Description of a novel Ligia species from Nihoa, a remote island in the Papahānaumokuākea Marine National Monument
- Published
- Accepted
- Received
- Academic Editor
- Stefano Kaburu
- Subject Areas
- Biodiversity, Entomology, Evolutionary Studies, Taxonomy, Zoology
- Keywords
- Oniscidea, Cryptic species, Ligiidae, Intertidal, Species description, Pacific biodiversity
- Copyright
- © 2025 Santamaria et al.
- Licence
- This is an open access article, free of all copyright, made available under the Creative Commons Public Domain Dedication. This work may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose.
- Cite this article
- 2025. Description of a novel Ligia species from Nihoa, a remote island in the Papahānaumokuākea Marine National Monument. PeerJ 13:e19373 https://doi.org/10.7717/peerj.19373
Abstract
Isopods in the genus Ligia have been shown to harbor deeply divergent genetic lineages that have, in some instances, been recognized as cryptic species. For instance, the use of molecular taxonomic approaches to characterize coastal Ligia from the Hawaiian Islands led to the redescription of Ligia hawaiensis, the sole endemic coastal species previously recognized in the region, and to the description of seven new species endemic to the region. These species appear to be highly restricted to rift zones within single islands, single islands, or previously connected islands, suggesting these species evolved in allopatry. These findings, coupled with the poor dispersal capabilities exhibited by Ligia isopods and the geology of the Hawaiian Islands, suggest that additional cryptic species may exist in highly isolated populations yet to be studied. Studies to date have characterized Ligia from throughout the younger Hawaiian Islands (e.g., Kaua‘ i, O‘ ahu, Moloka‘ i, Maui, Lanai, and Hawai‘ i); however, no endemic Ligia populations from the older islands and more remote islands that form part of the Papahānaumokuākea Marine National Monument (PMNM) have been studied. This region represents the largest marine conservation area in the USA, and includes at least three islands where L. hawaiensis have been previously reported from. Herein, we apply molecular taxonomic approaches to characterize Ligia specimens from Nihoa, a remote island in the PMNM. Results show that Ligia from Nihoa form a highly divergent that is reciprocally monophyletic lineage with other Hawaiian Ligia species. This lineage, described as Ligia barack sp. nov., adds to the known biodiversity of the PMNM and highlights the importance of continued exploration and conservation of this remote and highly biodiverse region.
Introduction
The Hawaiian Islands (hereafter HI) are a series of islands, atolls, islets, and rocky outcroppings of volcanic origin spanning ∼2,400-km of the northern Pacific Ocean. Islands in the archipelago are arranged in a relatively linear manner, with younger islands located towards the eastern end of the archipelago and older islands found in its western end. Younger islands include eight major islands, all of which have formed in the past 5 million years (in decreasing age: Ni‘ ihau, Kaua‘ i, O‘ ahu, Moloka‘ i, Maui, Lanai, Kaho‘ olawe, and Hawai‘ i). The older islands, found west of Kaua‘ i, include ten island groups ranging widely in size and elevation (in decreasing age: Kure Atoll, Midway Atoll, Pearl & Hermes Atoll, Lisianski, Laysan, Maro Reef, Gardner Pinnacles, French Frigate Shoals, Necker, and Nihoa). These islands are part of the Papahānaumokuākea Marine National Monument (hereafter “PMNM”), a protected area of the United States of America established by presidential decree on June 15, 2006 to protect natural and cultural resources from the region. The Monument initially protected 362,073 km2 of marine habitats; however, it was extended by President Barack H. Obama in 2016 to encompass 1,508,870 km2 of the Pacific Ocean. This makes the PMNM the largest contiguous fully protected conservation in the United States of America and one of the largest marine preserves in the world. The habitats of the PMNM support an incredible diversity of coral, fish, birds, marine mammals and other flora and fauna, many of which are endemic to the PMNM (Starr & Martz, 1999; Starr & Starr, 2008; Kane, Kosaki & Wagner, 2014). Nonetheless, recent descriptions of new species from the PMNM suggest additional new species may exist in this region (Stein & Drazen, 2014; Pyle, Greene & Kosaki, 2016; Sherwood et al., 2020; Alvarado et al., 2022; Sherwood et al., 2022).
Intertidal habitats of the PMNM are known to harbor Ligia isopods, a genus of poorly dispersing isopods shown to hold high levels of cryptic diversity (Taiti et al., 2003; Hurtado, Mateos & Santamaria, 2010; Eberl et al., 2013; Santamaria et al., 2013; Raupach et al., 2014; Santamaria, Mateos & Hurtado, 2014; Santamaria et al., 2017; Greenan, Griffiths & Santamaria, 2018; Santamaria, 2019). Currently, nine Ligia species are thought to be endemic to the HI: eight coastal species that inhabit rocky intertidal habitats, and a terrestrial species that inhabit terrestrial habitats at elevation in the islands of Kaua‘ i, O‘ ahu and Hawai‘ i. The eight coastal species were formerly recognized as L. hawaiensis Dana 1853; however, they were split into these species on the basis of molecular, morphological, and geographic distributional data (Santamaria, 2019). Despite reports of “L. hawaiensis” from the islands of Nihoa, Necker, and La Perouse Pinnacle in the PMNM (Taiti & Howarth, 1996), which are located 190 to 675-km from Kaua‘ i, no specimens from these islands were included in any of the molecular studies characterizing Ligia from the HI to date (Taiti et al., 2003; Santamaria et al., 2013; Santamaria, 2019). Given the limited dispersal potential exhibited of Ligia isopods and the long-term isolation of these oceanic islands, molecular characterizations of these populations are likely to uncover additional cryptic species of Ligia in the region.
In this study, we use molecular approaches to characterize Ligia isopods from the island of Nihoa, the easternmost island in the PMNM. Doing so, we aim to determine: (a) whether Ligia individuals from this highly remote island harbor any unique genetic lineages, (b) if so, what are the phylogenetic relationships of these lineages to other Ligia species previously reported from the HI, (c) whether these lineages are divergent enough to be considered a novel species, and if so (d) describe said lineages as a new species. We do so by incorporating phylogenetic reconstructions, and distance- and phylogeny-based molecular species delimitation methods on a multi-locus dataset comprised of all extant Ligia species from the Hawaiian Islands and newly collected specimens from Nihoa. Our results indicate Ligia from Nihoa represent a highly divergent genetic lineage that is reciprocally monophyletic with all other Ligia species from the HI. Given its genetic uniqueness and geographic isolation, we describe this lineage as Ligia barack sp. nov. on the basis of molecular characters. The formal description of this cryptic species adds to our understanding of the biodiversity of the PMNM.
Materials and Methods
Sample collection
Ligia specimens were collected from the splash zone of rocky coastlines of Hanaka‘ ie‘ ie (Adam’s Bay) in Nihoa during April of 2023. All individuals were caught by hand and field-preserved in 70% isopropanol. The collection of specimens from Nihoa was conducted under a permit granted to the Papahanaumokuakea Marine National Monument Co-Trustees, which include the US Fish and Wildlife Service, by the State of Hawai‘ i Board of Land and Natural Resources (Permit Number PMNM-2022-001). Once in the laboratory, specimens were transferred to 70% ethanol. We identified male individuals as members of the L. hawaiensis cryptic species complex by inspecting the morphology of the distal process of the endopod of the 2nd pleopod and comparing to previous reports (Taiti et al., 2003). Specimens from Nihoa were then inspected under an AmScope SM-4TZ-144 3.5X-90X Trinocular Zoom Stereo Microscope equipped with a 20 MP imaging system. Drawings of characters were made electronically from images produced in the above mentioned system.
Molecular laboratory methods
We used Zymo Research’s Quick g-DNA MiniPrep Kit to extract total genomic DNA for six Ligia individuals collected in Nihoa. DNA was extracted from 2–3 pereopods per individual using standard protocol instructions. We then used previously published primers and conditions to polymerase chain reaction (PCR) amplify the same four mitochondrial and three nuclear gene fragments used by Santamaria (2019) to conduct a taxonomic revision of L. hawaiensis: (a) a 658-bp segment of the Cytochrome Oxidase I gene using primers LCO-1490/HCO-2198 (hereafter COI, primers LCO1490/HCO2198; Folmer et al., 1994), (b) a ∼490-bp segment of the 16S rRNA gene using primers 16Sar/16Sbr (primers 16Sar/16Sbr; Palumbi, 1996), (c) a ∼495-bp segment of the 12S rDNA gene using primers crust-12Sf/crust-12Sr (primers crust-12Sf/crust-12Sr; Podsiadlowski & Bartolomaeus, 2005), (d) a 361-bp fragment of the Cytochrome-b gene using primers 144F and 270R to amplify (hereafter Cyt-b, primers 144F/151F and 270R/272R; Merritt et al., 1998), (e) a ∼1,000-bp segment of the 28S rDNA gene using primers 28SA/28SB (primers 28SA/28SB Whiting, 2002), (f) a 664-bp region of the alpha-subunit of the Sodium Potassium ATPase using primers NaK-forb/NaK-rev2 (hereafter NaK, primers NaK-forb/NaK-rev2; Tsang et al., 2008), and (g) a ∼328-bp fragment of the Histone H3 gene using primers H3AF/H3ARto amplify (primers H3AF/H3AR; Colgan et al., 1998). PCR products were visualized on 1% agarose gels, stained using Apex Safe DNA Gel Stain (Apex Bioresearch Products). Positive amplicons were sequenced at the Arizona Genetics Core.
Sequence alignment and model testing
Sequences were assembled, edited (i.e., had primers removed), and inspected for evidence indicative of heteroplasmy and/or heterozygosity (e.g., multiple peaks in chromatograms) in CodonCode Aligner v10.0.1. No evidence of heteroplasmy or heterozygosity was observed. Sequences produced in this study were then aligned and added to the aligned dataset produced by Santamaria in 2019 using the “—add” option of the MAFFT webserver (Katoh & Standley, 2013) using standard settings. This dataset includes all currently valid species of Ligia endemic to the Hawaiian Islands. Alignments for the three ribosomal genes included in this study (i.e., 28S rDNA, 16S rDNA, and 12S rDNA) were compared to those produced by Santamaria (2019) with poorly aligned sites removed. We inspected alignments of protein coding genes (i.e., COI, Cyt-b, NaK, H3A) and did not observe any evidence suggestive of pseudo-genes such as the presence of early stop codons or indels.
For each gene alignment, we selected the most appropriate model of nucleotide evolution from all available models in jModeltest v2.1 (Darriba et al., 2012) by evaluating their likelihood using a fixed BioNJ-JC tree under the Bayesian information criterion (BIC). Gene alignments were then concatenated using SequenceMatrix v.1.9 (Vaidya, Lohman & Meier, 2011). We used a similar approach as described above to select the most appropriate model of nucleotide evolution for the concatenated alignment. We also selected the most appropriate partition scheme to use in our phylogenetic reconstructions in PartitionFinder v2.1.1 (Lanfear et al., 2016) by evaluating different partitioning combinations of an a priori partitioning scheme that consisted of each ribosomal gene as a single partition with protein coding genes separated by gene and codon position. Partitioning schemes were evaluated under the BIC criterion and the following parameters: branch lengths = unlinked; models = all; model selection = BIC; search = greedy. Lastly, we estimated pairwise Kimura-2-parameter (K2P) distances in MEGA v11.0.13 (Kumar, Stecher & Tamura, 2016) for the COI dataset.
Phylogenetic reconstructions
We conducted phylogenetic reconstructions on the concatenated alignment of all gene fragments under both maximum likelihood and Bayesian inference approaches using two different partitioning approaches: by gene, and as determined by PartitionFinder. Maximum likelihood (ML) searches were conducted in RAxML-NG v1.1.0 (Kozlov et al., 2019) and consisted of 1,000 bootstrap replicates followed by a thorough ML search under the GTR +Γ model run with all other settings as default. Bayesian searches were conducted in MrBayes v3.2.7 (Ronquist & Huelsenbeck, 2003) and consisted of four separate runs consisting each of two chains, run for 20 × 106 generations, sampled every 1,000th generation. All other settings were as default. Bayesian searches were monitored to determine if they had reached and maintained stationarity using the following criteria: (a) stable posterior probability values; (b) high correlation between the split frequencies of independent runs as implemented in AWTY (Nylander et al., 2007); (c) small and stable average standard deviation of the split frequencies of independent runs; (d) potential scale reduction factor close to 1; and (e) an effective sample size (ESS) >200 for the posterior probabilities, as evaluated in Tracer v1.7.2 (Rambaut et al., 2018). For all searches, we calculated majority-rule consensus trees using the SumTrees command of DendroPy v3.10.1 (Sukumaran & Holder, 2010). For Bayesian analyses, samples prior to stationarity were discarded as burn-in.
Molecular species delimitation analyses (MSDAs)
We implemented both tree- and distance-based species delimitation analyses to determine whether our molecular dataset supports the identification of Ligia from Nihoa as a separate species. Tree-based molecular species delimination analyses (MSDAs) were carried out using the Poisson Tree Processes model as implemented in the PTP server (http://species.h-its.org/) and the General Mixed Yule Coalescent model (hereafter GMYC; Fujisawa & Barraclough, 2013). PTP analyses were carried out on all phylogenetic trees produced in RAxML and MrBayes. Settings used were as follows: 500,000 Markov chain Monte Carlo (MCMC) iterations; a burn-in of 0.10; and a thinning value of 100. As GMYC delineations require ultrametric trees as input, we estimated ultrametric trees for the unpartitioned concatenated mitochondrial dataset using BEAST v2.1.3 (Bouckaert et al., 2014) assuming a constant rate of evolution and speciation assuming a Yule process (i.e., constant speciation rate; Yule, 1925; Gernhard, 2008), and under a coalescent model of speciation assuming a constant population size (Kingman, 1982). Both searches were carried out for 50 million generations sampled every 1,000th generation using the most appropriate model of nucleotide evolution. Resulting trees were summarized using the SumTrees command with burn-in discarded and with edges set as per the mean-age option. Resulting ultrametric trees were analyzed using the GMYC approach as implemented by the ‘splits’ package (http://r-forge.r-project.org/projects/splits/) in R using default settings.
We conducted distance-based analyses using ASAP (Puillandre et al., 2012) on the COI gene dataset alone after masking ambiguous sites using the ASAP webserver (https://bioinfo.mnhn.fr/abi/public/asap/). ASAP analyses were carried out under the Kimura 2-parameter (K2P) nucleotide evolution model, with all other settings as default. We used KoT (Spöri et al., 2022) to estimate the K/θ ratio (Birky Jr et al., 2010; Birky Jr, 2013) between Ligia from Nihoa and their most closely related taxa identified by phylogenetic analyses. Analyses were carried on the concatenated dataset assuming a K/θ threshold of 4, a value that represents a >95% probability that sister clades have become reciprocally monophyletic (Birky Jr, 2013).
We evaluated the following criteria to determine whether Ligia from Nihoa represent a novel species in need of description: (1) did all phylogenetic reconstructions place all Nihoa Ligia individuals in a well-supported (bootstrap support (BS) >90%, Bayesian posterior probability (BPP) >95%) monophyletic clade that excluded all other Ligia from the Hawaiian Islands; (2) were pairwise COI K2P distances amongst Ligia Nihoa specimens <1.0%; (3) do comparisons between Ligia from Nihoa and its sister taxon produce a K/θ>4 (i.e., 4X rule; Birky Jr, 2013); (4) did most MSDAs separate Nihoa individuals as a putative species; (5) did this putative species exclude all other Ligia from the HI. As the answer for all these criteria was affirmative, we herein describe Ligia barack, a novel species of Ligia from Nihoa. We determined diagnostic nucleotide positions for this novel species using FASTACHAR v0.2.4 (Merckelbach & Borges, 2020) by comparing L. barack sp. nov. to all other Ligia species endemic to the HI included in the dataset used in this study.
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/. The LSID for this publication is: urn:lsid:zoobank.org:pub:6CE79D26-19BA-435D-94A8-5A822ADD42B0. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central and CLOCKSS.
Results
We successfully produced sequences for four mitochondrial and three nuclear genes for six Ligia specimens from Nihoa (hereafter L. barack sp. nov). Unique haplotypes have been deposited in GenBank and BOLD (see Table 1 for GenBank Accession Numbers). The addition of these sequences to the alignment produced by Santamaria (2019) produced a concatenated dataset 3,996-bp long prior to the removal of poorly aligned positions for the 16S, 12S, and 28S rDNA gene. This final alignment included 196 individuals from 40 localities in the HI including Nihoa (Fig. 1, Table 1). Removal of poorly aligned sites (43, 17, and 49 for the 16S, 12S, and 28S rDNA genes respectively) produced a final alignment 3,887-bp long containing 543 parsimony informative sites (COI = 185; Cyt-b: 120; 12S rDNA = 99; 16S rDNA = 91; 28S rDNA = 39; NaK = 6; H3A = 3). An annotated alignment is provided as Dataset S1.
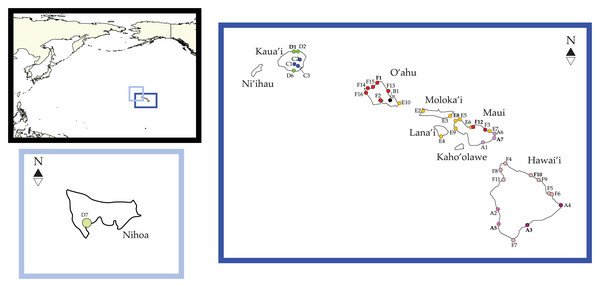
Figure 1: Ligia localities included in this study.
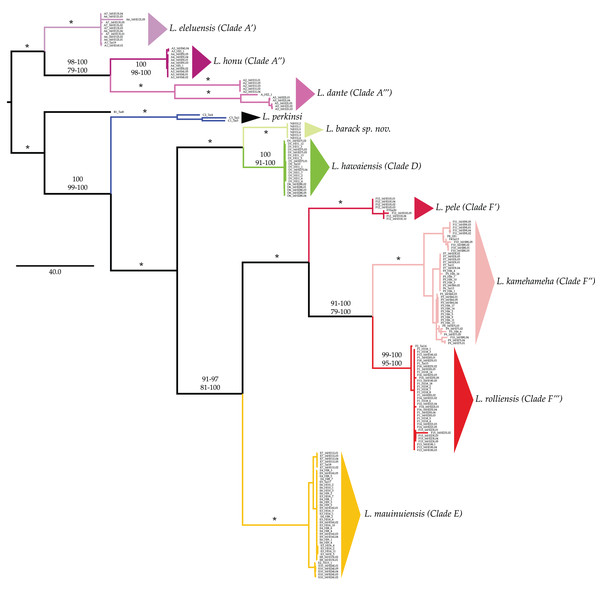
Labels and colors correspond with other figures and tables in this study and that of Santamaria et al. (2013) and Santamaria (2019). Detailed information for each locality is presented in Table 1. Localities of the suppralittoral Ligia included: Kaua‘ i: D1-Kalihiwai Beach, D2-Kauapea Beach, D6-Hoai Bay; D7- Hanaka‘ ie‘ ie (Adam’s Bay); O‘ ahu: E10-Wawamalu Beach Park, F1-Pupukea, F2-Pouhala Marsh, F13-Kahaluu, F14-Kaena Point (North), F15-Kaiaka Bay Beach Park, F16-Kaena Point (South); Moloka‘ i: E2-Papohaku Beach Park, E4-Manele Bay; Lana‘ i: E3-North of Puko‘ o; Maui: A1-Wai‘ Ōpae; A6-Waianapanapa State Park, A7-Koki Beach Park, E5-Poelua Bay, E6-Spreckelsville, E7-Keanae, E8-DT Fleming Beach Park, E9-Hanakao‘ o Park, F3-Honomanu Bay, F12-Baby Beach Spreckelsville Area; Hawai‘ i: A2-Kealakukea Bay, A3-Pu‘ unalu Beach Park, A4-Isaac Hale Beach Park, A5-Miloli Beach Park, F4-Keokea Beach, F5-Onekahakaha Beach Park, F6-Leleiwi Beach, F7-South Point, F8-Kapa‘ a State Park, F9-Kolekole Beach Park, F10-Laupahoehoe Beach Park, F11-Spencer Beach Park. Localities of the terrestrial L. perkinsi included are Kaua‘ i: C1-Mt Kahili, C2-Makaleha Mts, C3-Haupu Range; O‘ ahu: B1-Nu‘ uanu Pali. World map is edited from a public domain map produced by Colohisto. Original vector map is available at https://commons.wikimedia.org/wiki/File:BlankMap-World_1990.svg. Map of the Hawaiian Islands is reproduced from Santamaria (2019). Map is available at: https://doi.org/10.7717/peerj.7531/fig-1.All phylogenetic reconstructions completed in this study were similar to those reported by Santamaria et al. (2013) and Santamaria (2019), with the exception of L. barack sp. nov individuals (Fig. 2). These individuals were placed in a well-supported clade (BS = 100; PP = 100, Fig. 2) that excluded individuals from all other Ligia species endemic to the HI. Our phylogenetic reconstructions identified four highly divergent and reciprocally monophyletic lineages consisting solely of coastal Ligia endemic to the Hawaiian Islands: (a) Clade A (lavenders and purples in all figures; BS = 100; posterior probability (PP) = 100) which consisted of all L. dante (A2, A5 in Hawai‘ i), L. honu (A3–4 in Hawai‘ i) and L. eleluensis (A1, A6–7 in Maui) individuals; (b) Clade D (green in all figures; BS =100; PP =100) which included all L. hawaiensis individuals from Kaua‘ i (D1–2, D6) as well as L. barack sp. nov. (D7); (c) Clade E (oranges and yellows in all figures; BS = 100; PP = 100) consisting of all L. mauinuiensis individuals from O‘ ahu (E10), Moloka‘ i (E2, E3), Lana‘ i (E4), and Maui (E5–E9); and lastly (d) Clade F (reds in all figures; BS = 100; PP = 100) which included all L. kamehameha (F4–F11 in Hawai‘ i), L. rolliensis (F1–2 and F13–16 in O‘ ahu), and L. pele (F3, F12 in Maui) individuals. We also observed two lineages consisting of individuals of the terrestrial L. perkinsi: (a) Clade B (black in all figures) from O‘ ahu (B1), and (b) Clade C (blue in all figures; BS = 100; PP = 100) from Kaua‘ i (C1–3).
Figure 2: Majority rule consensus tree of bootstrap replicates produced by analyzing the concatenated mitochondrial and nuclear dataset of Ligia from the Hawaiian Islands in RAxML under the GTR +Γ under a “by gene” partitioning scheme.
Branches and clades are colored as per Santamaria et al. (2013) and Santamaria (2019). Values by nodes correspond with bootstrap support values observed in RAxML analyses (above) and posterior probabilities produced in MrBayes analyses (below). Asterisks (*) denote 100% support across all analyses.Clades D, E, and F were placed in a well-supported monophyletic group (BS = 100; PP = 100) with clades E and F identified as each other’s sister clade (BS = 81–100; PP = 91–97). The sister to the “D + E + F” clade was Clade C (BS = 99–100; PP = 100), which consisted of the terrestrial L. perkinsi from Kaua‘ i. Clade B, consisting of the terrestrial L. perkinsi from O‘ ahu, was identified as the sister clade to the large monophyletic group consisting of clades C, D, E, and F (BS = 100; PP = 100). The most basal group was Clade A, which consisted of coastal Ligia species from the islands of Maui and Hawai‘ i.
COI K2P distances between Ligia species from HI ranged between 3.0–17.8%, with comparisons between L. barack sp. nov and other species in the region ranging between 3.0–17.8% (Table 2). Genetic diversities produced when comparing L. barack sp. nov were low, ranging between 0.0–0.3% (Table 2).
| L. barack | L. dante | L. honu | L. eleluensis |
L. perkinsi (O‘ ahu) |
L. perkinsi (Kaua‘ i) |
L. hawaiiensis | L. mauinuiensis | L. rolliensis | L. kamehameha | |
|---|---|---|---|---|---|---|---|---|---|---|
| L. barack | 0.0–0.3 (0.2) |
|||||||||
| L. dante | 13.9–15.2 (14.5) |
0.0-4.6 (2.4) |
||||||||
| L. honu | 14.9–15.1 (15) |
5.8–7.5 (6.8) |
N/A | |||||||
| L. eleluensis | 17–17.8 (17.4) |
9.7–11.2 (10.7) |
10.9–11.3 (11.0) |
0.0–0.9 (0.5) |
||||||
|
L. perkinsi (O‘ ahu) |
14.8–15.0 (14.9) |
14.2–15.0 (14.5) |
15.1–15.1 (15.1) |
14.3–14.8 (14.6) |
N/A | |||||
|
L. perkinsi (Kaua‘ i) |
15.8–16.9 (16.4) |
11.9–14.3 (13.2) |
13.7–15.1 (14.1) |
12.5–13.9 (13.2) |
14.5–15.3 (14.9) |
1.0–2.7 (1.9) |
||||
| L. hawaiiensis | 3.0–4.0 (3.5) |
12.8–15.4 (14.4) |
15.0–16.9 (16.4) |
14.7–16.4 (15.8) |
14.8–15.8 (15.5) |
13.8–15.6 (15.1) |
0.0–2.2 (0.9) |
|||
| L. mauinuiensis | 11.5–13.2 (12.5) |
13.6–15.9 (14.9) |
14.5–15.3 (15.0) |
13.9–16.4 (15.3) |
16.0–16.6 (16.4) |
12.5–14.2 (13.0) |
10.3–12.7 (11.5) |
0.0–2.4 (0.7) |
||
| L. rolliensis | 12.5–14.3 (13.6) |
14.0–16.6 (15.3) |
14.9–16.2 (15.3) |
13.4–16.2 (14.5) |
13.7–14.7 (14.1) |
12.9–16.1 (13.9) |
12.5–14.8 (13.6) |
10.9–12.8 (11.6) |
0.0–7.1 (1.9) |
|
| L. kamehameha | 12.3–15.4 (13.5) |
13.8–16.4 (14.8) |
13.6–15.5 (14.6) |
13.8–15.4 (14.6) |
12.6–13.9 (13.2) |
11.1–15.0 (13.2) |
11.6–14.6 (12.9) |
8.7–13.1 (10.6) |
4.0–8.7 (5.3) |
0.0–5.4 (2.5) |
Molecular species delimitations consistently identified L. barack sp. nov as a separate and distinct species from other Ligia species endemic to the HI. ASAP analyses of the COI dataset placed all Nihoa specimens in a separate putative species containing no Ligia from other localities in nine of the ten best partitions produced by ASAP, with only the ninth best supported partition (p-value rank = 4; W rank = 18; threshold distance = 0.040249) grouping Nihoa Ligia with L. hawaiensis individuals (Kaua‘ i). All tree-based MSDAs carried out in PTP, bPTP, and GMYC recognized L. barack sp. nov as a separate species. Lastly, comparisons between this newly described species and L. hawaienesis, its sister taxon, in KoT produced a K/θ ratio of 9.912.
Taxonomy
Based on the long-term and geographical isolation for Nihoa, morphological comparisons, results of phylogenetic reconstructions and MSDAs, COI K2P pairwise distances reported herein, and K/θ ratio between it and its sister taxon, we describe Ligia barack sp. nov., a new species of Ligia from Nihoa. A holotype and three paratypes were deposited at the Florida Museum of Natural History (FLMNH) in Gainesville, FL, USA. We describe L. barack sp. nov. primarily using molecular characters and some diagnostic morphological characters. We include a broad description of the holotype that covers traits evaluated by previous authors (Taiti et al., 2003; Santamaria et al., 2013; Santamaria, 2019), a photograph of the holotype of L. barack (Fig. 3), and illustrations of diagnostic features of this species (Fig. 4). Other traits not mentioned below (e.g., pereopods 2–6) are as described and/or illustrated by Taiti et al. (2003), Taiti, Ferrara & Kwon (1992), and Jackson (1933).
Figure 3: Holotype of Ligia barack, a new species from Nihoa.
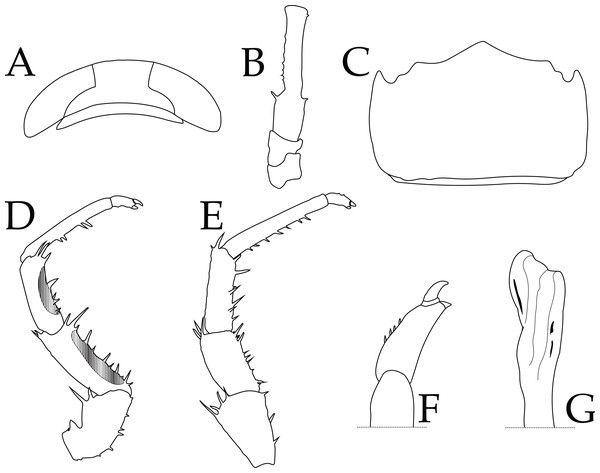
Holotype shown in this picture is deposited at the Florida Museum of Natural History (UFID 72496).Figure 4: Ligia barack nov. sp. holotype (UFID 72496).
(A) Cephalon; (B) peduncles 1–3 of antenna; (C) Pleotelson, dorsal view; (D) Pereopod 1; (E) Pereopod 6; (F) Closeup of dactylus of Pereopod 6; (G) apex of appendix masculina of second pleopod.Ligia barack nov. sp.
LSID: urn:lsid:zoobank.org:act:558494AB-37D7-47BA-BA54-4E532D7585C6.
BOLD BINs: AFQ9578.
Materials examined: six individuals from the island of Nihoa (D7). Both males and females were included. The holotype (♂; UFID 72496; Fig. 3), and three paratypes (♀♀♀; UFID 72497-72499) from the type locality have been deposited at the Florida Museum of Natural History (FLMNH) in Gainesville, FL, USA.
Type locality: Hanaka‘ ie‘ ie (Adam’s Bay), Nihoa, Hawai‘ i, USA. (D7; 23°03′30.3″N 161° 55′27.6″W).
Description of holotype: male individual that is 17.8 mm long and 6.7 mm wide at the widest point of the pereionite 4 (body length to width ratio of ∼2.7; Fig. 2). Eyes are large (eye length is ∼0.5 greatest width of cephalon) and closely spaced (inter-eye distance ∼0.5 times eye length) (Fig. 4A). Posterolateral processes of the pereionite 7 extend ∼ length of the pleonite 3. Antennae extends just past midbody, being about ∼0.6 times the total body length. Article 3 of the peduncle of the antenna is about 1.5X length of the peduncle article 2, with robust setae on either side of the distal end (Fig. 4B). Pleotelson shape similar to that of other Ligia in the Hawaiian Islands; however, the lateral posterior points are nearly parallel to the interior lateral posterior points (Fig. 4C). First three pereopods have papillar fields present, with those in the carpus and merus of the first pereopod present in over half of pereopod segment (e.g., pereopod 1 illustrated in Fig. 4D). The dactylus of the sixth and seventh pereopod have four small robust setae on the tergal margin (Fig. 4F). Apex of the appendix masculina in the endopod of the second pleopod is enlarged and slightly bilobed with distal end rounded (Fig. 4G).
The holotype is deposited in the FLMNH under UFID 72496. GenBank Accession numbers for sequences obtained from the holotype are: PP851829 (COI); PP852382 (16S rDNA); PP852387 (12S rDNA); PP856001 (Cyt-b); PP852394; (28S rDNA); PP856007 (NaK); and PP861092 (H3A).
Remarks: the herein described L. barack sp. nov can be distinguished from other coastal Ligia species endemic to the Hawaiian Islands by the absence of a tuft of very long thin setae on the tergal margin of the dactylus of the sixth and seventh pereopods (Figs. 4E–4F; see Fig. 4 in Taiti et al., 2003). The appendix masculina of the endopod of the second pleopod also differs between L. barack sp. nov and other species in the region (Taiti et al., 2003). The appendix masculina of L. barack is slightly bilobed with the distal end being rounded, which contrasts with the obliquely truncate morphology seen in coastal Ligia from the Hawaiian Islands (see Fig. 4G; contrast with Fig. 6 in Taiti et al., 2003). Lastly, L. barack exhibits slight differences in eye shape and size. This species has large eyes (eye length is ∼0.5 greatest width of cephalon) which is similar to other coastal Ligia species endemic to the Hawaiian Islands; however, the eyes appear to be the most distantly spaced of all these species. The inter-eye distance of L. barack is about ∼0.5 times the eye length, while the smallest ratio for other nearby species is ∼0.7 (seen in L. honu, L. hawaiiensis, L. kamehameha, L, mauiniensis, L. rolliensis; Santamaria, 2019). Lastly L. barack can also be distinguished using molecular characters listed below.
Diagnostic molecular characters:
COI: 1-C; 31-A; 94-C; 526-C
16S: 288(316)-T.
Cyt-b: 181-G; 223-C; 262-G; 265-G; 354-G
12S: 380(398)-G.
Distribution: rocky intertidal habitats of Nihoa.
Hawaiian common name: Pokipoki o Hanaka‘ ie‘ ie. Pokipoki is the Hawaiian name for terrestrial isopods and similar creatures inhabiting aquatic and terrestrial habitats. Meanwhile, Hanaka‘ ie‘ ie refers to the traditional name for Adam’s Bay of Nihoa Island. Thus, this name broadly translates to “the isopod from Adam’s Bay of Nihoa Island.”
Etymology: this species is named after Barack H. Obama, the former President of the United States of America, who was born in the island of O‘ ahu and who is responsible for the expansion of the Papahānaumokuākea Marine National Monument to its current size.
Discussion
The Hawaiian Islands (HI) were previously thought to harbor a single endemic coastal Ligia species: Ligia hawaiensis. This species, first described by Dana (1853), was determined to represent a cryptic species complex composed of allopatric species with distributional ranges largely limited to rift zones within a single island, single islands, or previously connected islands (Santamaria, 2019). Despite previous reports of L. hawaiensis from the remote and older islands found in the Papahānaumokuākea Marine National Monument (Taiti & Howarth, 1996), none of the molecular studies conducted on Hawaiian Ligia to date have included populations from these islands. This has left unanswered whether Ligia populations from the older and highly remote islands in the PMNM harbor highly divergent genetic lineages and/or novel species in need of description. By using similar molecular approaches to those used by Santamaria (2019) to describe highly genetically divergent yet largely morphologically cryptic lineages of Ligia in the HI as new species, we herein describe L. barack sp. nov from Nihoa.
Our molecular characterizations of Ligia individuals collected in Nihoa show this population to be highly divergent and isolated from other Ligia lineages and species found in the HI. We observed no sharing of haplotypes between L. barack sp. nov individuals and other Ligia populations in the HI at any of the four mitochondrial genes studied (e.g., COI, Cyt-b, 16S and 12S rDNA). Instead, Nihoa specimens harbored unique and private haplotypes that form a well-supported monophyletic group that excludes all other Ligia species from the HI and that are highly divergent from other ones found to date in Hawaiian Ligia. COI K2P divergences between Nihoa Ligia and other Ligia species from Hawaii ranged from 3.0–17.8%, values that are similar to other amongst species comparisons (Table 2). Meanwhile, within species COI K2P divergences amongst L. barack sp. nov individuals ranged from 0.0–0.7%. Not surprisingly, the K/θ ratio between L. barack sp. nov and its sister taxon (L. hawaiensis; K/θ = 9.912), greatly exceeds the K/θ ratio of 4 at which there is a 95% probability that two separate species are being compared (Rosenberg, 2003).
The phylogenetic placement of L. barack sp. nov is of interest, as our analyses recovered with high support both the monophyly of this newly described species and its sister relationship to L. hawaiensis (Fig. 4). The latter is a coastal species of Ligia whose distributional range is thought to be limited to the island of Kaua‘ i, the closest island to Nihoa. These islands are separated by ∼240 km of open ocean and have never been connected. This suggests that oceanic dispersal led to the colonization of these islands by Ligia. Nihoa’s older age (7.5 My) suggests the ancestor to L. hawaiensis in Kaua‘ i may have originated from Nihoa; however, additional work is necessary to establish the origins of these species as back-dispersals appear to have occurred in Ligia from the HI.
Despite consisting of the Ligia from the two oldest islands in our analyses (Kaua‘ i: 5 My, Nihoa 7.5 My), the monophyletic group consisting of L. hawaiensis + L. barack sp. nov clade was not found in a basal position in any of our analyses (Fig. 3). Instead, the most basal clade in all analyses was one comprised of Ligia species found in Maui and Hawai‘ i, the two youngest islands in the archipelago (<1.5 My). These findings are consistent with previous studies of Ligia from the HI (Santamaria et al., 2013; Santamaria, 2019) and suggest that the evolution of Ligia in the region have been shaped by colonization, extinction, and back-dispersal events.
Our description of L. barack sp. nov from the island of Nihoa underscores the importance of molecular approaches in conservation efforts in the PNMN. Future studies of Ligia from other islands in the PNMN are likely to uncover additional highly divergent genetic lineages likely representing new species in need of description. These studies may also help further elucidate the evolutionary history of Ligia in the HI. Meanwhile, molecular characterizations of other poorly dispersing organisms may similarly uncover new species or genetic lineages in other taxa and thus increase our understanding of the biodiversity of these highly remote and isolated islands. Molecular tools may also aid in the monitoring of the spread of alien species, a critical threat to the fauna and flora of the PMNM (DeFelice et al., 1998; Selkoe, Halpern & Toonen, 2008). Ligia exotica has been shown to occur in Midway Atoll, an island within the PMNM (Santamaria et al., 2022). This species of Asian origin known to have been introduced to manmade coastal habitats around the world and is a potential competitor to endemic coastal Ligia (Hurtado et al., 2018). The use of genetic tools such as COI barcoding and eDNA may be useful to monitor the presence of this species in other regions of the PMNM without extensive field-work.
Conclusion
The use of both mitochondrial and nuclear gene fragments to characterize Ligia isopods from Nihoa uncovered a highly divergent lineage of Hawaiian Ligia not previously reported from other localities in the HI. Phylogenetic and species delimitation approaches provide evidence that this lineage represents a new species of Ligia, which we describe as Ligia barack sp. nov. To our knowledge, this species is the first intertidal crustacean that is described from and likely solely endemic to the island of Nihoa. This discovery underscores the unique biodiversity of the PMNM and the need for additional studies of poorly dispersing taxa within it. Our findings also further provide evidence of Ligia isopods as an example of in-situ speciation of a Hawaiian marine animal.