Detection and drug resistance profile of Escherichia coli from subclinical mastitis cows and water supply in dairy farms in Saraburi Province, Thailand
- Published
- Accepted
- Received
- Academic Editor
- Paul Tulkens
- Subject Areas
- Microbiology, Infectious Diseases
- Keywords
- Subclinical bovine mastitis, Escherichia coli, Antibiotic resistance, Extend spectrum beta-lactamase
- Copyright
- © 2017 Hinthong et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2017. Detection and drug resistance profile of Escherichia coli from subclinical mastitis cows and water supply in dairy farms in Saraburi Province, Thailand. PeerJ 5:e3431 https://doi.org/10.7717/peerj.3431
Abstract
Subclinical mastitis is a persistent problem in dairy farms worldwide. Environmental Escherichia coli is the bacterium predominantly responsible for this condition. In Thailand, subclinical mastitis in dairy cows is usually treated with various antibiotics, which could lead to antibiotic resistance in bacteria. E. coli is also a reservoir of many antibiotic resistance genes, which can be conveyed to other bacteria. In this study, the presence of E. coli in milk and water samples was reported, among which enteropathogenic E. coli was predominant, followed by enteroaggregative E. coli and enterohemorrhagic E. coli, which was found only in milk samples. Twenty-one patterns of antibiotic resistance were identified in this study. Ampicillin- and carbenicillin-resistant E. coli was the most common among the bacterial isolates from water samples. Meanwhile, resistance to ampicillin, carbenicillin, and sulfamethoxazole-trimethoprim was the pattern found most commonly in the E. coli from milk samples. Notably, only the E. coli from water samples possessed ESBL phenotype and carried antibiotic resistance genes, blaTEM and blaCMY-2. This indicates that pathogenic E. coli in dairy farms is also exposed to antibiotics and could potentially transfer these genes to other pathogenic bacteria under certain conditions.
Introduction
In dairy farms, mastitis is a persistent problem resulting in economic losses and premature culling of cows. Staphylococcus aureus (S. aureus) is considered a major causative pathogen which is a threat to farmers, although easily identifiable, whereas other gram negative bacteria is overlooked or not considered to be a cause for concern by farmers. Subclinical mastitis, which is defined as a somatic cell count (SCC) of >200,000 cells/mL in milk, is usually caused by gram negative bacteria, such as Escherichia coli (E. coli), Klebsiella pneumoniae, and Serratia marcescens (Schukken et al., 2012; Azevedo et al., 2016). These bacteria are commonly found in environmental settings, such as bedding, clothes, farmers’ hands, and water used on farms (Perkins et al., 2009; Iraguha, Hamudikuwanda & Mushonga, 2015; Azevedo et al., 2016). Among gram negative bacteria, E. coli is the most notable cause of mastitis. E. coli was found to usually infected mammary gland of cows parturition and early lactation period which could lead to local and acute mastitis (Burvenich et al., 2003). In a study in Portugal, E. coli was found to be the second most common bacteria after non-coagulative staphylococci found in bulk tank milk (Azevedo et al., 2016). In Uruguay, E. coli was second only to S. aureus in bovine subclinical mastitis cases (Gianneechini et al., 2002), whereas in China, it was one of the leading types of coliform bacteria found in milk from cows with subclinical mastitis (Memon et al., 2013; Wang et al., 2015).
The treatment of bovine subclinical mastitis usually depends on the severity of the symptoms. In Thailand, the disease is usually treated with antibiotics or the infected cows are culled. Antibiotics are also used for prevention in some farms. However, this can lead to bacteria developing resistance to them. For example, increased resistance to antibiotics in S. aureus in the form of oxacillin- or gentamicin-resistant strains was reported in Thailand due to their excessive use (Suriyasathaporn, 2011; Suriyasathaporn et al., 2012). Despite this background, there is little information on antibiotic resistance and drug resistance genes in other bacteria related to bovine mastitis in Thailand. E. coli can be antibiotic-resistant as it is also exposed to antibiotics from wastewater from farms. Furthermore, E. coli that carries resistance genes can transfer those genes to other pathogenic bacteria (Hu et al., 2016). The discovery of antibiotic resistance in E. coli isolates from farms could possibly show the trend or specific characteristic of antibiotic resistance and facilitate better prevention or the more effective treatment for mastitis on dairy farm. This study was thus conducted to detect E. coli from water sources and milk from cows with subclinical mastitis, and their antibiotic resistance patterns.
Materials and Methods
Sample collection
All procedures performed in this study are in accordance with the ethical standards of the Faculty of Tropical Medicine–Animal Care and Use Committee (FTM-ACUC), Mahidol University, Thailand (protocol no. 002-2016). Water and milk samples were collected from 17 dairy farms in Saraburi Province, Thailand, where agriculture and livestock are the main source of income of the people. A total of 35 water samples were collected in 500-ml sterile bottles from drinking water for cows in a milking area and also from washing water. Thirty-eight milk samples were collected in sterile falcon tubes from cows with subclinical mastitis, which had an SCC of >200,000 cells/ml in milk, after the teats had been disinfected with 70% ethanol and 4–5 streams of milk had been removed. Both water and milk samples were stored at 4 °C and transported to the laboratory within 24 h for the experiment.
Bacterial isolation
Each water and milk sample was centrifuged at 6,000 rpm for 10 min, and the precipitant was subjected to 10-fold dilution and spread on MacConkey agar (Becton, Dickinson, and Company). Suspected E. coli lactose-fermenting colonies (pink colonies) were subjected to gram staining and standard biochemical tests, including triple sugar iron agar, lysine decarboxylase, ornithine decarboxylase/deaminase, motility, and indole production tests.
Antibiotic susceptibility tests
All E. coli isolates were subjected to antibiotic susceptibility tests following the Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2016). The antimicrobial disks used in the experiment included 10 µg ampicillin (≤13 mm), 100 µg piperacillin (≤17 mm), 10 µg carbenicillin (≤17 mm), 20 µg amoxicillin-clavulanic acid (≤13 mm), 30 µg cefepime (≤14 mm), 30 µg cefotaxime (≤22 mm), 30 µg ceftriaxone (≤19 mm), 30 µg ceftazidime (≤17 mm), 75 µg cefoperazone (≤15 mm), 30 µg cefuroxime (≤14 mm), 10 µg imipenem (≤19 mm), 10 µg meropenem (≤19 mm), 10 µg gentamicin (≤12 mm), 30 µg amikacin (≤14 mm), 15 µg tigecycline (≤14 mm), 5 µg ciprofloxacin (≤15 mm), 10 µg norfloxacin (≤12 mm), and 23.75 µg trimethoprim–sulfamethoxazole (≤10 mm) (Oxoid). E. coli strain ATCC 25922 was used as a control in this experiment.
Extended spectrum β-lactamase (ESBL) production was tested by double disk synergy (DDS) method modified from Clinical and Laboratory Standards Institute (2012). The test uses 30 µg antibiotic disks of cefepime, cefotaxime ceftriaxone, ceftazidime, and cefuroxime. The antibiotic disks were placed on the E. coli spreaded MHA culture plate, 30 mm (center to center) from the amoxicillin-clavulanic acid (30 µg) disk. Plates were incubated at 37 °C overnight and observed for the presence of an extended spectrum beta-lactamase (ESBL) phenotype by an extension of the edge of inhibition zone of antibiotic disks toward the amoxicillin-clavulanic acid.
Gene detection by polymerase chain reaction
All E. coli isolates from both water and milk samples were determined using specific gene and plasmid, and the isolates that showed resistance to antibiotics were selected and subjected to PCR to investigate their drug resistance genes. The bacteria were cultured in 1.5 ml of tryptic soy broth (Oxoid) and incubated overnight; they were then harvested and centrifuged for 10 min at 6,000 rpm. The pellet was resuspended with 800 µl of sterile distilled water, boiled for 10 min, centrifuged at 6,000 rpm for 10 min, and then the supernatant was collected for use as a DNA template in PCR. PCR primers, conditions, and positive control strains for the detection of target gene and drug resistance genes are presented in Tables 1 and 2. All PCR reactions with a total volume of 25 µl were performed in 1× Taq buffer, 1 mM MgCl2, 0.2 mM dNTP, 1 µM of each of the forward and reverse primers, and 2 units of Taq DNA polymerase (Thermo Scientific). The PCR amplicon was subjected to 1.5% agarose gel electrophoresis in TAE buffer. For gene amplification with no reference control, the PCR product from positive samples was subjected to nucleotide sequencing and sequence analysis for gene confirmation.
| Target genes | Positive control | Sequences (5′–3′) | Annealing temperature (°C) | Product size (bp) | References |
|---|---|---|---|---|---|
| Heat-labile toxin (lt) | ETEC | tctctatgcatacggag ccatactgattgccgcaatt | 55 | 322 | Deng et al. (1996) |
| Hest-stable toxin (st) | ETEC | tgctaaaccagtagagtcttcaaaa gcaggcttacaacacaattcacagcag | 55 | 138 | Mercado et al. (2011) |
| Shiga-like enterotoxins 1 (evt) | EHEC | caacactggatgatctcag ccccctcaactgctaata | 55 | 349 | Khan et al. (2002) |
| Shiga-like enterotoxins 2 (evs) | EHEC | atcagtcgtcactcactggt ctgctgtcacagtgacaaa | 55 | 110 | Khan et al. (2002) |
| Transcriptional activator of the aggregative adherence fimbriae (aggR) | EAEC 17-2 | ctaattgtacaatcgatgta atgaagtaattcttgaat | 55 | 308 | Nataro et al. (1994) |
| pCVD432 plasmid | EAEC 17-2 | ctggcgaaagactgtatcat caatgtatagaaatccgctgtt | 55 | 630 | Aranda, Fagundes-Neto & Scaletsky (2004) |
| Intimin (eaeA) | Plasmid-eaeA | aaacaggtgaaactgttgcc tctcgcctgatagtgtttggta | 55 | 350 | Yu & Kaper (1992) |
| Bundle-forming pilus (bfpA) | – | aatggtgcttgcgcttgctgc gccgctttatccaacctggta | 57 | 326 | Zhang et al. (2016) |
Notes:
- ETEC
-
enterotoxigenic E. coli
- EHEC
-
enterohemorrhagic E. coli
- EAEC
-
enteroaggregative E. coli
| Drug resistance genes | Positive control | Sequences (5′–3′) | Annealing temperature (°C) | Product size (bp) | References |
|---|---|---|---|---|---|
| Beta-lactams | |||||
| blaTEM | – | ttaactggcgaactacttac gtctatttcgttcatccata | 60 | 247 | Kozak et al. (2009) |
| blaSHV | – | aggattgactgccttttg atttgctgatttcgctcg | 60 | 393 | Kozak et al. (2009) |
| blaCMY-2 | – | gacagcctctttctccaca tggacacgaaggctacgta | 60 | 1,000 | Kozak et al. (2009) |
| Aminoglycosides | |||||
| aac(3)-IIa | – | cggaaggcaataacggag tcgaacaggtagcactgag | 60 | 740 | Soleimani et al. (2014) |
| aac(3)-IV | – | gtgtgctgctggtccacagc agttgacccagggctgtcgc | 60 | 627 | Maynard et al. (2004) |
| aadA | – | cccctggagagagcgagatt cgtggctggctcgaagatac | 60 | 152 | Our study |
| aadB | – | gaggagttggactatggatt cttcatcggcatagtaaaag | 60 | 208 | Kozak et al. (2009) |
| Quinolone | |||||
| qnrA | – | agaggatttctcacgccagg tgccaggcacagatcttgac | 60 | 580 | Cattoir et al. (2007) |
| qnrB | – | ggcattgaaattcgccactg tttgctgctcgccagtcgaa | 60 | 264 | Cattoir et al. (2007) |
| qnrS | – | gcaagttcattgaacagggt tctaaaccgtcgagttcggcg | 60 | 428 | Cattoir et al. (2007) |
Serotyping
E. coli isolates with virulence genes were serotyped using Serosystem (Serosystem, Clinag, Thailand) to identify O and H antigens present on the surface of the pathogenic E. coli isolates with slide agglutination test. The experiment was performed following the manufacturer’s protocol. EAEC, EHEC, EPEC, and ETEC strains were used as positive control in the experiment.
Results
E. coli isolation and antibiotic resistance patterns
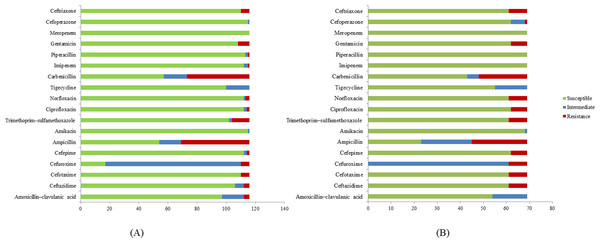
A total of 185 E. coli isolates were collected from water (116 isolates) and milk (69 isolates) samples and subjected to antibiotic susceptibility tests. Among these isolates, a total of 77 (51 isolates from water and 26 isolates from milk samples) showed resistance to at least one of the antibiotics use in the experiment. Penicillin-resistant E. coli (71/77, 92.2%) was found to be the largest group in this study followed by folate pathway inhibitor-resistant E. coli (20/77, 26%). E. coli resistant to cephems (14/77, 18.2%), aminoglycosides (14/77, 18.2%), β-lactamase inhibitor combination (4/77, 5.2%), fluoroquinolone (12/77, 14.3%), and carbapenem (1/77, 1.3%) were also found. Among antibiotic resistant E. coli, 84.31% (43/51) of E. coli found in water samples are multidrug resistance and 84.61% (22/26) in milk samples (Table 3). The antibiotic patterns could be divided into 21 types, as shown in Table 3. We also found the ESBL phenotype (12/185, 6.5%) in six E. coli isolates each from water and milk samples. The antibiotics that the E. coli strains are susceptible to are as shown in Fig. 1.
| Resistance pattern | Phenotypic resistance | Number of resistant E. coli isolates | |
|---|---|---|---|
| Water samples (n = 51) | Milk samples (n = 26) | ||
| I | AMC | 1 (1.9%) | 0 (0%) |
| II | AMC, AMP, CAR | 2 (3.9%) | 0 (0%) |
| III | AMC, AMP, CAR, IPM | 1 (1.9%) | 0 (0%) |
| IV | AMP | 3 (5.8%) | 2 (7.6%) |
| V | AMP, CAR | 26 (50.9%) | 0 (0%) |
| VI | AMP, CAR, CAZ, CN, CRO, CTX, CXM | 2 (3.9%) | 0 (0%) |
| VII | AMP, CAR, CAZ, CN, CRO, CTX, CXM, FEP | 0 (0%) | 6 (23.0%) |
| VIII | AMP, CAR, CAZ, CN, CRO, CTX, CXM, FEP, SCF | 0 (0%) | 1 (3.8%) |
| IX | AMP, CAR, CAZ, CN, CRO, CTX, CXM, FEP, SXT | 1 (1.9%) | 0 (0%) |
| X | AMP, CAR, CAZ, CN, CRO, CTX, CXM, SXT | 1 (1.9%) | 0 (0%) |
| XI | AMP, CAR, CIP, CN, CRO, CTX, CXM, FEP, SXT | 1 (1.9%) | 0 (0%) |
| XII | AMP, CAR, CIP, NOR | 0 (0%) | 7 (26.9%) |
| XIII | AMP, CAR, CIP, NOR, SXT | 1 (1.9%) | 0 (0%) |
| XIV | AMP, CAR, CN, CRO, CTX, CXM, SXT | 1 (1.9%) | 0 (0%) |
| XV | AMP, CAR, NOR | 1 (1.9%) | 0 (0%) |
| XVI | AMP, CAR, SXT | 6 (11.7%) | 7 (26.9%) |
| XVII | AMP, CAZ, CRO, CTX, CXM | 0 (0%) | 1 (3.8%) |
| XVIII | CN | 1 (1.9%) | 0 (0%) |
| XIX | NOR | 1 (1.9%) | 1 (3.8%) |
| XX | SXT | 1 (1.9%) | 1 (3.8%) |
| XXI | TZP | 1 (1.9%) | 0 (0%) |
Notes:
- AMP
-
ampicillin
- TZP
-
piperacillin
- CAR
-
carbenicillin
- AMC
-
amoxicillin–clavulanic acid
- FEP
-
cefepime
- CTX
-
cefotaxime
- CRO
-
ceftriaxone
- CAZ
-
ceftazidime
- SCF
-
cefoperazone
- CXM
-
cefuroxime
- IPM
-
imipenem
- MEM
-
meropenem
- CN
-
gentamicin
- AK
-
amikacin
- TGC
-
tigecycline
- CIP
-
ciprofloxacin
- NOR
-
norfloxacin
- SXT
-
trimethoprim–sulfamethoxazole
Specific gene and drug resistance gene detection and serotyping
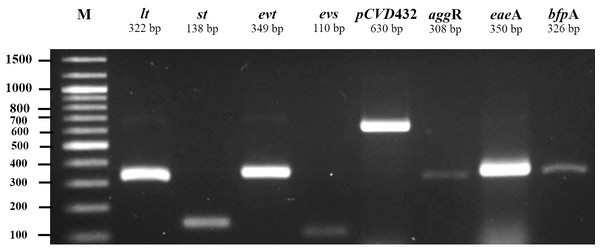
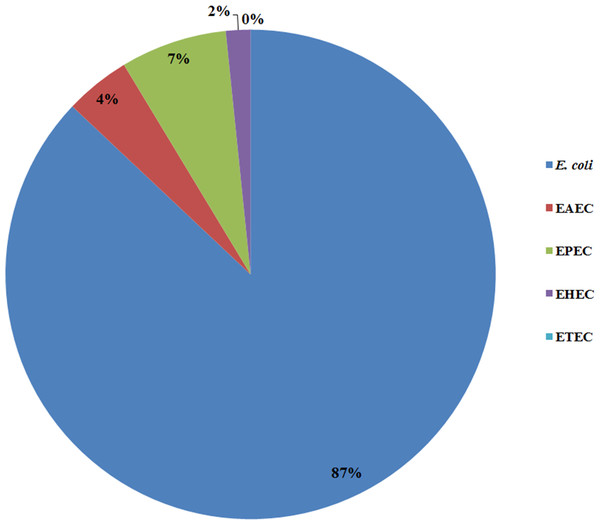
All 185 E. coli isolates from both water and milk samples were also subjected to an analysis of the virulence genes and plasmid for EAEC, EHEC, EPEC, and ETEC (agg R and pCVD432, evt and evs, eaeA and bfpA, and lt and st, Fig. 2). Among the bacterial isolates, 24 (24/185, 12.97%) showed positive results for gene detection by PCR, with bfpA positive isolates, EPEC forming the majority (13/185, 7.02%) followed by pCVD432 positive isolates, EAEC (8/185, 4.32%) and evt positive isolates, EHEC (3/185, 1.62%) (Fig. 3). All EPEC E. coli isolates were from water samples. Among them, the bacteria presented different serotypes, namely, O124:K62 (2/13, 15.4%), O111:K58 (2/13, 15.4%), O128:K67 (1/13, 7.7%), O78:K80 (1/13, 7.7%), and O86:K61 (1/13, 7.7%). EAEC isolated from milk samples possessed the O18aO18C:K77 serotype (6/8, 75%) and those from water samples possessed the O112aO112c:K66 serotype (1/8, 12.5%), whereas one isolate could not be serotyped. EHEC isolates from milk samples possessed the O114:K serotype (2/3, 66.7%), whereas positive isolates from water could not be serotyped (Table 4).
The bacterial antibiotic-resistant isolates (77 isolates) were investigated for drug resistance genes (β-lactam: blaTEM, blaSHV, blaCMY-2; aminoglycoside: aac(3)-IIa, aac(3)-IV, aadA, aadB; quinolone: qnrA, qnrB, and qnrS) using PCR. The results showed that one pCVD432 positive E. coli and one bfpA positive E. coli isolates possessed blaCMY-2 and blaTEM, respectively. We also found one pCVD432 positive isolate with the ESBL phenotype that carried both blaTEM and blaCMY-2. In non-pathogenic E. coli isolates, 43 (43/77, 55.9%) isolates were found to possess various antibiotic resistance genes (Table 5). The most common resistant gene found was blaTEM (38/62, 61.3%) followed by blaCMY-2 (16/62, 25.8%) and aac(3)IIa (3/62, 4.9%). Other resistance genes carried by non-pathogenic isolates were aadA (2/62, 3.3%) and blaSHV(2/62, 3.3%). None of the E. coli isolates carried quinolone resistance genes.
Figure 1: Antibiotic susceptibility test results.
(A) E. coli isolates from water samples. (B) E. coli isolates from milk samples.Figure 2: Agarose gel electrophoresis of 1% agarose of the amplification products of virulence genes and plasmid for ETEC (lt, st), EHEC (evt, evs), EAEC (pCVD432, aggR), and EPEC (eaeA, bfpA).
M, DNA Marker.Figure 3: Prevalence of pathogenic E. coli detected from water and milk samples.
| Isolate number | Origin | Target gene | Serotype | Antibiotic resistance pattern | Resistance gene profile | ESBL phenotype |
|---|---|---|---|---|---|---|
| M-W910-1 LF1 | Water | bfpA | a | * | − | − |
| M-W910-1 LF3 | Water | bfpA | a | * | − | − |
| M-W1010-1 LF1 | Water | bfpA | a | V | blaTEM | − |
| M-W1110-1 LF3 | Water | bfpA | a | XX | − | − |
| M-W1110-1 LF5 | Water | bfpA | O124:K62 | * | − | − |
| M-W1110-1 LF6 | Water | bfpA | O111:K58 | * | − | − |
| M-W12UD LFB6 | Water | bfpA | O111:K58 | I | − | − |
| M-W13UD LFB6 | Water | bfpA | O128:K67 | * | − | − |
| M-W13UD LFB7 | Water | bfpA | O124:K62 | * | − | − |
| M-W13UD LFB10 | Water | bfpA | O78:K80 | * | − | − |
| M-W15UD LFB4 | Water | bfpA | a | V | − | − |
| M-W22UD LF9 | Water | pCVD432 | a | VI | blaCMY-2, aac(3)IIa | − |
| M-W23UD LF2 | Water | pCVD432 | O112aO112c:K66 | VI | blaTEM, blaCMY-2, aac(3)IIa, aadA | + |
| M-W32UD LF1 | Water | bfpA | a | V | − | − |
| M-W33UD LF1 | Water | bfpA | O86:K61 | * | − | − |
| M-M10UD LFB4 | Milk | evt | O114:K | * | − | − |
| M-M10UD LFB5 | Milk | evt | O114:K | * | − | − |
| M-M35UD LFB2 | Milk | pCVD432 | O18aO18c:K77 | VII | − | + |
| M-M35UD LFB3 | Milk | pCVD432 | O18aO18c:K77 | VII | − | + |
| M-M35UD LFB4 | Milk | pCVD432 | O18aO18c:K77 | VII | − | + |
| M-M35UD LFB5 | Milk | pCVD432 | O18aO18c:K77 | VIII | − | + |
| M-M35UD LFB6 | Milk | pCVD432 | O18aO18c:K77 | VII | − | + |
| M-M35UD LFB7 | Milk | pCVD432 | O18aO18c:K77 | VII | − | + |
| M-M37UD LFB4 | Milk | evt | a | XVI | − | − |
| Isolate number | Origin | Target gene | Serotype | Antibiotic resistance pattern | Resistance gene profile | ESBL phenotype |
|---|---|---|---|---|---|---|
| M-W610-1 LF1 | Water | − | b | V | blaTEM | − |
| M-W910-1 LF2 | Water | − | b | IV | blaCMY-2 | − |
| M-W1110-1 LF9 | Water | − | b | XVI | blaTEM | − |
| M-W12UD LFB6 | Water | − | b | I | blaCMY-2 | − |
| M-W16UD LF1 | Water | b | V | blaTEM, blaSHY, blaCMY-2 | − | |
| M-W16UD LF2 | Water | − | b | V | blaTEM, blaCMY-2 | − |
| M-W1910-1 LF6 | Water | − | b | V | blaTEM | − |
| M-W20UD LF6 | Water | − | b | V | blaTEM, blaCMY-2 | − |
| M-W20UD LF9 | Water | − | b | V | blaTEM, blaCMY-2 | − |
| M-W22UD LF3 | Water | − | b | XIV | blaTEM, blaCMY-2 | + |
| M-W22UD LF7 | Water | − | b | X | blaTEM, blaCMY-2, aac(3)IIa | + |
| M-W24UD LF1 | Water | − | b | V | blaTEM | − |
| M-W24UD LF3 | Water | − | b | XI | blaTEM, blaCMY-2, aac(3)IIa, aadA | + |
| M-W24UD LF4 | Water | − | b | V | blaTEM | − |
| M-W24UD LF5 | Water | − | b | XVI | blaTEM | − |
| M-W24UD LF7 | Water | − | b | * | blaTEM, blaCMY-2 | − |
| M-W26UD LF8 | Water | − | b | IV | blaTEM | − |
| M-W27UD LF4 | Water | − | b | XVI | blaTEM | − |
| M-W28UD LF1 | Water | − | b | XV | blaTEM | − |
| M-W28UD LF5 | Water | − | b | V | blaSHV, blaCMY-2 | − |
| M-W28UD LF7 | Water | − | b | V | blaTEM, blaCMY-2 | − |
| M-W29UD LF1 | Water | − | b | IX | blaTEM, blaCMY-2, aac(3)IIa, aadA | + |
| M-W29UD LF10 | Water | − | b | XVI | blaTEM | − |
| M-W31UD LF6 | Water | − | b | V | blaTEM | − |
| M-W33UD LF2 | Water | − | b | V | blaTEM | − |
| M-W33UD LF6 | Water | − | b | V | blaTEM | − |
| M-W33UD LF9 | Water | − | b | V | blaTEM | − |
| M-W33UD LF10 | Water | − | b | V | blaTEM | − |
| M-W34UD LF3 | Water | − | b | V | blaTEM | − |
| M-W34UD LF7 | Water | − | b | XIII | blaTEM | − |
| M-M1610-1 LFB2 | Milk | − | b | IV | blaTEM | − |
| M-M37UD LFB1 | Milk | − | b | XVI | blaTEM | − |
| M-M37UD LFB3 | Milk | − | b | XVI | blaTEM | − |
| M-M37UD LFB5 | Milk | − | b | XVI | blaCMY-2 | − |
| M-M37UD LFB6 | Milk | − | b | XVI | blaTEM, blaCMY-2 | − |
| M-M37UD LFB8 | Milk | − | b | XVI | blaCMY-2 | − |
| M-M38UD LFB1 | Milk | − | b | XII | blaTEM | − |
| M-M38UD LFB2 | Milk | − | b | XII | blaTEM | − |
| M-M38UD LFB3 | Milk | − | b | XII | blaTEM | − |
| M-M38UD LFB4 | Milk | − | b | XII | blaTEM | − |
| M-M38UD LFS2 | Milk | − | b | XII | blaTEM | − |
| M-M38UD LFS3 | Milk | − | b | XII | blaTEM | − |
| M-M38UD LFS4 | Milk | − | b | XII | blaTEM | − |
Discussion
E. coli is known to be the most common gram negative bacteria that potentially causes subclinical mastitis and exhibits antibiotic resistance. However, pathogenic E. coli in the environment has often been overlooked. Many studies have reported the presence of E. coli among subclinical mastitis cases in dairy farms in many regions of the world, particularly in developing countries, such as Uruguay, Turkey, Brazil, Ethiopia, Mexico, and China (Gianneechini et al., 2002; Guler & Gunduz, 2007; Fernandes et al., 2011; Haftu et al., 2012; Abera et al., 2012; Olivares-Perez et al., 2015; Wang et al., 2015). This study demonstrated the existence of pathogenic E. coli in environmental sources and also in milk from cows with subclinical mastitis by detecting specific genes associated with the pathogenic types of this species. bfpA-positive E. coli was found to be the most common strain of pathogenic E. coli residing in water sources. pCVD432-positive isolate was found in both water and milk samples. evt-positive E. coli was the least common and was only identified in milk samples; it was not present in any of the water samples. In this study, EPEC possessed only bfpA, which encodes bundle-forming pili that are a specific characteristic of EPEC (Cleary et al., 2004). The presence of EPEC in water sources in dairy farms could lead to intramammary infection of cows. A study by Dopfer, Nederbragt & Almeida (2001) also reported the isolation of bfpA-positive EPEC from persistent cases of bovine mastitis (Dopfer, Nederbragt & Almeida, 2001). Although none of the E. coli isolates was positive for eaeA in this study, there are reports of the presence of eaeA-positive EPEC among E. coli found in cows with mastitis in Brazil and Turkey (Correa & Marin, 2002; Guler & Gunduz, 2007). However, in Iran, eaeA-positive E. coli was not found in clinical mastitis cases (Ghanbarpour & Oswald, 2010). This indicates that bfpA- and eaeA-positive EPEC may be distributed unevenly across the globe. EAEC (pCVD432-positive isolates) was found in both water and milk samples. However, the serotypes of those isolates differed. This may indicated different sources of EAEC in water and infected cows. The results also designated that EAEC may be an epidemic strain in dairy farms in Saraburi Province, and EAEC and EPEC could be causative agents of mastitis considering their potential infection through water in farms. EHEC was the least common group found only in milk samples in this study and positive only for evt (shiga-toxin 1-encoding gene). These results raise concerns regarding the bacterial distribution to nearby areas via the contaminated water which workers should be aware of. Studies by Lira, Macedo & Marin (2004) and Momtaz (2010) also reported shiga-toxin 1-producing E. coli from cases of subclinical mastitis in cows in Brazil and Iran. Momtaz et al. (2012) later reported that shiga-toxin 1-producing E. coli was the most common type of E. coli in milk samples from cows with subclinical mastitis in Iran (Momtaz et al., 2012). These results also correlated with many studies on clinical cases of bovine mastitis. For example, Momtaz et al. (2012) reported the presence of EHEC with shiga-toxin 1-encoding gene as the most common virulence gene in milk samples from cases with subclinical and clinical mastitis (Momtaz et al., 2012), which also correlated with the study by Suojala et al. (2011), in which shiga-toxin 1-encoding gene was among the most common virulence genes found in clinical cases of bovine mastitis (Suojala et al., 2011).
Among the 21 antibiotic resistance patterns, the most common pattern found in E. coli from water sources was pattern V (ampicillin and carbenicillin resistance), followed by pattern XVI (ampicillin, carbenicillin, gentamicin, ceftriaxone, cefotaxime, and trimethoprim/sulfamethoxazole resistance). Among the antibiotic patterns in the E. coli from milk, pattern XII (ampicillin, carbenicillin, ciprofloxacin, and norfloxacin resistance) and XVI were the most common. This may indicate that E. coli in milk could potentially derive from water or other environmental sources. A study by Sayah et al. (2005) also reported the difference in antibiotic resistance patterns between E. coli isolated from farm water and fecal samples (Sayah et al., 2005). Our results call for a more cautious approach with antibiotics usage in dairy farms in the Saraburi province area, since the antibiotics that the E. coli isolates were susceptible to are from the high generation cephalosporin and β-lactam classes which are normally used for the treatment of drug-resistance bacteria.
In another study, Geser, Stephan & Hachler (2012) reported on ESBL-positive E. coli in milk from cows with mastitis Geser, Stephan & Hachler (2012), and ESBL-producing E. coli was shown to be able to spread from infected animals to the environment, such as air and slurry, as reported in a pig farm in Germany (Von Salviati et al., 2015). In this study, EAEC was the only pathogenic strain that possessed the ESBL phenotype. Notably, only the ESBL-producing EAEC isolates from water samples contained antibiotic resistance genes (blaTEM and blaCMY-2). The results also correlate with the study by Franz et al. (2015), who reported that EAEC found in surface water and wastewater dominates over other strains of pathogenic E. coli in terms of possessing the ESBL phenotype (Franz et al., 2015). In this study, we found that non-pathogenic E. coli isolates carried ESBL-associated genes (blaTEM, blaSHV, and blaCMY-2. However, only four isolates (M-W22UD LF3, M-W22UD LF7, M-W24UD LF3, and M-W29UD LF1) presented the ESBL phenotype and all of these carried blaTEM and blaCMY-2. These results imply that the presence of drug-resistant strains of non-pathogenic E. coli isolates from the environment is possible. This can pose a threat to mastitis management programs for farm since one study also reported that non-pathogenic E. coli can serve as a reservoir of antibiotic resistance genes and could possibly transfer the genes to other pathogenic E. coli if conditions are suitable (Hu et al., 2016).
Conclusion
This study provides evidence that E. coli isolates from cows with subclinical mastitis and from water at dairy farms in Saraburi Province of Thailand consisted of pathogenic E. coli strains that are resistant to many groups of antibiotics, including the fluoroquinolone group, which should raise concerns regarding the improper use of antibiotics in this area. However, the information on which antibiotics are being used on the farms is very limited. Identification of the ESBL phenotype and β-lactamase genes was also a concern as these can be transferred to other E. coli strains, including pathogenic strains, and bacterial species. This could lead to more serious problems associated with antibiotic resistance in the future. It should be recommended that farms prevent mastitis by promote clean environments for cows such as frequently changing bedding at the stalls and milking areas by cleaning the areas thoroughly. The use of dry and clean cloths to clean the teats before milking and effective teat dips should reduce mastitis on farms. The use of antibiotics, mastitis control programs, and milking hygiene should be considered and supervised by veterinarians to improve mastitis status and treatment in this area.