistic normalize functions in Algorithms 3 and 4 to be needed for the proposed algorithms’ pseudocodes (Algorithms 5 and 6).
| Input: |
| Output: |
| 1: function normalize(a) |
| 2: |
| 3: if then |
| 4: |
| 5: else |
| 6: ones(m, n) |
| 7: end if |
| 8: return |
| 9: end function |
| Input: , λ |
| Output: |
| 1: function inormalize |
| 2: |
| 3: for to m do |
| 4: for to n do |
| 5: |
| 6: |
| 7: end for |
| 8: end for |
| 9: return |
| 10: end function |
| Input: , , and |
| Output: |
| 1: function AIFPIFSC1(train, C, test) |
| 2: |
| 3: |
| 4: Compute using and |
| 5: Compute feature fuzzification of Dtrain and Dtest, namely and |
| 6: Compute and |
| 7: for i from 1 to tm do |
| 8: Compute test ifpifs-matrix using and |
| 9: for j from 1 to em do |
| 10: Compute train ifpifs-matrix using and |
| 11: |
| 12: |
| 13: |
| 14: |
| 15: |
| 16: |
| 17: |
| 18: end for |
| 19: for j from 1 to do |
| 20: |
| 21: end for |
| 22: |
| 23: |
| 24: |
| 25: |
| 26: |
| 27: end for |
| 28: end function |
| Input: , , and |
| Output: |
| 1: function AIFPIFSC2(train, C, test) |
| 2: |
| 3: |
| 4: Compute using and |
| 5: Compute feature fuzzification of Dtrain and Dtest, namely and |
| 6: Compute and |
| 7: for i from 1 to tm do |
| 8: Compute test ifpifs-matrix using and |
| 9: for j from 1 to em do |
| 10: Compute train ifpifs-matrix using and |
| 11: |
| 12: |
| 13: |
| 14: |
| 15: |
| 16: |
| 17: |
| 18: end for |
| 19: for j from 1 to do |
| 20: |
| 20: end for |
| 21: |
| 22: |
| 23: |
| 24: |
| 25: |
| 26: end for |
| 27: end function |
In the algorithm AIFPIFSC1, parameters and are determined based on the dataset’s characteristics. Then, ifw is computed by measuring the Pearson correlation coefficient between each feature and the class labels. These weights are utilized to construct two ifpifs-matrices: one for training data and the other for testing data. Feature fuzzification of the training data features is obtained using the ifw. For each testing sample, a comparison matrix is constructed by calculating pseudo-similarities between the testing ifpifs-matrix and training ifpifs-matrix. This comparison matrix serves as the basis for parameter weights’ computation. Parameter weights are determined by calculating the standard deviation of each column in the comparison matrix. A comparison ifpifs-matrix is then generated by combining these parameter weights with the comparison matrix. Next, we apply the algorithm isMBR01 to this comparison ifpifs-matrix to identify the optimum training sample. The class label of this optimum training sample is assigned to the corresponding testing sample. These steps are repeated for all testing samples to obtain the predicted class labels.
The similar steps in Algorithm 6 are repeated for the algorithm AIFPIFSC2. In the last part, we apply the algorithm iCCE10 to the aforesaid comparison ifpifs-matrix to determine the optimum training sample.
Finally, an illustrative example is presented below to enhance the clarity of the AIFPIFSC1 algorithm concerning its pseudocode. In this study, we use -fold cross-validation to avoid the possibility of the algorithm’s results being overly optimistic in a single run.
Illustrative Example:
A data matrix from the “Iris” dataset (Fisher, 1936) is provided below for implementing AIFPIFSC1. The matrix contains 10 samples divided into three classes ( ), with the class labels in the last column. Class one consists of three samples, class two has three samples, and class three includes four. In the first iteration of five-fold cross-validation, , , , and are obtained as follows:
, C, and are inputted to AIFPIFSC1. After the classification task, the ground truth class matrix T is utilized to calculate the Acc, Pre, Rec, Spe, MacF, and MicF rates of AIFPIFSC1.
Secondly, and are calculated as and , respectively.
Thirdly, the ifw matrix is calculated via and C concerning the Pearson correlation coefficient. The main goal of obtaining the ifw matrix is to construct the train and test ifpifs-matrices in the following phases.
Thirdly, feature fuzzifications of and are computed as follows:
Fourthly, intuitionistic feature fuzzifications of and are computed as follows:
In the next steps, and are required to constructing the train and test ifpifs-matrices.
For , the following steps are performed:
Fifthly, for all , test ifpifs-matrix and train ifpifs-matrix are constructed by utilizing and . For instance ( ), for and , test ifpifs-matrix and -train ifpifs-matrix are constructed as follows:
The first rows of and are the intuitionistic feature weights obtained in the third step. Second rows of and are the first row (first sample) of and first row (first sample) of , respectively.
Sixtly, , , , , , , and of CM are computed by employing the pseudo-similarities of the aforesaid and as follows:
Then, CM is calculated as follows:
Seventhly, the standard deviation matrix and second weight matrix are computed as follows:
and
Eighthly, the decision matrix DM is obtained by employing CM and as follows:
Ninthly, the intuitionistic decision matrix is computed as follows:
Tenthly, the soft decision-making algorithm isMBR01 based on ifpifs-matrices is applied to and determines the optimum train sample.
Finally, the label of the optimum train sample is assigned to . Because the optimum train sample is the fifth, its label is assigned to .
If is calculated as is, then the predicted class matrix is attained as . According to these results, the Acc rate of the proposed algorithm is for the considered example.
Simulation and performance comparison
The current section provides a comprehensive overview of the 15 datasets within the UCI-MLR for classification tasks. Six performance metrics that can be used to compare performances are then presented. Subsequently, a simulation is conducted to illustrate that AIFPIFSC1 and AIFPIFSC2 outperform Fuzzy kNN, FPFS-kNN, FPFS-AC, FPFS-EC, FPFS-CMC, IFPIFSC, and IFPIFS-HC in terms of classification performance. Furthermore, the section conducts statistical analyses on the simulation results using the Friedman test, a non-parametric test, and the Nemenyi test, a post hoc test.
UCI datasets and features
This subsection provides the characteristics of the datasets utilized in the simulation, as outlined in Table 1. The datasets in Table 1 can be accessed from the UCI-MLR (Kelly, Longjohn & Nottingham, 2024).
| No. | Ref. | Name | #Instance | #Attribute | #Class | Balanced/Imbalanced |
|---|---|---|---|---|---|---|
| 1 | Nakai (1996) | Ecoli | 336 | 7 | 8 | Imbalanced |
| 2 | Silva & Maral (2013) | Leaf | 340 | 14 | 36 | Imbalanced |
| 3 | Dias, Peres & Bscaro (2009) | Libras | 360 | 90 | 15 | Balanced |
| 4 | Higuera, Gardiner & Cios (2015) | Mice | 1,077 | 72 | 8 | Imbalanced |
| 5 | Lichtinghagen, Klawonn & Hoffmann (2020) | HCV Data | 589 | 12 | 5 | Imbalanced |
| 6 | Barreto & Neto (2005) | Column3C | 310 | 6 | 3 | Imbalanced |
| 7 | Quinlan (1986) | NewThyroid | 215 | 5 | 3 | Imbalanced |
| 8 | Deterding, Niranjan & Robinson (1988) | Vowel | 990 | 13 | 11 | Imbalanced |
| 9 | Fisher (1936) | Iris | 150 | 4 | 3 | Imbalanced |
| 10 | Charytanowicz et al. (2010) | Seeds | 210 | 7 | 3 | Balanced |
| 11 | Bhatt (2017) | Wireless | 2,000 | 7 | 4 | Balanced |
| 12 | Aeberhard & Forina (1992) | Wine | 178 | 13 | 3 | Imbalanced |
| 13 | Cardoso (2013) | WholesaleR | 440 | 6 | 3 | Imbalanced |
| 14 | JP & Jossinet (1996) | Breast Tissue | 106 | 9 | 6 | Imbalanced |
| 15 | Breiman et al. (1984) | Led7Digit | 500 | 7 | 7 | Imbalanced |
Note:
# represents “the number of”.
The main problems related to each dataset in Table 1 are as follows: In “Ecoli” predicting the localization site of Ecoli proteins based on biological attributes. In “Leaf”, different species of plants are classified using leaf shape and texture features. In “Libras, ” accelerometer data recognizes hand movements in Libras (a Brazilian sign language). In “Mice”, gene expression data is analyzed to distinguish between mouse strains. In “HCV Data”, detect Hepatitis C Virus (HCV) infection stages using blood test data. In “Column 3C”, diagnosing spinal disorders by analyzing biomechanical features of vertebrae. In “NewThyroid”, thyroid conditions (e.g., normal or hypothyroid) are classified based on medical attributes. In “Vowel”, spoken vowels are classified using speech acoustics data. In “Iris”, identify iris species based on flower petal and sepal dimensions. In “Seeds”, classify types of wheat seeds based on their geometric properties. In “Wireless”, predict indoor locations based on Wi-Fi signal strength features. In “Wine”, distinguish wine varieties based on their chemical compositions. In “Wholesale”, predict customer segments based on annual spending on various product categories. In “Breast Tissue”, types of breast tissue samples (e.g., healthy or tumor) are classified using bioimpedance data. In “Led7Digit”, human errors in recognizing handwritten digits are simulated on a seven-segment display.
Performance metrics
This subsection outlines the mathematical notations for six performance metrics (Erkan, 2021; Fawcett, 2006; Nguyen et al., 2019), namely Acc, Pre, Rec, Spe, MacF, and MicF, used to compare the mentioned classifiers. Let , , , and be samples for classification, ground truth class sets of the samples, prediction class sets of the samples, and the number of classes for the samples, respectively. Here,
where the numbers for true positive, true negative, false positive, and false negative for the class are, respectively, , , , and , respectively. Their mathematical notations are as follows:
Here, the notation denotes the cardinality of a set.
Simulation results
This subsection conducts a comparative analysis between AIFPIFSC1 and AIFPIFSC2 and well-established classifiers that rely on fuzzy and soft sets, such as Fuzzy kNN, FPFS-kNN, FPFS-AC, FPFS-EC, FPFS-CMC, IFPIFSC, and IFPIFS-HC. The comparison is performed using MATLAB R2021b (The MathWorks, Natick, NY, USA) on a laptop with an Intel(R) Core(TM) i5-10300H CPU @ 2.50 GHz and 8 GB RAM. The mean performance results of the classifiers are derived from random independent runs based on -fold cross-validation (Stone, 1974). In each cross-validation iteration, the dataset is randomly divided into five parts, whose four parts are used for training and the remaining part for testing (for further details on k-fold cross-validation, refer to Stone (1974)). Table 2 presents the average Acc, Pre, Rec, Spe, MacF, and MicF results for AIFPIFSC1, AIFPIFSC2, Fuzzy kNN, FPFS-kNN, FPFS-AC, FPFS-EC, FPFS-CMC, IFPIFSC, and IFPIFS-HC across the datasets.
| Datasets | Classifiers | Acc | Pre | Rec | Spe | MacF | MicF |
|---|---|---|---|---|---|---|---|
| Ecoli | Fuzzy kNN | 92.1824 | 53.5997 | 60.2825 | 95.6846 | 66.3808 | 68.7296 |
| FPFS-kNN | 94.4340 | 72.9682 | 65.3519 | 96.4741 | 74.9499 | 81.1010 | |
| FPFS-AC | 94.2172 | 72.7755 | 68.6908 | 96.3172 | 75.8222 | 79.3758 | |
| FPFS-EC | 94.2085 | 70.1592 | 67.0879 | 96.3468 | 74.7286 | 79.2559 | |
| FPFS-CMC | 94.0483 | 69.1430 | 66.2550 | 96.2559 | 73.5161 | 78.6602 | |
| IFPIFSC | 92.2406 | 63.4527 | 60.4435 | 95.1167 | 68.6435 | 72.5285 | |
| IFPIFS-HC | 92.6453 | 64.7535 | 62.4497 | 95.4705 | 69.8072 | 73.6901 | |
| AIFPIFSC1 | 94.6825 | 76.8664 | 71.1918 | 96.6138 | 78.6652 | 81.3745 | |
| AIFPIFSC2 | 94.4892 | 76.1900 | 70.1926 | 96.4886 | 77.8141 | 80.6892 | |
| Leaf | Fuzzy kNN | 96.2016 | 32.5347 | 32.0444 | 98.0528 | 62.0025 | 32.7941 |
| FPFS-kNN | 97.8353 | 73.3337 | 67.4556 | 98.8794 | 73.8272 | 67.5294 | |
| FPFS-AC | 97.8569 | 71.8471 | 67.6889 | 98.8910 | 74.4906 | 67.8529 | |
| FPFS-EC | 97.8039 | 71.8566 | 66.9333 | 98.8633 | 73.7364 | 67.0588 | |
| FPFS-CMC | 97.7686 | 71.2570 | 66.5444 | 98.8451 | 73.4777 | 66.5294 | |
| IFPIFSC | 97.7608 | 71.0219 | 65.9667 | 98.8419 | 72.6249 | 66.4118 | |
| IFPIFS-HC | 97.7176 | 70.1101 | 65.4611 | 98.8193 | 72.3277 | 65.7647 | |
| AIFPIFSC1 | 98.0784 | 75.5176 | 70.7778 | 99.0055 | 75.9554 | 71.1765 | |
| AIFPIFSC2 | 98.0843 | 75.2774 | 70.9778 | 99.0086 | 76.2688 | 71.2647 | |
| Libras | Fuzzy kNN | 95.8963 | 74.0815 | 69.2600 | 97.8017 | 70.1990 | 69.2222 |
| FPFS-kNN | 96.7037 | 79.3345 | 75.2267 | 98.2343 | 75.3681 | 75.2778 | |
| FPFS-AC | 97.2815 | 82.1165 | 79.6200 | 98.5435 | 79.3057 | 79.6111 | |
| FPFS-EC | 97.0667 | 80.8036 | 78.0400 | 98.4288 | 78.0580 | 78.0000 | |
| FPFS-CMC | 96.9481 | 79.8471 | 77.1400 | 98.3655 | 77.2585 | 77.1111 | |
| IFPIFSC | 95.9704 | 73.5069 | 69.9000 | 97.8417 | 70.0397 | 69.7778 | |
| IFPIFS-HC | 96.1889 | 74.5579 | 71.4867 | 97.9589 | 71.0616 | 71.4167 | |
| AIFPIFSC1 | 97.3444 | 82.8129 | 80.1467 | 98.5777 | 79.7321 | 80.0833 | |
| AIFPIFSC2 | 97.2593 | 82.6086 | 79.5267 | 98.5321 | 79.3585 | 79.4444 | |
| Mice | Fuzzy kNN | 99.8608 | 99.4865 | 99.4429 | 99.9200 | 99.4470 | 99.4431 |
| FPFS-kNN | 100 | 100 | 100 | 100 | 100 | 100 | |
| FPFS-AC | 100 | 100 | 100 | 100 | 100 | 100 | |
| FPFS-EC | 100 | 100 | 100 | 100 | 100 | 100 | |
| FPFS-CMC | 100 | 100 | 100 | 100 | 100 | 100 | |
| IFPIFSC | 100 | 100 | 100 | 100 | 100 | 100 | |
| IFPIFS-HC | 100 | 100 | 100 | 100 | 100 | 100 | |
| AIFPIFSC1 | 100 | 100 | 100 | 100 | 100 | 100 | |
| AIFPIFSC2 | 100 | 100 | 100 | 100 | 100 | 100 | |
| HCV Data | Fuzzy kNN | 97.0934 | 55.1835 | 48.1424 | 96.0803 | 64.1767 | 92.7334 |
| FPFS-kNN | 96.9913 | 56.7283 | 38.1858 | 90.3967 | 84.7871 | 92.4782 | |
| FPFS-AC | 97.7862 | 70.9108 | 53.5449 | 94.7062 | 74.2838 | 94.4654 | |
| FPFS-EC | 97.0867 | 59.1956 | 46.0910 | 91.5718 | 77.9663 | 92.7168 | |
| FPFS-CMC | 97.0798 | 62.4317 | 47.7072 | 91.6225 | 75.4864 | 92.6994 | |
| IFPIFSC | 97.7930 | 67.9679 | 55.7708 | 95.5895 | 75.1353 | 94.4824 | |
| IFPIFS-HC | 97.8271 | 67.3802 | 54.1163 | 95.8067 | 76.3137 | 94.5677 | |
| AIFPIFSC1 | 98.1460 | 77.3406 | 62.4411 | 95.6001 | 77.2945 | 95.3649 | |
| AIFPIFSC2 | 98.0916 | 77.7644 | 62.1039 | 95.1745 | 76.7595 | 95.2289 | |
| Column3c | Fuzzy kNN | 80.4086 | 75.9644 | 61.3111 | 82.1301 | 63.4923 | 70.6129 |
| FPFS-kNN | 84.0430 | 73.2282 | 71.4111 | 87.7623 | 71.6751 | 76.0645 | |
| FPFS-AC | 82.1290 | 69.5714 | 68.6000 | 86.5810 | 68.5903 | 73.1935 | |
| FPFS-EC | 82.1720 | 69.6658 | 68.6444 | 86.6150 | 68.6118 | 73.2581 | |
| FPFS-CMC | 81.8710 | 69.1010 | 68.1556 | 86.3683 | 68.1437 | 72.8065 | |
| IFPIFSC | 85.0538 | 72.2589 | 71.4000 | 89.1168 | 71.3761 | 77.5806 | |
| IFPIFS-HC | 84.3441 | 71.8038 | 70.3111 | 88.4744 | 70.4134 | 76.5161 | |
| AIFPIFSC1 | 85.7204 | 73.5563 | 72.8556 | 89.6705 | 72.7302 | 78.5806 | |
| AIFPIFSC2 | 85.6344 | 73.7498 | 72.4556 | 89.5333 | 72.3830 | 78.4516 | |
| NewThyroid | Fuzzy kNN | 95.0388 | 90.7974 | 87.8857 | 95.0752 | 87.9835 | 92.5581 |
| FPFS-kNN | 97.0853 | 96.7062 | 91.2889 | 95.7410 | 93.1413 | 95.6279 | |
| FPFS-AC | 96.3721 | 93.8027 | 90.9937 | 95.3598 | 91.9502 | 94.5581 | |
| FPFS-EC | 96.4961 | 93.7248 | 91.3365 | 95.5977 | 92.0784 | 94.7442 | |
| FPFS-CMC | 96.6202 | 93.8859 | 91.6921 | 95.7696 | 92.4125 | 94.9302 | |
| IFPIFSC | 97.6124 | 96.1328 | 94.4222 | 97.0730 | 94.9601 | 96.4186 | |
| IFPIFS-HC | 97.8605 | 97.0014 | 94.8095 | 97.2816 | 95.5104 | 96.7907 | |
| AIFPIFSC1 | 98.0775 | 97.2697 | 95.1175 | 97.4475 | 95.8640 | 97.1163 | |
| AIFPIFSC2 | 97.9845 | 97.1825 | 94.8000 | 97.2937 | 95.6615 | 96.9767 | |
| Vowel | Fuzzy kNN | 99.2140 | 95.8955 | 95.6768 | 99.5677 | 95.6417 | 95.6768 |
| FPFS-kNN | 99.3352 | 96.5957 | 96.3434 | 99.6343 | 96.3159 | 96.3434 | |
| FPFS-AC | 99.7337 | 98.6159 | 98.5354 | 99.8535 | 98.5312 | 98.5354 | |
| FPFS-EC | 99.6474 | 98.1818 | 98.0606 | 99.8061 | 98.0533 | 98.0606 | |
| FPFS-CMC | 99.6125 | 98.0247 | 97.8687 | 99.7869 | 97.8614 | 97.8687 | |
| IFPIFSC | 98.9513 | 94.5523 | 94.2323 | 99.4232 | 94.2318 | 94.2323 | |
| IFPIFS-HC | 99.2507 | 96.1076 | 95.8788 | 99.5879 | 95.8730 | 95.8788 | |
| AIFPIFSC1 | 99.7906 | 98.9147 | 98.8485 | 99.8848 | 98.8459 | 98.8485 | |
| AIFPIFSC2 | 99.7723 | 98.8142 | 98.7475 | 99.8747 | 98.7437 | 98.7475 | |
| Iris | Fuzzy kNN | 97.3778 | 96.4257 | 96.0667 | 98.0333 | 96.0430 | 96.0667 |
| FPFS-kNN | 97.5111 | 96.5657 | 96.2667 | 98.1333 | 96.2529 | 96.2667 | |
| FPFS-AC | 97.4222 | 96.4202 | 96.1333 | 98.0667 | 96.1199 | 96.1333 | |
| FPFS-EC | 97.4222 | 96.4202 | 96.1333 | 98.0667 | 96.1199 | 96.1333 | |
| FPFS-CMC | 97.3778 | 96.3697 | 96.0667 | 98.0333 | 96.0521 | 96.0667 | |
| IFPIFSC | 91.5111 | 88.0210 | 87.2667 | 93.6333 | 87.1189 | 87.2667 | |
| IFPIFS-HC | 92.8000 | 90.1718 | 89.2000 | 94.6000 | 89.0407 | 89.2000 | |
| AIFPIFSC1 | 97.5111 | 96.5434 | 96.2667 | 98.1333 | 96.2543 | 96.2667 | |
| AIFPIFSC2 | 97.4222 | 96.4337 | 96.1333 | 98.0667 | 96.1199 | 96.1333 | |
| Seeds | Fuzzy kNN | 90.4444 | 87.2996 | 85.6667 | 92.8333 | 85.4920 | 85.6667 |
| FPFS-kNN | 92.8571 | 89.7479 | 89.2857 | 94.6429 | 89.1821 | 89.2857 | |
| FPFS-AC | 93.7143 | 91.0210 | 90.5714 | 95.2857 | 90.4056 | 90.5714 | |
| FPFS-EC | 93.1429 | 90.0824 | 89.7143 | 94.8571 | 89.5837 | 89.7143 | |
| FPFS-CMC | 93.3016 | 90.3377 | 89.9524 | 94.9762 | 89.8107 | 89.9524 | |
| IFPIFSC | 92.9524 | 89.7158 | 89.4286 | 94.7143 | 89.2777 | 89.4286 | |
| IFPIFS-HC | 92.7302 | 89.5061 | 89.0952 | 94.5476 | 88.8379 | 89.0952 | |
| AIFPIFSC1 | 94.3175 | 91.7485 | 91.4762 | 95.7381 | 91.4237 | 91.4762 | |
| AIFPIFSC2 | 94.7619 | 92.3747 | 92.1429 | 96.0714 | 92.1077 | 92.1429 | |
| Wireless | Fuzzy kNN | 99.0725 | 98.1721 | 98.1450 | 99.3817 | 98.1485 | 98.1450 |
| FPFS-kNN | 95.2525 | 90.6801 | 90.5050 | 96.8350 | 90.5397 | 90.5050 | |
| FPFS-AC | 95.5500 | 91.2345 | 91.1000 | 97.0333 | 91.0944 | 91.1000 | |
| FPFS-EC | 94.6725 | 89.4593 | 89.3450 | 96.4483 | 89.3541 | 89.3450 | |
| FPFS-CMC | 94.3875 | 88.8904 | 88.7750 | 96.2583 | 88.7865 | 88.7750 | |
| IFPIFSC | 98.2200 | 96.4930 | 96.4400 | 98.8133 | 96.4425 | 96.4400 | |
| IFPIFS-HC | 98.2675 | 96.6149 | 96.5350 | 98.8450 | 96.5404 | 96.5350 | |
| AIFPIFSC1 | 99.0100 | 98.0371 | 98.0200 | 99.3400 | 98.0204 | 98.0200 | |
| AIFPIFSC2 | 99.0875 | 98.1902 | 98.1750 | 99.3917 | 98.1746 | 98.1750 | |
| Wine | Fuzzy kNN | 82.4910 | 74.0454 | 72.4131 | 86.5646 | 72.5250 | 73.7365 |
| FPFS-kNN | 97.0063 | 95.6477 | 96.2381 | 97.8322 | 95.6542 | 95.5095 | |
| FPFS-AC | 95.4011 | 93.8981 | 94.1831 | 96.6168 | 93.3926 | 93.1016 | |
| FPFS-EC | 97.3090 | 96.1400 | 96.6000 | 98.0425 | 96.0995 | 95.9635 | |
| FPFS-CMC | 97.1947 | 95.9917 | 96.4635 | 97.9589 | 95.9366 | 95.7921 | |
| IFPIFSC | 98.0921 | 97.5585 | 97.6127 | 98.5773 | 97.3807 | 97.1381 | |
| IFPIFS-HC | 98.3915 | 97.8120 | 97.9841 | 98.8081 | 97.7592 | 97.5873 | |
| AIFPIFSC1 | 98.1979 | 97.3359 | 97.7556 | 98.6979 | 97.4052 | 97.2968 | |
| AIFPIFSC2 | 98.5005 | 97.7649 | 98.1333 | 98.9283 | 97.8170 | 97.7508 | |
| WholesaleR | Fuzzy kNN | 64.5455 | 32.7706 | 32.1678 | 67.0562 | 36.7254 | 46.8182 |
| FPFS-kNN | 77.2273 | 34.0997 | 32.5751 | 66.0077 | 56.2602 | 65.8409 | |
| FPFS-AC | 70.6061 | 33.4320 | 33.2949 | 66.6446 | 39.5025 | 55.9091 | |
| FPFS-EC | 70.5909 | 33.1335 | 33.1258 | 66.3717 | 39.7357 | 55.8864 | |
| FPFS-CMC | 70.5000 | 33.4180 | 33.2277 | 66.7968 | 39.1309 | 55.7500 | |
| IFPIFSC | 70.5455 | 33.4457 | 33.4324 | 65.9316 | 38.5804 | 55.8182 | |
| IFPIFS-HC | 70.1970 | 32.6805 | 32.7455 | 65.7756 | 39.0998 | 55.2955 | |
| AIFPIFSC1 | 71.0303 | 35.9047 | 35.8277 | 67.0121 | 38.4634 | 56.5455 | |
| AIFPIFSC2 | 71.0303 | 35.9170 | 35.9634 | 67.1598 | 38.1598 | 56.5455 | |
| Breast Tissue | Fuzzy kNN | 84.3146 | 55.5706 | 51.1056 | 90.6527 | 57.0971 | 52.9437 |
| FPFS-kNN | 88.8066 | 67.3580 | 65.0500 | 93.3213 | 69.8296 | 66.4199 | |
| FPFS-AC | 90.1833 | 70.8637 | 69.6167 | 94.1207 | 72.5957 | 70.5498 | |
| FPFS-EC | 88.6768 | 67.5787 | 64.8278 | 93.2355 | 71.9591 | 66.0303 | |
| FPFS-CMC | 88.3968 | 66.5644 | 63.6722 | 93.0684 | 70.6789 | 65.1905 | |
| IFPIFSC | 89.2323 | 69.9230 | 67.0278 | 93.5696 | 68.1063 | 67.6970 | |
| IFPIFS-HC | 89.9942 | 71.4022 | 68.9444 | 94.0127 | 71.6861 | 69.9827 | |
| AIFPIFSC1 | 89.7792 | 71.5443 | 68.5667 | 93.8846 | 72.6934 | 69.3377 | |
| AIFPIFSC2 | 89.5613 | 71.2471 | 67.7611 | 93.7502 | 72.2527 | 68.6840 | |
| Led7Digit | Fuzzy kNN | 92.8360 | 65.5106 | 64.3707 | 96.0143 | 63.8773 | 64.1800 |
| FPFS-kNN | 92.3760 | 63.1691 | 62.1362 | 95.7535 | 63.2381 | 61.8800 | |
| FPFS-AC | 92.8320 | 65.4730 | 64.4296 | 96.0116 | 64.0844 | 64.1600 | |
| FPFS-EC | 92.8320 | 65.4730 | 64.4296 | 96.0116 | 64.0844 | 64.1600 | |
| FPFS-CMC | 92.8280 | 65.4508 | 64.4010 | 96.0094 | 64.0602 | 64.1400 | |
| IFPIFSC | 92.8320 | 65.4730 | 64.4296 | 96.0116 | 64.0844 | 64.1600 | |
| IFPIFS-HC | 92.8640 | 65.5359 | 64.5760 | 96.0297 | 64.1747 | 64.3200 | |
| AIFPIFSC1 | 92.8480 | 65.3969 | 64.5455 | 96.0238 | 63.9295 | 64.2400 | |
| AIFPIFSC2 | 92.9000 | 65.6258 | 64.8141 | 96.0524 | 64.1907 | 64.5000 | |
| Mean | Fuzzy kNN | 91.1318 | 72.4892 | 70.2654 | 92.9899 | 74.6155 | 75.9551 |
| FPFS-kNN | 93.8310 | 79.0775 | 75.8214 | 93.9765 | 82.0681 | 83.3420 | |
| FPFS-AC | 93.4057 | 80.1321 | 77.8002 | 94.2688 | 80.6779 | 83.2745 | |
| FPFS-EC | 93.2752 | 78.7916 | 76.6913 | 94.0175 | 80.6779 | 82.6885 | |
| FPFS-CMC | 93.1957 | 78.7142 | 76.5281 | 94.0077 | 80.1741 | 82.4181 | |
| IFPIFSC | 93.2512 | 78.6349 | 76.5182 | 94.2836 | 79.2002 | 81.9587 | |
| IFPIFS-HC | 93.4052 | 79.0292 | 76.9062 | 94.4012 | 79.8964 | 82.4427 | |
| AIFPIFSC1 | 94.3023 | 82.5859 | 80.2558 | 95.0420 | 82.4851 | 85.0485 | |
| AIFPIFSC2 | 94.3053 | 82.6093 | 80.1285 | 95.0217 | 82.3874 | 84.9823 |
Note:
The best performance results are shown in bold.
Table 2 demonstrates that AIFPIFSC1 and AIFPIFSC2 precisely categorize the dataset “Mice Protein Expression” in the same manner as FPFS-kNN, FPFS-AC, FPFS-EC, FPFS-CMC, IFPIFSC, and IFPIFS-HC. Furthermore, according to all performance measures, AIFPIFSC1’s results for the datasets “NewThyroid”, “Vowel”, “Iris”, “Seeds”, “Wireless”, and “Wine” exceed the performance rates of , , , , , and , respectively. AIFPIFSC2’s results for the datasets “NewThyroid”, “Vowel”, “Iris”, “Seeds”, “Wireless”, and “Wine” exceed the performance rates of , , , , , and . Moreover, AIFPIFSC1 achieves the highest scores across all performance indicators in “Ecoli”, “Libras”, “HCV Data” (excluding Pre value), “Column3C” (excluding Pre value), “NewThyroid”, “Vowel”, and “Iris” (excluding Pre value). AIFPIFSC2 achieves the highest scores across all performance metrics in “Leaf” (excluding Pre value), “Seeds”, “Wireless”, “Wine” (excluding Pre value), “WholesaleR” (excluding Acc, MacF, and MicF value), and “Led7Digit”. As a result, the average performance outcomes presented in Table 2 suggest that AIFPIFSC1 and AIFPIFSC2 are more effective classifiers than other classifiers for the datasets under consideration.
Statistical analysis
This subsection conducts the Friedman test (Friedman, 1940), a non-parametric method, along with the Nemenyi test (Nemenyi, 1963), a post-hoc test, following the approach suggested by Demsar (2006). The process aims to evaluate all performance outcomes in terms of Acc, Pre, Rec, Spe, MacF, and MicF. The Friedman test creates a ranking based on performance for the classifiers across each dataset. Therefore, a rank of 1 indicates the top-performing classifier, followed by a rank of 2 for the next best, and so on. If classifiers exhibit identical performance, the test assigns them the mean of their potential ranks. Subsequently, it assesses the average ranks and computes , which follows a distribution with degrees of freedom, where represents the total number of classifiers. A post hoc analysis, such as the Nemenyi test, is applied to identify significant differences among the classifiers. Differences between any pair of classifiers that exceed the critical distance are regarded as statistically significant.
In the statistical analysis, the average ranking for each classifier is computed using the Friedman test. Nine classifiers, which means , including AIFPIFSC1 and AIFPIFSC2, are compared concerning 15 datasets (denoted as ) for each of the six performance criteria. The Friedman test yields the following statistics for Acc, Pre, Rec, Spe, MacF, and MicF: , , , , , and , respectively.
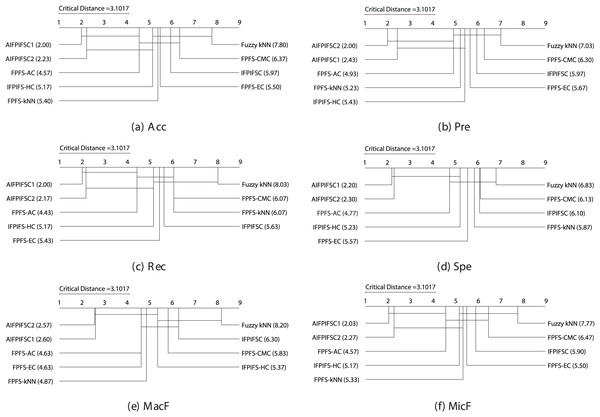
With and , the Friedman test indicates a critical value of at a significance level of (for additional details, refer to Zar (2010)). As the Friedman test statistics for Acc (59.27), Pre (48.50), Rec (62.13), Spe (46.78), MacF (52.15), and MicF (57.99) surpass the critical value of 15.51, it implies a notable distinction in the performances of the classifiers. Consequently, the null hypothesis stating “There are no performance differences between the classifiers” is dismissed, allowing the application of the Nemenyi test. For , , and , the critical distance is computed as following the Nemenyi test. Figure 1 illustrates the crucial diagrams generated by the Nemenyi test for each of the six performance metrics.
Figure 1: The essential diagrams resulting from the Friedman and Nemenyi tests at a significance level of 0.05 for the six performance criteria in the context of the classifiers mentioned above.
Figure 1 manifests that the performance differences between the average rankings of AIFPIFSC1 and those of IFPIFS-HC, FPFS-kNN, Fuzzy kNN, FPFS-CMC, IFPIFSC, and FPFS-EC, are more significant than the critical distance (3.1017). Besides, the performance differences between the average rankings of AIFPIFSC2 and those of FPFS-kNN, Fuzzy kNN, FPFS-CMC, IFPIFSC, and FPFS-EC are more significant than the critical distance (3.1017). Figure 1 shows that although the difference between the mean rankings of AIFPIFSC1 and FPFS-AC, as well as AIFPIFSC2 and FPFS-AC and IFPIFS-HC is less than the critical distance (4.0798), AIFPIFSC1 and AIFPIFSC2 outperforms them in terms of all performance measures. Therefore, AIFPIFSC1 and AIFPIFSC2 outperform them in all performance metrics.
Conclusion and future studies
This study introduced two adaptive machine learning algorithms that concurrently utilize seven pseudo-similarities over ifpifs-matrices, applying them to a data classification problem. Besides, it employed two soft decision-making methods, constructed by ifpifs-matrices to improve the aforesaid machine learning approaches. In this study, the input values and , previously provided externally in the algorithm IFPIFSC, are determined adaptively based on the number of classes and instances. Implementing the ifpifs-matrices in the proposed algorithms resulted in a noticeable enhancement in classification Acc. The adaptive values played a crucial role in fine-tuning the classification process, relying on the specificities of each dataset. The adaptive nature of values in AIFPIFSC1 and AIFPIFSC2 proved pivotal in managing the intricacies and variabilities inherent in real-world data. This adaptability ensured the suggested algorithms remained robust and effective across diverse datasets and scenarios. The proposed algorithms are compared with well-known and state-of-the-art fuzzy/soft-set-based classifiers such as Fuzzy kNN, FPFS-kNN, FPFS-AC, FPFS-EC, IFPIFSC, and IFPIFS-HC. The performance results were utilized for a fair comparison and were statistically analyzed. Therefore, the current investigation demonstrates that the suggested approaches yield superior performance outcomes, making it a highly convenient method in supervised learning.
By dynamically adjusting lambda values and integrating ifpifs-matrices into two classification algorithms, this study contributes to the theoretical aspects of soft decision-making and machine learning. It provides a practical framework that can be employed in various real-world applications. The adaptability and efficiency of the proposed methods make it a valuable addition to machine learning, especially in scenarios where data uncertainty and dynamism are predominant.
Although our proposed algorithms can classify problems with intuitionistic fuzzy values, no fuzzy or intuitionistic fuzzy uncertain data exists in standard machine learning databases. Therefore, to show that the mentioned algorithms can successfully work on data with intuitionistic fuzzy uncertainties, we subject the data in common databases to fuzzification and intuitionistic fuzzification processes, and then apply the proposed approaches. One of the most well-known examples of intuitionistic fuzzy uncertainties can be expressed to explain the usefulness of the methods: If a detector emits ten signals per second and produces six positive and four negative signals, this situation can be represented by the fuzzy value . Since intuitionistic fuzzy sets extend fuzzy sets, they can also model this scenario using the intuitionistic fuzzy values and . However, suppose the detector records six positive, three negative, and one corrupted signal. In that case, this cannot be described using fuzzy values alone but can be represented using intuitionistic fuzzy values as and . These examples illustrate the superiority of intuitionistic fuzzy sets over traditional fuzzy sets. Furthermore, soft sets are required to address the challenge of determining the optimal location for constructing a wind turbine when processing data from detectors at various locations. As seen in this example, problems with intuitionistic fuzzy uncertainties are among the types of uncertainties we can encounter daily. Classical machine learning methods cannot work on data with such uncertainty, but the proposed machine learning approaches can efficiently work on issues such as those presented in the study. Moreover, there is no data with such uncertainty in known databases. Therefore, to compare the classical methods with our proposed approaches, we can obtain the performance results of classical machine learning methods and our proposed methods by converting these data into fuzzy and intuitionistic fuzzy values using the same classical data. The abilities and advantages of the proposed approach compared to the others can be summarized in Table 3.
| Classifer | Ref. | Crisp value | Fuzzy value | Int fuzzy value | Cla metric or sim | fpfs-p metric or sim | ifpifs-p metric or sim | fpfs DM | ifpifs DM | ADA |
|---|---|---|---|---|---|---|---|---|---|---|
| Fuzzy-kNN | Keller, Gray & Givens (1985) | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ |
| FPFS-kNN | Memiş, Enginoğlu & Erkan (2022b) | ✓ | ✓ | ✗ | ✓ | ✓ | ✗ | ✗ | ✗ | ✗ |
| FPFS-AC | Memiş, Enginoğlu & Erkan (2022c) | ✓ | ✓ | ✗ | ✓ | ✓ | ✗ | ✓ | ✗ | ✗ |
| FPFS-EC | Memiş, Enginoğlu & Erkan (2021) | ✓ | ✓ | ✗ | ✓ | ✓ | ✗ | ✗ | ✗ | ✗ |
| FPFS-CMC | Memiş, Enginoğlu & Erkan (2022a) | ✓ | ✓ | ✗ | ✓ | ✓ | ✗ | ✓ | ✗ | ✗ |
| IFPIFSC | Memiş et al. (2023) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ |
| IFPIFS-HC | Memiş et al. (2021) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ |
| AIFPIFSC1 | Proposed | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| AIFPIFSC2 | Proposed | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
Note:
Int, Cla, Sim, fpfs-p, ifpifs-p, DM, and ADA represent intuitionistic, classical, similarity, fpfs-pseudo, ifpifs-pseudo, decision making, and adaptive, respectively.
While the proposed algorithms demonstrate significant improvements in classification Acc by effectively incorporating ifpifs-matrices and adaptive values, an aspect warranting further attention is their time complexity. Comparative analysis reveals that our robust and adaptable algorithms perform slower than some benchmarked classifiers. This observation underscores the need for optimization in computational efficiency. Future research could focus on refining the algorithmic structure to enhance processing speed without compromising the Acc gains observed. Potential avenues might include integrating more efficient data structures, algorithmic simplification, or parallel processing techniques. Addressing this time complexity issue is crucial for practical applications, especially in real-time data processing scenarios. Further exploration of the scalability of the proposed methods for handling larger datasets could also be a valuable direction for subsequent studies. Moreover, future works can extend this research by integrating other forms of fuzzy and soft sets, e.g., bipolar soft sets (Mahmood, 2020) and picture fuzzy parameterized picture fuzzy soft sets (Memiş, 2023a) into machine learning (Li, 2024), further enhancing the capabilities and applications of adaptive machine learning methods based on soft decision-making.