Genome-wide identification of bHLH gene family and its response to cadmium stress in Populus × canescens
- Published
- Accepted
- Received
- Academic Editor
- Muhammad Aamer Mehmood
- Subject Areas
- Bioinformatics, Plant Science, Soil Science, Environmental Contamination and Remediation, Forestry
- Keywords
- bHLH gene family, Populus canescens, Cd stress
- Copyright
- © 2024 Yao et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits using, remixing, and building upon the work non-commercially, as long as it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2024. Genome-wide identification of bHLH gene family and its response to cadmium stress in Populus × canescens. PeerJ 12:e17410 https://doi.org/10.7717/peerj.17410
Abstract
The basic helix-loop-helix (bHLH) gene family is integral to various aspects of plant development and the orchestration of stress response. This study focuses on the bHLH genes within Populus × canescens, a poplar species noted for its significant tolerance to cadmium (Cd) stress. Through our comprehensive genomic analysis, we have identified and characterized 170 bHLH genes within the P. canescens genome. These genes have been systematically classified into 22 distant subfamilies based on their evolutionary relationships. A notable conservation in gene structure and motif compositions were conserved across these subfamilies. Further analysis of the promoter regions of these genes revealed an abundance of essential cis-acting element, which are associated with plant hormonal regulation, development processes, and stress response pathway. Utilizing quantitative PCR (qPCR), we have documented the differential regulation of PcbHLHs in response to elevated Cd concentrations, with distinct expression patterns observed across various tissues. This study is poised to unravel the molecular mechanism underpinning Cd tolerance in P. canescens, offering valuable insights for the development of new cultivars with enhanced Cd accumulation capacity and tolerance. Such advancements are crucial for implementing effective phytoremediation strategies to mitigate soil pollution caused by Cd.
Introduction
The eukaryotic domain is replete with basic helix-loop-helix (bHLH) proteins, which constitute one of the largest transcription factor family in plants, second only to the MYB family (Ledent & Vervoort, 2001; Riechmann & Ratcliffe, 2000). The bHLH proteins are characterized by a conserved 60 amino acids domain, which is divisible into two distinct functional regions (Jones, 2004). The N-terminal basic region is critical for DNA binding (Atchley, Terhalle & Dress, 1999), while the C-terminal helix-loop-helix facilitates protein-protein interactions, essential for the formation of homodimeric or heterodimeric complex (Ferre-D’Amare et al., 1994; Murre, McCaw & Baltimore, 1989).
In plants, bHLH transcription factors are implicated in a myriad of biological processes, including growth, development, and stress responses (Liu, Peng & Dai, 2014; Meraj et al., 2020). Notably, certain bHLH proteins have been identified as key mediators of heavy metal, particularly cadmium (Cd), tolerance (Wu et al., 2012; Xu et al., 2017; Yuan et al., 2008). Populus × canescens, a poplar species with remarkable Cd accumulation capacity, is of particular interest for its potential in phytoremediation—a strategy for mitigating soil contamination using plants (He et al., 2011).
This study marks the first comprehensive characterization of the bHLH gene family in P. canescens, revealing 170 genes distributed across the genome and classified into 22 subfamilies. Our analysis encompassed gene structures, conserved motifs, DNA-binding capability and cis-acting elements in promoters, alongside gene duplications, synteny, and phylogenetic relationships.
Furthermore, we conducted a detailed analysis of tissue-specific and Cd-induced expression profiles of selected PcbHLH genes, laying the groundwork for future research into the functional and regulatory mechanisms of these genes in response to Cd stress. This work not only contributes to our understanding of Cd tolerance in P. canescens but also identifies potential candidate genes for breeding new germplasm with enhanced Cd pollution resistance.
Materials and Methods
Identification of the basic/helix-loop-helix family genes in P. canescens
The genomic sequence data of P. canescens were sourced from the Aspen database (https://www.aspendb.org/downloads and the annotation version was sPta717alba_v2). bHLH transcription factors of Arabidopsis thaliana (AtbHLHs) were obtained from Carretero-Paulet et al. (2010), and those of Populus trichocarpa (PtbHLHs) were retrieved from NCBI’s database. Hidden Markov model (HMM) files for the bHLH domain (PF0010) were acquired from interProScan (https://www.ebi.ac.uk/interpro/entry/pfam/PF00010/), which were then utilized to identify bHLH proteins in P. canescens using the SPDE software (Xu et al., 2021). BLASTp searches were also performing using AtbHLHs against the P. canescens amino acid sequence data. To confirm the presence of the bHLH domain (E-value <1e−5), CD-search (https://www.ncbi.nlm.nih.gov/) and SMART (http://smart.embl-heidelberg.de/) were employed to analyze the identified PcbHLH sequences. The molecular weights (kDa) and isoelectric points (pI) of the PcbHLHs were calculated using the SPDE software.
Chromosomal locations of PcbHLHs
Chromosomal locations of the PcbHLH genes were determined using the P. canescens database (https://www.aspendb.org/downloads), and a distribution map was created with TBtools (Chen et al., 2023).
Multiple sequence alignments of PcbHLHs
Multiple sequence alignments were conducted using ClustalX in MEGA7, followed by visualization with Jalview (version 1.8.3). The variable sequences at the N- and C-terminal regions were excised, preserving the conserved domains in the central region. Sequence logos for the bHLHs were created by submitting the multiple alignment sequences to a specialized online platform (https://weblogo.berkeley.edu/logo.cgi). Employing the criteria delineated by Massari & Murre (2000), we classified the PcbHLH proteins into two principal categories based on the sequence information within the N-terminal region of the bHLH domains: DNA binding and Non-DNA binding (containing fewer than four basic amino acids). The DNA binding bHLHs were further categorized into two groups: E-box binding and Non-E-box binding (based on the presence of only Glu12 or Arg15). Consequently, E-box binding were further subdivided into two subgroups: G-box binding (characterized by His/Lys8, Glu12 and Arg16) and non-G-box binding (defined by the presence of only Glu12 and Arg16).
Phylogenetic analysis of PcbHLHs
To elucidate the evolutionary relationships between P. canescens and Arabidopsis bHLHs, we performed multiple sequences alignment of 170 complete PcbHLH sequences and 167 complete AtbHLH sequences using MEGA7. Subsequently, a Maximum Likelihood phylogenetic tree was constructed in MEGA7, with a bootstrap value set to 1,000 replicates for reliability. The phylogenetic tree was visualized using Itol (https://itol.embl.de/), which provided a clear depiction of the bHLHs’ evolutionary history.
Gene structure and protein motifs analysis
The exon/intron organization and splicing phase of the predicted PcbHLHs were analyzed using the Gff3 annotation files from P. canescens genome. This data were then graphically represented using TBtools. To identify conserved motifs within the PcbHLH proteins, we employed MEME (https://meme-suite.org/meme/tools/meme, version 5.5.4) to identify a maximum of ten motifs with an optimal width ranging from 10 to 100 amino acids. The resulting phylogenetic trees, gene structures, and conserved motifs were integrated and visualized within TBtools for comprehensive analysis.
Cis-acting element analysis in PcbHLH promoters
Promoter regions of PcbHLH genes, defined as 2000bp upstream sequences, were analyzed for cis-acting elements using PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). TBtools facilitated the visualization of these elements, and a heat map was generated using Rstudio (version 4.2.2; RStudio Team, 2024) to represents the distribution of various cis-acting elements, providing insight into the regulatory mechanism of these genes.
Gene duplication and synteny analysis of PcbHLH genes
Gene duplication and synteny analysis provided insights into the evolutionary dynamics of bHLH genes in poplar and other plants. This analysis was performed using the Multiple Collinearity Scan toolkit (MCScanX), with results visualized in TBtools. Additionally, the nonsynonymous (ka) and synonymous (ks) substitution rates of bHLH gene pairs were calculated, offering further evidence of the evolutionary pressures acting on these genes.
Plant growth conditions and and Cd treatment
P. canescens seedlings, derived from micropropagation (Li et al., 2023), were cultured hydroponically at 25 °C under a photoperiod of 16 h light and 8 h dark within an artificial climate chamber. After acclimatization in half-strength Hoagland nutrient medium for one month, seedlings were treated with 10 µM CdCl2 to induce Cd stress for 168 h. A concentration of 10 µM CdCl2 was sufficient to elicit a transcription factor response in P. cansecens without causing harm. Samples of root, stems and leaves from individual P. canescens plants were collected at 0, 6, 12, 24, 48, 96, and 168 h post-treatment, with all samples collected simultaneously to minimize the impact of diurnal rhythms. All samples were collected in triplicate to negate the impact of biological variability.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from root, stem, and leaf samples of P. canescens seedlings using the RNAprep Pure Plant Plus Kit (TIANGEN, Beijing, China). First-strand cDNA synthesis was performed using the PrimeScript™ RT Master Mix (TaKaRa, Dalian, China). Quantitative primers were designed using TBtools and qPCR was conducted with the 2 × Q3 SYBR qPCR Master Mix (TOLOBIO, Shanghai, China) on a 7300 Real-Time PCR System (Applied Biosystems, CA, United States). Technical replicates were conducted three times for each sample. Relative expression levels were quantified using the 2−ΔΔCT method with EF1B (accession number FJ372570) gene serving as a reference (Wildhagen et al., 2010).
Results
Identification and classification of bHLH protein in P. canescens
The initial bioinformatic screening of P. canescens using HMMER with hidden Markov models and BLASTp yielded a total of 179 predicted bHLH proteins. Subsequent filtering, employing CD-Search and SMART to exclude proteins with incomplete domains, culminated in the identification of 170 putative bHLH proteins (Table S1). These proteins were designated as PcbHLH1 to PcbHLH170, based on their chromosome locations. An analysis of their biochemical properties (Table S2) indicated a range of amino acid lengths from 90 for PcbHLH43 and PcbHLH154 to 740 for PcbHLH46. The molecular weights spanned from 10.29 kDa to 71.95 kDa, and the predicted isoelectric points ranged from 4.63 to 10.18. All proteins exhibited a grand average of hydropathy values below zero, indicative of their hydrophilic nature.
Chromosomal localization of PcbHLH genes
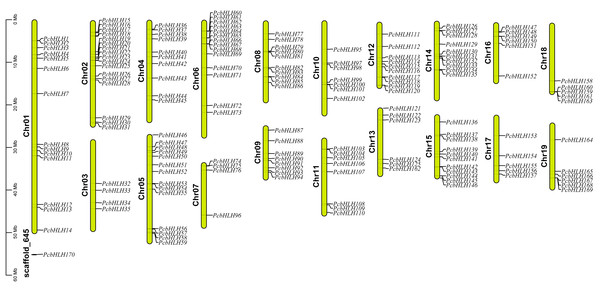
Distribution mapping revealed that 169 of the 170 PcbHLH genes are unevenly distributed across the P. canescens genome, with Chr2 harboring the highest number (17 genes). In contrast, only four PcbHLH genes were mapped to Chr3 and Chr7 (Fig. 1). Notably, PcbHLH170 remained unassigned to any chromosomes, which may be attributed to incomplete genome assembly.
Figure 1: Distribution of P. canescens bHLH genes on chromosomes.
The names of chromosomes and genes are shown on the left and right, respectively. The scale on the left is in megabases (Mb).Multiple sequence alignments of PcbHLHs
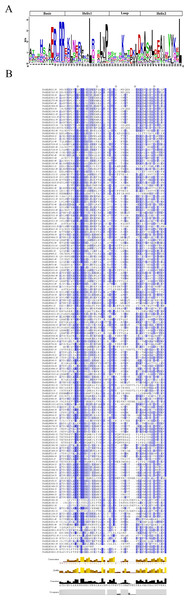
A comprehensive multiple sequence alignment of the 170 PcbHLH protein sequences was performed, revealing that the bHLH domain encompasses four conserved regions: a basic region, two helical regions, and a loop region. The basic region is characterized by specific residues, including His-5, Glu-9, Arg-10, Arg-12, and Arg-13, while the first helix region is defined by Ile-16, Leu-23, Leu-27, Val-28, and Pro-29. The loop region is predominantly composed of Lys-35 and Asp-41, and the second helix region comprises Ala-43, Leu-46, Glu-48, Ala-49, Ile-50, Tyr-52, and Leu-56. Notably, Fig. 2 demonstrated the high conservation of Leu-23 across the 170 PcbHLH amino acid sequences, underscoring its pivotal role in facilitating dimerization among PcbHLH proteins.
Figure 2: Schematic diagrams of the conserved amino acids and multiple alignment of the PcbHLH domains.
(A) Sequence logo for the PcbHLH domains drawn by WebLogo. The overall height of each stack represents conservation of the sequence at a position. (B) Multiple sequence alignment of the PcbHLH domains.The basic region of the PcbHLH proteins is crucial for DNA-binding. As per Massari’s classification, these proteins are divided into two main groups based on sequence information within the basic region of the bHLH domains. The majority, comprising 165 PcbHLH proteins, were identified as DNA binders, while a smaller group of five was classified as non-DNA binders (Massari & Murre, 2000) (Table 1). The DNA-binding proteins are further classified into E-box binders (including G-box binder) and non-E-box binders. Residues conservation suggested that 144 PcbHLHs are likely to bind E-boxs, with 109 of these identified as G-box binders, contingent on the presence of His/Lys-5, Glu-9, and Arg-13 in the basic region.
| G-box binding | DNA-binding | Non-G-box binding | DNA-binding | Non-E-box binding | DNA-binding | Non-DNA binding | DNA-binding |
|---|---|---|---|---|---|---|---|
| PcbHLH42 | HERR | PcbHLH159 | NERR | PcbHLH126 | QARR | PcbHLH154 | QPTD |
| PcbHLH155 | HERR | PcbHLH160 | NERR | PcbHLH9 | QARR | PcbHLH114 | QSSD |
| PcbHLH25 | HERR | PcbHLH65 | TERR | PcbHLH91 | QARR | PcbHLH43 | QPTD |
| PcbHLH18 | HERR | PcbHLH68 | NERR | PcbHLH69 | QARR | PcbHLH139 | QSND |
| PcbHLH127 | HERR | PcbHLH34 | QERR | PcbHLH22 | QARR | PcbHLH107 | SSKR |
| PcbHLH54 | HERR | PcbHLH163 | NERR | PcbHLH152 | QARR | ||
| PcbHLH129 | HERR | PcbHLH124 | RERR | PcbHLH157 | QARR | ||
| PcbHLH79 | HERR | PcbHLH94 | NERR | PcbHLH49 | QARR | ||
| PcbHLH121 | HERR | PcbHLH149 | RERR | PcbHLH40 | QARR | ||
| PcbHLH46 | HERR | PcbHLH84 | NERR | PcbHLH53 | QARR | ||
| PcbHLH133 | HERR | PcbHLH20 | NERR | PcbHLH24 | QARR | ||
| PcbHLH56 | HERR | PcbHLH66 | NERR | PcbHLH128 | QARR | ||
| PcbHLH31 | HERR | PcbHLH67 | NEKR | PcbHLH7 | QARR | ||
| PcbHLH101 | HERR | PcbHLH161 | NERR | PcbHLH14 | QARR | ||
| PcbHLH138 | HERR | PcbHLH135 | RERR | PcbHLH77 | KPRS | ||
| PcbHLH148 | HERR | PcbHLH64 | RERR | PcbHLH108 | PFRK | ||
| PcbHLH99 | HERR | PcbHLH145 | RERR | PcbHLH12 | SFRK | ||
| PcbHLH81 | HERR | PcbHLH143 | NERR | PcbHLH110 | QARR | ||
| PcbHLH71 | HERR | PcbHLH165 | RERR | PcbHLH23 | RKRA | ||
| PcbHLH57 | HERR | PcbHLH118 | NERR | PcbHLH36 | PFRK | ||
| PcbHLH17 | HERR | PcbHLH170 | TERR | PcbHLH104 | PFRK | ||
| PcbHLH102 | HERR | PcbHLH38 | RERR | ||||
| PcbHLH78 | HERR | PcbHLH89 | RERR | ||||
| PcbHLH125 | HERR | PcbHLH105 | RERR | ||||
| PcbHLH61 | HERR | PcbHLH120 | RERR | ||||
| PcbHLH166 | HERR | PcbHLH2 | RERR | ||||
| PcbHLH62 | HERR | PcbHLH87 | TERR | ||||
| PcbHLH111 | HERR | PcbHLH116 | RERR | ||||
| PcbHLH26 | HERR | PcbHLH88 | SERR | ||||
| PcbHLH151 | HERR | PcbHLH168 | RERR | ||||
| PcbHLH130 | HERR | PcbHLH169 | RERR | ||||
| PcbHLH10 | HERR | PcbHLH167 | RERR | ||||
| PcbHLH60 | HERR | PcbHLH162 | RERR | ||||
| PcbHLH136 | HERR | PcbHLH164 | RERR | ||||
| PcbHLH144 | HERR | PcbHLH141 | RERR | ||||
| PcbHLH33 | HERR | ||||||
| PcbHLH3 | HERR | ||||||
| PcbHLH75 | HERR | ||||||
| PcbHLH6 | HERR | ||||||
| PcbHLH27 | HERR | ||||||
| PcbHLH4 | HEQR | ||||||
| PcbHLH35 | HERR | ||||||
| PcbHLH83 | KERR | ||||||
| PcbHLH131 | HERR | ||||||
| PcbHLH73 | HERR | ||||||
| PcbHLH70 | HERR | ||||||
| PcbHLH153 | HERR | ||||||
| PcbHLH74 | HERR | ||||||
| PcbHLH11 | HERR | ||||||
| PcbHLH122 | HERR | ||||||
| PcbHLH8 | HERR | ||||||
| PcbHLH90 | HERR | ||||||
| PcbHLH19 | HERR | ||||||
| PcbHLH158 | HERR | ||||||
| PcbHLH106 | HERR | ||||||
| PcbHLH39 | HERR | ||||||
| PcbHLH47 | HERR | ||||||
| PcbHLH51 | HERR | ||||||
| PcbHLH13 | KERR | ||||||
| PcbHLH97 | KERR | ||||||
| PcbHLH59 | HERR | ||||||
| PcbHLH72 | HERR | ||||||
| PcbHLH15 | HERR | ||||||
| PcbHLH5 | HERR | ||||||
| PcbHLH109 | KERR | ||||||
| PcbHLH147 | HERR | ||||||
| PcbHLH50 | HERR | ||||||
| PcbHLH52 | KERR | ||||||
| PcbHLH28 | HERR | ||||||
| PcbHLH96 | HERR | ||||||
| PcbHLH103 | KERR | ||||||
| PcbHLH37 | KERR | ||||||
| PcbHLH86 | HERR | ||||||
| PcbHLH95 | HERR | ||||||
| PcbHLH93 | KERR | ||||||
| PcbHLH140 | HERR | ||||||
| PcbHLH132 | HERR | ||||||
| PcbHLH85 | HERR | ||||||
| PcbHLH45 | KERR | ||||||
| PcbHLH119 | HERR | ||||||
| PcbHLH58 | HERR | ||||||
| PcbHLH113 | HERR | ||||||
| PcbHLH16 | HERR | ||||||
| PcbHLH41 | HERR | ||||||
| PcbHLH115 | HERR | ||||||
| PcbHLH117 | HERR | ||||||
| PcbHLH123 | HERR | ||||||
| PcbHLH156 | HERR | ||||||
| PcbHLH48 | HERR | ||||||
| PcbHLH1 | HERR | ||||||
| PcbHLH92 | HERR | ||||||
| PcbHLH63 | HERR | ||||||
| PcbHLH150 | HERR | ||||||
| PcbHLH30 | HERR | ||||||
| PcbHLH21 | HERR | ||||||
| PcbHLH55 | HERR | ||||||
| PcbHLH142 | HERR | ||||||
| PcbHLH134 | HERR | ||||||
| PcbHLH29 | HERR | ||||||
| PcbHLH80 | HERR | ||||||
| PcbHLH44 | HERR | ||||||
| PcbHLH100 | HERR | ||||||
| PcbHLH76 | HERR | ||||||
| PcbHLH112 | HERR | ||||||
| PcbHLH137 | HERR | ||||||
| PcbHLH82 | HERR | ||||||
| PcbHLH98 | HERR | ||||||
| PcbHLH32 | KEKR | ||||||
| PcbHLH146 | KEKR |
Phylogenetic analysis of PcbHLH genes
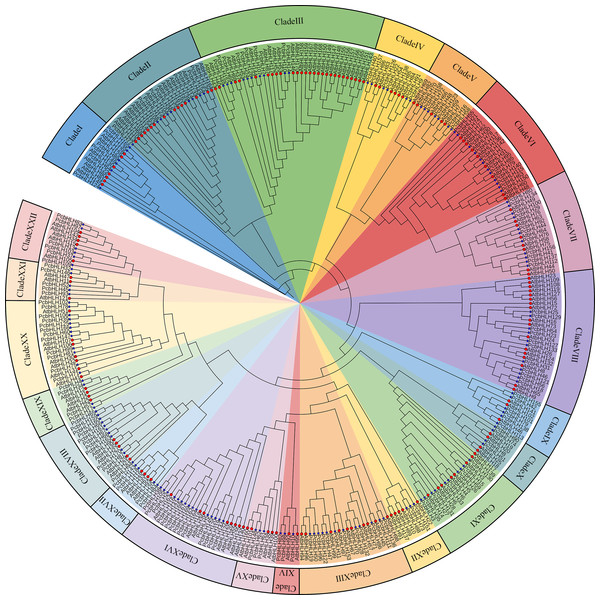
To elucidate the evolutionary relationships between PcbHLHs and the AtbHLHs, a maximum-likelihood phylogenetic tree was constructed. The analysis of clade support values and tree topology led to the identification of 22 subfamilies. As shown in Fig. 3, each subfamily includes representatives from both A. thaliana and P. canescens, indicating a high degree of conservation in the bHLH domains through their evolution. The largest subfamily, Clade III, contains 37 members, whereas the smallest, Clade XIV, consists of only five members.
Figure 3: Phylogenetic tree constructed from the Maximum Likelihood method using the bHLH conserved domains in P. canescens and A. thaliana.
The bHLH proteins were grouped into 22 distinct clades, which are indicated by colored branches. Genes on branch ends from P. canescens and A. thaliana are denoted by blue colored stars and red colored circles, respectively.Analysis of gene structure and conserved motif of bHLH family
Motif analysis using MEME identified ten types of putative conserved protein motifs within the PcbHLH family, furthering our understanding of their conservation and diversity. As depicted in Fig. 4, members within the same subfamily exhibit similar motif structure, suggesting shared structural and functional characteristics. Gene structure analysis provided insights into the evolutionary relationships among PcbHLH family members. Of the 170 family members, 11 were found to lack introns and were clustered within two subfamilies. The remaining 159 members, which contained introns, showed significant structural similarities, reinforcing the notion of close evolutionary ties within their respective subfamilies.
Figure 4: Phylogenetic relationships, conserved protein motif and gene structure of PcbHLH genes.
(A) Phylogenetic tree constructed by MEGA7 with maximum likelihood classification. (B) Conserved motif of PcbHLH genes sorted based on the results of MEME analysis. (C) Exon-intron structure of PcbHLH genes. The horizontal black lines, the yellow and green boxes represent introns, exons and UTRs, respectively. The scale at the bottom represents the base length.Analysis of cis-acting elements in PcbHLH promoters
Cis-acting elements within the promoter regions are crucial for classifying subfamilies and functional characterizing members of the PcbHLH family. A 2,000 base pair (bp) upstream of the transcriptional start sites were identified as the promoter regions for this analysis. Several functionally significant cis-acting elements were identified and categorized into three groups: TC-rich repeats (cis-acting element involved in defense and stress responsiveness), LTR (cis-acting element involved in low-temperature responsiveness), MBS (MYB binding site involved in drought-inducibility) belong to stress-responsive. GCN4_motif (cis-regulatory element involved in endosperm expression), CAT-box (cis-acting regulatory element related to meristem expression), O2-site (cis-acting regulatory element involved in zein metabolism regulation) belong to plant development-related. AuxRR-core (cis-acting regulatory element involved in auxin responsiveness), P-box (gibberellin-responsive element), CGTCA/TGACG-motif (cis-acting regulatory element involved in the MeJA-responsiveness), ABRE (cis-acting element involved in the abscisic acid responsiveness), SARE (cis-acting element involved in salicylic acid responsiveness) belong to phytohormone responsive. Figure 5 illustrates that the majority of the PcbHLH members contain phytohormone-related cis-acting elements, implying their potential roles in various abiotic stress responses.
Figure 5: Analysis of cis-acting elements in PcbHLHs promoter regions.
Heatmap of number of cis-acting elements in 2 kb promoter region of each PcbHLH gene. The gradient colors in the red grid represent the number of cis-acting elements in PcbHLHs. Distribution of cis-acting elements in promoters. The different types of cis-acting elements were shown in different colors.Gene duplication and synteny analysis of PcbHLHs
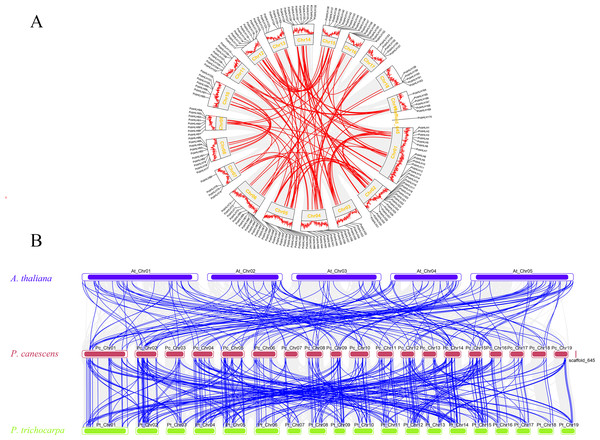
To discern the primary evolutionary forces shaping the PcbHLH gene family, gene duplication events within P. canescens were analyzed using TBtools. Figure 6A displays 92 segmental duplication gene pairs and eight tandem duplication gene pair (PcbHLH65/PcbHLH66, PcbHLH66/PcbHLH67, PcbHLH119/PcbHLH120, PcbHLH144/PcbHLH145, PcbHLH159/PcbHLH160, PcbHLH160/PcbHLH161, PcbHLH161/PcbHLH163, PcbHLH167/PcbHLH168). These findings suggest that gene duplication has been a significant mechanism in the evolution of PcbHLH genes. The Ka/Ks ratio for these duplicated genes, as detailed in Table S3, was consistently below 0.8, indicating a predominant purifying selection acting on the bHLH genes.
Figure 6: Gene duplication and synteny analysis of PcbHLH gens.
(A) Schematic representations of the chromosomal distribution and interchromosomal relationships of PcbHLHs. The gray and colourful connecting genes show all collinearity blocks and the fragment doubling event. The outermost layer of the circle represents the genes density of the chromosome. (B) The synteny analysis of bHLH genes in P. canescens with A. thaliana and P. trichocarpa. The blue, red and yellow rectangles represent the chromosomes of A. thaliana, P. canescens and P. trichocarpa, respectively. The gray lines represent synteny blocks within A. thaliana, P. canescens and P. trichocarpa, whereas the blue lines represent the collinearity of bHLH gene pairs.Comparative synteny analysis across species
To further elucidate the phylogenetic mechanisms underlying the PcbHLH family, we constructed syntenic maps comparing P. canescens with P. trichocarpa and Arabidopsis. A total of 144 orthologous bHLH gene pairs were identified between P. canescens and P. trichocarpa, and 93 orthologs between P. canescens and A. thaliana (Fig. 6B). The higher number of orthologous pairs between P. canescens and P. trichocarpa suggests a closer phylogenetic relationship compared to A. thaliana.
PcbHLH genes expression patterns in response to Cd stress
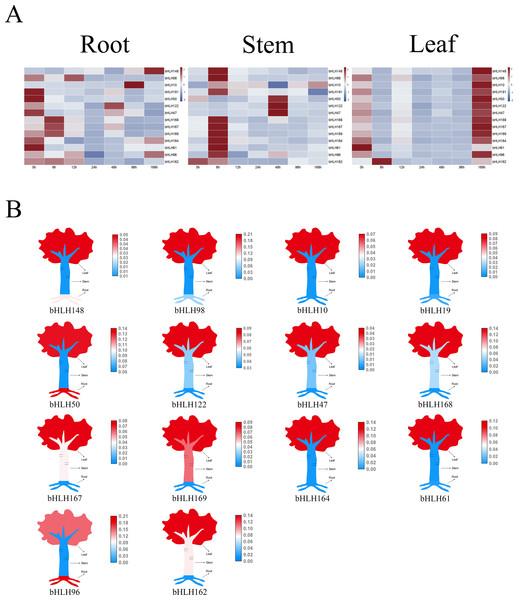
Prior research has established the role of certain clade IVc and Ib members as key regulators of Cd stress response in Arabidopsis (Hao et al., 2021). Leveraging these findings, we conducted a phylogenetic analysis of AtbHLHs and PcbHLHs to explore the functions of PcbHLH proteins. We meticulously selected 14 PcbHLH genes based on their functional homologs within the same subfamily for further analysis. Following Cd treatment, we observed transcriptional changes in these genes (Fig. 7A). In roots, genes such as PcbHLH148 and PcbHLH98 exhibited upregulated expression, while others like PcbHLH19 and PcbHLH50 showed downregulated expression. In stems, PcbHLH162 displayed decreased expression initially, with other genes upregulated up to 6 h. In leaves, all but PcbHLH61 exhibited upregulated expression, with a notable positive regulatory response observed for most PcbHLH genes, particularly in leaf tissues (Fig. 7B); PcbHLH96 was an exception, showing high expression levels in roots.
Figure 7: Relative expression analysis of the PcbHLH genes under cadmium stress conditions in different tissues.
(A) Expression profiles of PcbHLH genes in root, stem and leaf of P. canescens under Cd stress. The range of fold change in expression in the heat map is indicated by the colour bar. (B) Tissue expression patterns of PcbHLH genes.Discussion
Numerous plant species have been thoroughly investigated to characterize their bHLH families, which has highlighted the bHLH transcription factor family as one of the largest in eukaryotes (Ledent & Vervoort, 2001; Riechmann & Ratcliffe, 2000), and thus, it is of particular significance. The proliferation of plant genomic data has been instrumental, offering essential sequence resources that enable comprehensive identification of bHLH genes across plant species. This study identified and characterized 170 bHLH genes in P. canescens (Fig. 3). Surpassing the number found in A. thaliana (167) and tomato (159), but fewer than those in rice (177) and P. trichocarpa (183) (Carretero-Paulet et al., 2010). Relative to genome size, P. canescens possesses a higher ratio of PcbHLH genes compared to tomato but a lower ratio than Arabidopsis. These results indicate that the number of bHLH genes is variable across plant species and does not strictly correlate with genome size. Through phylogenetic analysis, we categorized the 170 PcbHLH genes into 22 distant subfamilies. The presence of both AtbHLHs and PcbHLHs in each subfamily suggests that the PcbHLH genes diversified prior to the evolution of two species (Atchley & Fitch, 1997). In contrast, animal genomes harbor only six subfamilies. But in plants, the bHLH gene family was divided into 24 subfamilies in tomato (Wang et al., 2015), 17 in Ginkgo biloba (Zhou et al., 2020), 20 in Camellia sinensis (Cui et al., 2018), 21 in A. thaliana (Toledo-Ortiz, Huq & Quail, 2003). This highlights a significant divergence in the classification of the bHLH gene family between plants and animals.
Analysis of the conserved protein motifs and gene structures, as illustrated in Fig. 4, reveals that subfamily members likely share a common evolutionary origin and are involved in analogous physiological processes (Ke et al., 2020). Our study identified a common conserved motif, motif 2, across 170 PcbHLH genes. This suggests that motif 2 could represent the consensus motif within the bHLH domain. Furthermore, alterations in gene structure are identified as key factors contributing to functional diversity among genes. A total of 11 PcbHLH genes lack introns, which may be indicative of their evolution within P. canescens in response to environmental selective pressures (Lin et al., 2006; Yang, Zhu & Niu, 2013). This observation suggests a potential link between intron loss and adaptive evolution in this species. These variations are driven by three principal mechanisms: exon/intron gain or loss, exonization or pseudo-exonization, and insertion or deletion. Additionally, a striking uniformity in the number and arrangement of exons/introns was observed within subfamily members. Notably, prior research has documented a range of 0 to 10 exons/introns in sesame bHLH genes (Kazemitabar, Faraji & Najafi-Zarrini, 2020). Similarly, Andrographis paniculata presents a variation of 0 to 14 exons/introns (Xu et al., 2022), and P. canescens exhibits a similar range from 0 to 14. These observatiobs are consistent with the aforementioned results.
The expansion of the PcbHLH gene family in P. canescen s is likely attributed to various gene duplication events, including tandem, fragment, whole-genome duplication, and transposition (Flagel & Wendel, 2009; Zhang, 2003). Tandem duplication typically results in two or more genes on the same chromosome, whereas segmental duplication occurs across different chromosomes (Schlueter et al., 2007). Segmental and tandem duplications are the primary drivers of the plant gene family expansion throughout evolution. P. canescens exhibited eight tandem repeats and 92 segmental duplications (Fig. 6A), indicating that these duplications have significantly contributed to the gene family’s expansion. Comparative syntenic maps between P. canescens and both P. trichocarpa and A. thaliana further elucidate the origin and evolution of the PcbHLH gene family (Fig. 6B). A total of 144 and 93 syntenic gene pairs were identified between P. canescens and P. trichocarpa and A. thaliana, respectively, suggesting a closer phylogenetic relationship with P. trichocarpa. The Ka/Ks ratio analysis (Table S3) indicates that PcbHLH gene pairs have predominantly undergone purifying selection. The basic region’s key residues are critical for distinguishing variations in the hexanucleotide core sequence at the promoters of target genes, enabling the classification of plant bHLHs into distinct DNA-binding categories. As per the criteria by Massari & Murre (2000), the PcbHLH proteins are categorized into several categories (Table 1) with G-box binding proteins constituting the majority (64%). In contrast to A. thaliana, which has 18% Non-DNA binding proteins (Toledo-Ortiz, Huq & Quail, 2003). PcbHLHs exhibit a lower percentage (3%), although the DNA-binding activity of these non-DNA binding sequences warrants further investigation.
Cadmium, a toxic heavy metal, poses a significant threat to human health and living organisms. Prolonged exposure to Cd can lead to severe health issues, such as kidney disorders, neurotoxicity, and osteoporosis (Jarup & Akesson, 2009; Satarug et al., 2010). The accumulation of heavy metals in the human body through the consumption of contaminated crops is a concerning issue (Shimbo et al., 2001; Zahir et al., 2005). Phytoremediation, an emerging technology, utilizes hyperaccumulator plants to remediate contaminated soils. While most hyperaccumulators are herbaceous with limited biomass, P. canescens, a fast-growing woody species with an extensive root system, is deemed suitable for this purpose. The role of bHLH proteins in plants’ response to heavy metal stress is well-documented. For example, the heterologous expression of the soybean GmORG3 gene, a member of the bHLH family, has been shown to enhance Cd tolerance in yeast (Xu et al., 2017). In Arabidopsis, bHLH genes such as FIT/bHLH3 8 and FIT/bHLH39 have been identified as key regulators of the Cd stress response (Wu et al., 2012; Yuan et al., 2008). Our phylogenetic analysis revealed that PcbHLH proteins from specific clades are closely related to known heavy metal stress-responsive AtbHLH proteins. We selected 14 PcbHLH genes from Clade XV (FIT/bHLH38 and FIT/bHLH39), Clade XVI, Clade XIV and Clade XIII (neighboring subfamily) for further study and used qPCR to assess their transcript levels under Cd stress. The results from the qPCR experiments revealed a diverse range of expression patterns among the PcbHLH genes (Fig. 7B), highlighting their potential roles in the regulatory mechanisms underpinning cadmium stress responses in P. canescens. Some PcbHLH genes exhibited increased expression levels under cadmium stress, suggesting their possible involvement in the direct response to heavy metal toxicity or in the activation of detoxification processes. Conversely, the downregulation of other PcbHLH genes may indicate their participation in maintaining cellular homeostasis or in the adaptation mechanisms that allow the plant to tolerate or accumulate cadmium. The variation in the expression profiles of the PcbHLH genes across different tissues and time points further underscores the complexity of the plant’s response to cadmium. This temporal and spatial regulation of gene expression could be a strategic adaptation by the plant to optimize its resource allocation and stress response. The qPCR data, therefore, not only contribute to our understanding of the molecular basis of heavy metal stress in plants but also pave the way for future research aimed at identifying key regulators and potential targets for the development of plants with improved phytoremediation capabilities.
To examine the interact between transcription factors and cis-acting elements in regulating the expression of downstream genes, we analyzed the cis-elements from the transcriptional start site extending 2,000 bps upstream. The analysis of cis-acting element indicated that PcbHLH family genes participate in numerous physiological processes, including plant growth and development, hormone responses, and stress responses. Furthermore, a multitude of stress and hormone response-related elements are prevalent in the promoter regions of PcbHLH gene (Fig. 5), underscoring the crucial role of PcbHLHs in the mediation of plant response to abiotic stress (Yamaguchi-Shinozaki & Shinozaki, 2005). Prior research has established the involvement of these elements in modulating plant responses to various abiotic stressors, including drought, salinity, and temperature fluctuations (Saidi & Hajibarat, 2019). For instance, both ABA-dependent and ABA-independent regulatory mechanisms contribute to stress-responsive gene expression (Shinozaki, Yamaguchi-Shinozaki & Seki, 2003; Thomashow, 1999; Zhu, 2002). The cis-acting element analysis of the PcbHLH promoters reveal a wide range of physiological processes and stress responses, highlighting the pivotal role of PcbHLHs in mediating abiotic stress in plants.
The bHLH family, one of the largest transcription factor families in plants, remains poorly understood, with many members’ functions yet to be explored. The comprehensive analysis of PcbHLH genes presented herein aims to enhance our fundamental knowledge and provide a theoretical foundation for developing new germplasms resistant to Cd pollution. The ultimate goal is to cultivate P. canescens materials with high Cd concentration and tolerance, offering a potential solution for soil remediation.