Molecular signature of the ontogenic development of the prawn Macrobrachium tenellum
- Published
- Accepted
- Received
- Academic Editor
- María Ángeles Esteban
- Subject Areas
- Aquaculture, Fisheries and Fish Science, Developmental Biology, Molecular Biology, Freshwater Biology
- Keywords
- Ontogeny, Prawn, Proteomic, Macrobrachium, Molecular pathway
- Copyright
- © 2023 Mateos Guerrero et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Molecular signature of the ontogenic development of the prawn Macrobrachium tenellum. PeerJ 11:e16344 https://doi.org/10.7717/peerj.16344
Abstract
The prawn Macrobrachium tenellum shows aquaculture potential due to its well-defined reproductive cycle linked to female nutritional requirements. Significant changes occur in egg composition during the 16 to 17-day embryo development. Understanding the ontogenic proteins is crucial for developmental insights and controlled reproduction. We employed free-label quantitative proteomics to analyze egg peptides at the initial and final stages of wild females. Using the emPAI protocol and Proteome Discoverer 2.0, we identified 89 differentially expressed proteins in M. tenellum eggs. Of these, 27 were exclusive to early-stage development and three to late-stage. Abundant proteins included Vitellogenin, glyceraldehyde-3-phosphate dehydrogenase, histone 4, beta-actin, and hemocyanin. Gene Ontology analysis revealed 518 terms across molecular functions, biological processes, and cellular components using the GoRetriever tool of AgBase and the CateGOrizer tool of the Animal Genome Research Program. Carbohydrate metabolism was significant in early-stage development, with glyceraldehyde-3-phosphate dehydrogenase being the second most abundant protein. Proteins involved in ATP synthesis and cytoplasmic proteins associated with catalytic and binding activities related to primary metabolism were also detected. Our study elucidates the role of Vitellogenin in lipid transport activity and its potential involvement in the juvenile hormone feedback pathway. This pathway includes farnesoic acid O-methyltransferase and juvenile hormone epoxide oxidase, regulating protein biosynthesis, molt cycles (including chitinase activity), and potentially influencing controlled reproduction. Our proteomic analysis provides insights into the molecular mechanisms driving Ontogenic development in Macrobrachium tenellum, with implications for controlled reproduction strategies and advancements in aquaculture practices.
Introduction
Macrobrachium is one of the most diverse, abundant, and widely distributed genres of Decapoda (Hendrickx, 1995; Jayachandran, 2001; New, 2005; Espinosa-Chaurand et al., 2011; Briones-Fourzán & Hendrickx, 2022). The river prawns Macrobrachium tenellum and M. americanum are common freshwater prawns from the Americas’ tropical and subtropical Pacific continental side (Jayachandran, 2001; Arzola-González & Flores-Campaña, 2008; De los Santos Romero et al., 2021); they have different biological attributes and are currently threatened by human impact on their ecosystems. Both species provide different quality products for human consumption and are intensively exploited without proper management. Despite this, due to their adaptive capacity for either polyculture or co-culture with Tilapia in rustic ponds and lagoons, they have a strong potential for farming since it has fast growth in a broad range of temperature, salinity, altitude, density, and biochemical parameters (Vega-Villasante et al., 2011; García-Guerrero et al., 2013).
As in Penaeidae, M. tenellum larval development is closely related to the nutritional requirements of females during maturation (Racotta, Palacios & Ibarra, 2003; García-Ulloa, Rodríguez & Ogura, 2004); four phases of courtship, mating, and impregnation have been reported, after mating, females lay the eggs under the abdomen, attached to the pleopods by secretions produced by the cementing glands. M. tenellum eggs are elliptical with an average diameter of 0.55 mm; embryo development lasts 16 to 17 days, during which eggs exhibit color changes (García-Ulloa, Rodríguez & Ogura, 2004).
As a palaemonid, the principal energetic metabolites in eggs are carbohydrates and lipids (García-Guerrero, 2009; Guerrero & Sandoval, 2012), while proteins are mainly structural components (Holland, 1978; García-Guerrero, Racotta & Villarreal, 2003; García-Guerrero, Villarreal & Racotta, 2003). Some reports demonstrated that protein and lipid content in yolk varies according to the maturation stage of the eggs (García-Guerrero, 2009). In Macrobrachium japonicus eggs, protein content increases during early embryonic development, apparently as a source for tissue differentiation for early structures. In contrast, lipid content decreases since it is used as an energy source. In late embryonic development, protein decreases after being used as an energy source (after all structures are formed), while lipid turns into anatomical structures (Zhao, Zhao & Zeng, 2007). This very same pattern was observed in Macrobrachium idella idella, where total amino acids in eggs increase during the I–IV developmental stages, particularly in five essential amino acids (glutamine, asparagine, glycine, cysteine, and alanine) (Varadharajan, 2012). In M. occidentale, embryonic development correlates with proteins on day three, simultaneously with gastrulation (Bliss, 1982).
Proteins seem to play a central role in in the ontogeny of Macrobrachium. Therefore, information about its proteome would give a better understanding of the development process for possible controlled reproduction. Quantitative Proteomics identifies and quantifies proteins from complex samples using liquid chromatography, resulting in large datasets related to the proteome. Free Label Quantitative Proteomics is an automated, label-free methodology to identify and quantify changes in the proteome of complex mixtures using liquid chromatography-mass spectrometry (Wiener et al., 2004; Zhu, Smith & Huang, 2010); its applications on aquatic organisms are limited, despite its relatively low-cost and comprehensive output.
In this study, we used Free Label Quantitative Proteomics to identify and quantify the proteins related to metabolism in two different stages of egg development and to provide the necessary information for a better understanding of the successful transition from yolk-dependent nutrition (lecithotrophy) to life as a free-feeding organism.
Materials & methods
Animals
Adult females of M. tenellum were purchased from the local market in Santa Maria Colotepec, Oaxaca, Mexico. Organisms were transported in darkness and with oxygen supplementation into aquaculture facilities. Organisms were kept in 1,200 L tanks (T = 25–28 °C, 12:12 h of light/darkness, constant aeration, in a closed carbon-UV filtered water system) until use. Prawns were fed a commercial diet twice daily (08:00 and 18:00 h) at 5% of body weight daily. Ovigerous females were moved to unique aquariums individually and continuously monitored from day 1 to 16; egg mass was extensively washed using distilled water and carefully removed in a biosafety cabinet; afterward, eggs were classified and measured according to sub-stages of stage III of reproduction by using an optical microscope with an eyepiece graticule (40×). Only eggs from stages A (n = 8) and E (n = 11) were used.
Egg lysate and protein quantification
On the obtention of protein lysate from eggs, we used RIPA buffer (Sigma Chemicals, St. Louis, MO, USA) and mechanical mortar grinding at −20 °C. Frozen eggs in 1× filtered Phosphate-Buffered Saline (PBS: NaCl 137 mM/L, KH2PO4 1.8 mM/L, KCl 2.7 mM/L, Na2HPO4 10 mM/L, pH 7.4) placed in a sterile mortar previously cooled to −20 °C, 300 μl of protease inhibitor RIPA (−4 °C) were added, ground to a homogeneous suspension of lysate and later centrifuged at 12,000g (10 min, 0 °C); the supernatant transferred to a sterile microtube, washed twice using sterile PBS by centrifugation (12,000g, 10 min, 0 °C) and frozen at −80 °C until use.
Soluble protein from egg lysates was measured using a Pierce BCA protein microassay kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer, using Bovine Seric Albumin as standard; absorbance was measured at 570 nm in an ELISA reader (Thermo Scientific, Waltham, MA, USA). Values were calculated using a cubic equation of the BSA standard curve.
Label-free based quantitative proteomic analysis
To verify the integrity of the sample, the protein from the eggs lysate (50 µg) was separated electrophoretically using a 12.5% polyacrylamide gel in the presence of 0.1% sodium dodecyl sulfate (SDS-PAGE), with the Laemmli (1970) buffer system. Gels were stained using Coomassie brilliant blue G-250 (Sigma, Burlington, MA, USA).
Lysate samples (A and E, early and late stage of development, respectively) were lyophilized and shipped to Creative Proteomics (Shirley, NY, EUA), where samples were digested by trypsin and quantified using a nanoLC-MS/M.S. platform, as follows:
Chemicals and instrumentation
DL-dithiothreitol (DTT), iodoacetamide (IAA), formic acid (F.A.), acetonitrile (ACN), and methanol were purchased from Sigma (St. Louis, MO, USA); trypsin from bovine pancreas from Promega (Madison, WI, USA). Ultrapure water was obtained from a Millipore purification system (Billerica, MA, USA). The essays were done in a Dionex Ultimate 3000 Nano LC system coupled with a Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) with an ESI nano-spray source.
Sample preparation
We transferred 5 mg of protein dissolved in 200 μl of 50 mM ammonium bicarbonate into Microcon devices YM-10 (Millipore) and centrifuged at 12,000g at 4 °C for 10 min, subsequently added 200 μl of ammonium bicarbonate (50 mM), followed by centrifugation and repeated once. Afterward, it was reduced with ten mM DTT at 56 °C for one h and alkylated with 20 mM IAA at room temperature in the dark for 1 h; the device was centrifuged at 12,000g at 4 °C for 10 min and washed once with 50 mM ammonium bicarbonate. Then we added 100 μl of 50 mM ammonium bicarbonate and free trypsin into the protein solution at a ratio of 1:50 and incubated at 37 °C overnight. Finally, the device was centrifuged at 12,000g at 4 °C for 10 min. Next, 100 μl of 50 mM ammonium bicarbonate was added, centrifuged, and repeated once. Extracted peptides were lyophilized. All peptides were resuspended in 2–20 μl of 0.1% formic acid before LC-MS/MS analysis.
Nanoflow UPLC
Liquid chromatography was carried out on an easy-nLC1000 (Thermo Fisher Scientific, Waltham, MA, USA) using a Nanocolumn (100 μm × 10 cm in-house made column packed with a reversed-phase ReproSil-Pur C18-AQ resin, 3 μm, 120 Å, Dr. Maisch GmbH, Germany); loaded sample volume was 5 μl; mobile phase was A: 0.1% formic acid in water; B: 0.1% formic acid in acetonitrile. The total flow rate was adjusted to 600 nL/min. L.C. linear gradient ranged from 6% to 9% B for 8 min, from 9% to 14% B for 16 min, from 14% to 30% B for 36 min, from 30% to 40% B for 15 min, and from 40% to 95% B for 3 min, eluting with 95% B for 7 min.
Q exactive mass spectrometry
For mass spectrometry, we used a spray voltage of 2.2 kV; the capillary temperature was 270 °C; M.S. parameters were resolution: 60,000 at 400 m/z, precursor m/z range: 300.0–1,650.0; MS/MS parameters were product ion scan range, start from m/z 100. Data-dependent MS/MS was up to the top five most intense peptide ions from the preview scan in the Orbitrap.
Data analysis
The mass spectrometry files (M.S.) were analyzed and searched against the Macrobrachium (Crustacea, Decapoda) protein databases using Proteome Discoverer 2.0 (Thermo Fisher Scientific, Waltham, MA, USA). Parameters were protein modifications: carbamidomethylation (C) (fixed), oxidation (M) (variable); enzyme specificity: trypsin; maximum missed cleavages: 2; precursor ion mass tolerance: 10 ppm; MS/MS tolerance: 0.6 Da. PSM, q-Value, and Score of Sequest HT were used to determine and choose only highly confident identified peptides for downstream protein identification analysis. We used the Exponentially Modified Protein Abundance Index (emPAI) (Ishihama et al., 2005) as a component of the abundance of proteins; we calculated the ratio of abundance (RA) compared with protein abundance in late/early stages. If RA > 1, the protein is considered relatively more abundant in the late stage, and if RA < 1, the protein is relatively more abundant in the early stage.
The Gene Ontology term (GO) and ID were obtained for each protein (separated as early and late stages of egg development) using the respective UniProt ID number on the GoRetriever tool from AgBase (McCarthy et al., 2011; Pillai et al., 2012). Afterward, GO Terms were classified (Classification: Go-Slim2) using the CateGOrizer tool of the Animal Genome Research Program (www.animalgenome.org/tools/catego) (Hu, Bao & Reecy, 2008; Na, Son & Gsponer, 2014), on cellular components, molecular function, and biological process. Finally, the REVIGO web-based tool visualized the GO categorization (http://revigo.irb.hr/) (Supek et al., 2011).
Results
Eggs in the early stage of development had 4.23 mg/mL (sd = 0.49, n = 8) protein content; late-stage egg protein content was 1.17 mg/mL (sd = 0.52, n = 11). Electrophoresis showed a broad-spectrum pattern of proteins (10–250 kDa).
Quantitative proteomic analysis
Abundance of proteins in eggs
After the search of M.S. data on Macrobrachium databases, we identified 89 different proteins in M. tenellum eggs (Table 1). The most abundant protein in eggs (emPAI > 15) was Vitellogenin (Prot. Id: Q95P34 and A0A0E3JBN6), identified from those in M. nipponense and from a translated genetic sequence from M. rosenbergii; it is a 285 kDa protein (pI 8.9), in which 173 and 157 peptides covered 49.7% and 49.0% of the reference proteins, respectively. Gliceraldehyde-3-phosphate dehydrogenase (Prot. Id: A0A088BEN6) was the 2nd most abundant protein found, with 82% homologous to those reported in M. rosenbergii; this is a 13.6 kDa protein, with a 6.9 calculated isoelectric point. Histone H4 (Prot. Id: A0A0D6DQV4) was the 3rd most abundant protein found in M. tenellum eggs, being 51.4% homologous to the 11.4 kDa translated sequence from M. rosenbergii. Beta-actin (Prot. Id: Q58ZF3) was the 4th most abundant protein found in M. tenellum eggs, and it showed 56.1% homology to the 41.84 kDa protein from M. rosenbergii. Hemocyanin (Prot. Id: F5CEX2) was the 5th most abundant protein found in M. tenellum eggs, being 34.1% homologous to the 76 kDa translated sequence from M. nipponense. On the contrary, the less scarce proteins found were Retinoid X receptor (Prot. Id: V9PNQ4), Dorsal protein (Prot. Id: A0A173G7X5), Prophenoloxidase (Prot. Id: Q58HZ8), the Very large inducible GTPase 1 (VLG1; Prot. Id: A0A173GTS1) and the Very large inducible GTPase 2 (VLG2; Prot. Id: A0A173GTR2) (Table 1).
| Accession | Description | Species | Origin | emPAI | S sequest HT | A. ratio |
|---|---|---|---|---|---|---|
| Q95P34 | Vitellogenin | M. rosenbergii | GN | 89.852 | 14,270.07 | 1.54 |
| A0A0E3JBN6 | Vitellogenin | M. nipponense | PE | 76.668 | 14,829.93 | 1.76 |
| A0A088BEN6 | Glyceraldehyde-3-phosphate dehydrogenase | M. rosenbergii | PE | 55.234 | 122.00 | 0.50 |
| A0A0D6DQV4 | Histone H4 | M. rosenbergii | GN | 45.416 | 158.31 | 0.20 |
| Q58ZF3 | Beta-actin | M. rosenbergii | PE | 29.079 | 510.07 | 0.26 |
| F5CEX2 | Hemocyanin | M. nipponense | PE | 25.367 | 1,357.99 | 0.19 |
| W6E8R7 | Slow-tonic S2 tropomyosin | M. nipponense | PE | 16.783 | 189.02 | 0.51 |
| A0A0A0PM26 | Hemocyanin | M. nipponense | PE | 16.783 | 1,068.84 | 0.30 |
| E2JE77 | Arginine kinase 1 | M. rosenbergii | GN | 15.876 | 272.61 | 0.26 |
| Q6S4R6 | Heat shock protein 70 | M. rosenbergii | PE | 15.037 | 416.18 | 0.46 |
| D3XNR9 | Tropomyosin | M. rosenbergii | PE | 12.895 | 207.14 | 0.10 |
| A0A0A6Z676 | Actin (Fragment) | M. rosenbergii | PE | 12.335 | 248.17 | 0.00 |
| A0A142G5I8 | Adenine nucleotide translocase | M. rosenbergii | GN | 9 | 126.05 | 1.96 |
| A0A142G5I9 | Cathepsin-D | M. rosenbergii | GN | 9 | 191.15 | 0.64 |
| K9J9V3 | DEAD(Asp-Glu-Ala-Asp) box polypeptide 39 | M. nipponense | GN | 9 | 289.82 | 0.16 |
| A0A126TKR7 | Elongation factor 1 alpha (Fragment) | M. amazonicum | PE | 9 | 86.57 | 7.80 |
| A0A0A6ZEG8 | Heat shock protein 70 | M. nipponense | PE | 7.886 | 365.20 | 0.62 |
| A0A088BES7 | Elongation factor 1-alpha (Fragment) | M. rosenbergii | PE | 7.577 | 198.97 | 0.41 |
| A0A168SIW1 | Nucleoside diphosphate kinase | M. nipponense | PE | 7.111 | 168.15 | 0.33 |
| M4IQR3 | Hemocyanin subunit 1 | M. nipponense | PE | 6.263 | 771.93 | 0.21 |
| A0A0B4L994 | Peroxiredoxin | M. nipponense | PE | 6.197 | 105.20 | 0.32 |
| W6FEW7 | Acyl-CoA-binding protein | M. nipponense | GN | 6.197 | 40.29 | 0.41 |
| A0A172Q3Z0 | GTP-binding nuclear protein | M. rosenbergii | GN | 5.579 | 68.14 | 0.12 |
| B8ZXC6 | Histone 3 (Fragment) | M. aff. pilimanus | GN | 5.31 | 44.17 | 0.24 |
| R4JXM8 | Enolase (Fragment) | M. nipponense | PE | 5.31 | 49.54 | 0.16 |
| G0Z048 | Cytosolic manganese superoxide dismutase | M. nipponense | PE | 5.105 | 94.30 | 0.49 |
| D2KK94 | Glutamate dehydrogenase (Fragment) | M. rosenbergii | GN | 3.642 | 30.89 | 3.01 |
| Q58HZ6 | Actin (Fragment) | M. rosenbergii | PE | 3.642 | 19.95 | 0.00 |
| G8H336 | Ribosomal protein L10 (Fragment) | M. acanthurus | PE | 3.642 | 8.93 | 0.00 |
| A0A140AZ38 | Pyruvate kinase | M. nipponense | PE | 3.549 | 163.62 | 0.53 |
| A0A0B4UBV8 | Rab1A | M. rosenbergii | PE | 2.875 | 61.12 | 0.34 |
| A0A0F7RQ78 | Heat shock protein 90 | M. rosenbergii | GN | 2.728 | 78.82 | 0.31 |
| B5A8X6 | Putative uncharacterized protein | M. rosenbergii | PE | 2.162 | 13.46 | 1.41 |
| B8LG57 | Male reproductive-related LIM protein | M. rosenbergii | PE | 2.162 | 6.80 | 0.00 |
| J9PNK3 | Chaperonin | M. rosenbergii | PE | 2.065 | 81.21 | 0.11 |
| E5DHS2 | Heat shock protein 90 | M. nipponense | PE | 1.907 | 136.98 | 0.70 |
| H6UXP2 | Farnesoic acid O-methyltransferase | M. nipponense | PE | 1.848 | 13.43 | 0.00 |
| C0M152 | Cytochrome c oxidase subunit 2 | M. lanchesteri | GN | 1.783 | 11.07 | 0.26 |
| A0A0B4UBB1 | Rab11A | M. rosenbergii | PE | 1.683 | 26.35 | 0.46 |
| I1SSS7 | Cathepsin L | M. nipponense | PE | 1.637 | 92.35 | 0.12 |
| I7BBT8 | Cytochrome c oxidase subunit 3 (Fragment) | M. rosenbergii | PE | 1.512 | 8.58 | 0.00 |
| A0A0A7AD33 | HSP60 | M. nipponense | PE | 1.477 | 58.15 | 0.00 |
| A0A0B4UBA7 | Rab5C | M. rosenbergii | PE | 1.424 | 41.44 | 0.31 |
| H9BC94 | Fatty acid binding protein 10 | M. nipponense | PE | 1.371 | 21.24 | 0.14 |
| A0A0D6DQG1 | Histone H2A | M. rosenbergii | GN | 1.371 | 26.86 | 0.79 |
| A0A0M5DID3 | Histone 1 | M. rosenbergii | GN | 1.154 | 11.76 | 0.08 |
| A0A0U2DW78 | ATP synthase subunit a | M. bullatum | GN | 1.154 | 2.03 | 0.00 |
| G9BRM2 | Apoptosis inhibitor | M. rosenbergii | PE | 1.054 | 31.84 | 0.49 |
| M9UTQ1 | Glutathione S-transferases | M. nipponense | PE | 1.054 | 38.00 | 0.29 |
| Q6J1F2 | Peroxinectin (Fragment) | M. rosenbergii | PE | 0.931 | 3.40 | 0.00 |
| I0B8W2 | Alpha2-macroglobulin | M. nipponense | GN | 0.905 | 16.63 | 0.00 |
| A0A0B4UCH1 | Rab7A | M. rosenbergii | PE | 0.896 | 14.41 | 0.45 |
| Q3Y596 | Superoxide dismutase | M. rosenbergii | PE | 0.874 | 12.12 | 0.17 |
| I1SSS8 | Cathepsin B | M. nipponense | PE | 0.778 | 31.42 | 0.25 |
| A7UL77 | Superoxide dismutase [Cu-Zn] | M. rosenbergii | PE | 0.778 | 2.62 | 0.00 |
| A4Z4V4 | Crustacyanin-like lipocalin | M. rosenbergii | PE | 0.668 | 15.78 | 0.23 |
| A0A168SIY7 | Ribosomal protein L24 | M. nipponense | PE | 0.668 | 6.33 | 0.00 |
| I6LTQ0 | Mitochondrial prohibitin | M. rosenbergii | PE | 0.624 | 18.39 | 0.11 |
| A0A193CGZ5 | ATP-dependent 6-phosphofructokinase | M. nipponense | PE | 0.613 | 29.78 | 0.33 |
| I1VWN9 | Ribosomal protein L26 | M. rosenbergii | GN | 0.585 | 7.36 | 0.50 |
| G4XX74 | PL10-like protein | M. nipponense | PE | 0.48 | 37.51 | 0.11 |
| A0A0B4UC48 | Rab6A | M. rosenbergii | PE | 0.468 | 20.30 | 0.35 |
| H8XYP6 | Catalase | M. rosenbergii | PE | 0.45 | 10.89 | 26.36 |
| A0A140AZ39 | Pyruvate carboxylase subunit B | M. nipponense | PE | 0.448 | 28.82 | 0.00 |
| A0T1M1 | Alpha-2-macroglobulin | M. rosenbergii | PE | 0.434 | 28.23 | 0.22 |
| M9MSS8 | Mago nashi 1 | M. nipponense | PE | 0.425 | 9.44 | 0.65 |
| A0A0U1W4T3 | Putative clotting protein | M. rosenbergii | PE | 0.425 | 63.33 | 0.74 |
| K9JA10 | DEAD(Asp-Glu-Ala-Asp) box polypeptide 5 | M. nipponense | GN | 0.417 | 13.84 | 0.13 |
| C7EYW3 | Vacuolar H+-ATPase (Fragment) | M. amazonicum | PE | 0.389 | 1.73 | 0.38 |
| A0A140AZ37 | Malate dehydrogenase | M. nipponense | PE | 0.389 | 16.59 | 0.37 |
| A0A0U2DYD0 | Cytochrome b | M. bullatum | GN | 0.389 | 2.18 | 0.00 |
| A0A0B4UC52 | Rab14 | M. rosenbergii | PE | 0.359 | 24.13 | 0.00 |
| A0A0B4UCH7 | Rab18 | M. rosenbergii | PE | 0.359 | 1.74 | 0.00 |
| P00031 | Cytochrome c | M. malcolmsonii | PE | 0.334 | 4.25 | 0.00 |
| M9MSS7 | RNA-binding protein 8A | M. nipponense | PE | 0.334 | 2.35 | 0.00 |
| G0YP40 | M. nipponense hyperglycemic hormone protein | M. nipponense | GN | 0.334 | 0.00 | * |
| D4N5G6 | SUMO-conjugating enzyme | M. nipponense | GN | 0.259 | 6.34 | 0.32 |
| B9W2R6 | Glutathione peroxidase | M. rosenbergii | PE | 0.212 | 6.09 | * |
| A0A140AZ43 | Glycogen synthase kinase 3 beta | M. nipponense | PE | 0.202 | 5.13 | 0.00 |
| W0LYS5 | Calcium-calmodulin-dependent protein kinase I | M. nipponense | GN | 0.145 | 2.35 | 0.00 |
| W8PC51 | Chitinase 4 | M. nipponense | PE | 0.129 | 0..00 | * |
| W8PFL4 | Chitinase 3C | M. nipponense | PE | 0.122 | 0.00 | 0.00 |
| I3UJK1 | Cyclin-dependent kinases 2 | M. rosenbergii | GN | 0.116 | 3.80 | 0.00 |
| A0A0H3WF09 | Juvenile hormone epoxide hydrolase | M. rosenbergii | GN | 0.110 | 1.68 | 0.00 |
| V9PNQ4 | Retinoid X receptor | M. nipponense | GN | 0.105 | 1.73 | 0.00 |
| A0A173G7X5 | Dorsal | M. rosenbergii | PE | 0.075 | 2.56 | 0.00 |
| Q58HZ8 | Prophenoloxidase (Fragment) | M. rosenbergii | PE | 0.053 | 2.69 | 0.00 |
| A0A173GTS1 | VLIG2 | M. rosenbergii | PE | 0.020 | 1.71 | 0.00 |
| A0A173GTR2 | VLIG1 | M. rosenbergii | PE | 0.019 | 7.05 | 0.24 |
Note:
Proteins are listed by the abundance of proteins using the emPAI protocol. Accession numbers are in UniProt format. Repeated proteins were identified separately according to accession numbers. Species refers to the species where the protein (P.E.) or gene (G.N.) was reported. Proteins with A ratio cero are those exclusively found in the early stage, while those marked with an asterisk (*) were solely found in the late stage. We used the SEQUEST HT score to control the quality of the peptides and proteins identified. Relative Abundance Ratio E/A compares the average area of late-stage and those of early-stage.
From the 89 proteins identified, 30 proteins were found in only one stage of egg development: 27 proteins in the early stage (Table 1, proteins in blue) and 3 in the late stage (Table 1, proteins in red).
From those proteins exclusively found in the early stage, the most abundant (emPAI > 1) was actin (Prot. Id: A0A0A6Z676), which is a globular, multi-functional protein; likewise, in the late stage, only three proteins were exclusively found: glutathione peroxidase (Prot. Id. B9W2R6), hyperglycemic hormone (Prot. Id. G0YP40) and chitinase four (Prot. Id. W8PC51). Additionally, in the late stage, the proteins exclusively found were the ribosomal L-10 (Prot. Id: G8H336), male-reproductive related LIM protein (Prot. Id: B8LG57), farnesoic acid O-Methyl transferase (Prot. Id: H6UXP2), HSP-60 (Prot. Id: A0A0A7AD33), cytochrome-c oxidase (Prot. Id: I7BBT8) and ATP synthase (Prot. Id: A0A0U2DW78).
Relative abundance of proteins by the developmental stage of eggs
During the embryonic development of the organism, we found 59 proteins, classified according to their ratio as relatively more abundant in the late stage (RA > 1) and relatively more abundant in the early stage (RA < 1). Most of these proteins were abundant in the early stage, with only 10% being most abundant in the late stage. For the early stage, we identified 53 proteins; the most importantly downregulated were a putative clotting protein (Prot. Id. A0A0U1W4T3), cathepsin-D (Prot. Id. A0A142G5I9), heat shock protein-70 (Prot. Id. A0A0A6ZEG8), pyruvate kinase (Prot. Id. A0A140AZ38) and hemocyanin (Prot. Id. A0A0A0PM26). For proteins relatively more abundant in the late stage, our data suggest that catalase (Prot. Id. H8XYP6) is the most upregulated protein since its relative abundance is 26-fold greater at the late than at the early stage; Elongation factor one alpha (Prot. Id. A0A126TKR7), glutamate dehydrogenase (Prot. Id. D2KK94), DEAD box polypeptide 39 (Prot. Id. K9J9V3) and Vitellogenin (Prot. Id. A0A0E3JBN6 and Q95P34) were also found upregulated.
Gene ontology analysis of proteins
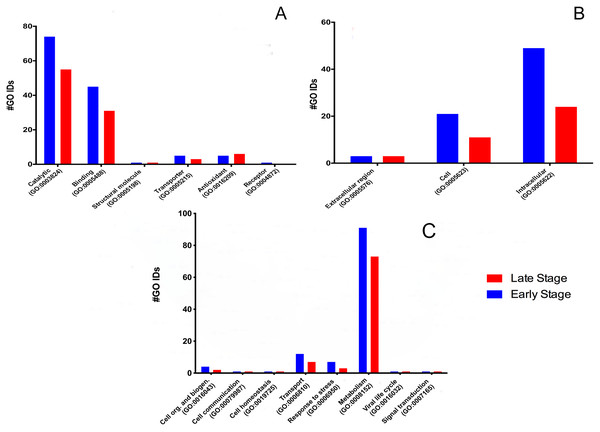
To determine the protein’s biological meaning, we identified 518 GO terms from 84 proteins (five proteins have no GO term associated); 73 GO terms were classified into three main categories: 135 in six main molecular functions (Fig. 1A), 116 cellular components (Fig. 1B), and 120 in eight main biological processes (Fig. 1C).
Figure 1: Molecular GO Slim2 analysis.
It is presented as the number of Gene Ontology IDs identified on the significant categories molecular function (A), cellular component (B), and biological process (C) for proteins from M. tenellum eggs. Analysis was performed using the Slim2 classification system. All proteins identified in the late and early stages are red and blue, respectively.Molecular function
We found six molecular functions at each stage: catalytic (GO:0003824), binding (GO:0005488), structural molecule activity (GO:0005198), transporter (GO:0005215), antioxidant (GO:0016209) and receptor (GO:0004872) (Fig. 1A).
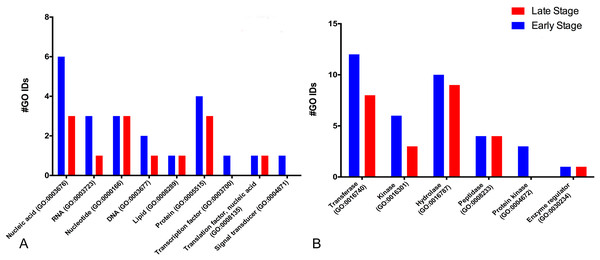
For the molecular function, many GO terms were found for the catalytic and binding activity in both stages. Still, these functions were more abundant in the early than the late stage. Binding activity was 1.45-fold more active earlier than in the late stage. Binding to nucleic acids, RNA, and DNA (GO:0003676, GO:0003723, GO:0003677) and proteins (GO:0005515) were the primary function found in the early stage; meanwhile, nucleic acid, nucleotide, and protein binding showed the same activity in the late stage. Transcription factor (GO:0003700) and signal transducer (GO:0004871) activities were only present in the early stage. (Fig. 2A). GO terms classified as catalytic were 1.34-fold more abundant in the early stage than in the late stage (74/55). We found more transferases (12/8) and kinases (6/3) during egg development (Fig. 2B).
Figure 2: Molecular GO terms for the most critical molecular functions: binding and catalytic activities.
Presented as the number of Gene Ontology IDs identified on the significant classes of (A) binding, (B) catalytic, for proteins from M. tenellum eggs. Analysis was done using the Slim2 classification system on February 17th, 2018. All proteins identified in the late and early stages are red and blue, respectively.For the catalytic activity-related proteins, in the early stage, transferases (GO:0016740) had the most relevant function, followed by hydrolases (GO:0016787), kinases (GO:0016301), peptidases (GO:0008233) and enzyme regulation (GO:0030234); meanwhile in the late stage, hydrolases were more active than kinases.
Cellular component
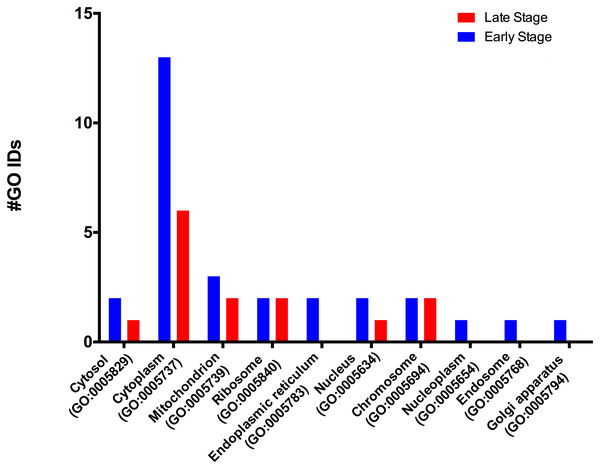
According to the GO-Slim2 analysis, the cellular components related to the cell were more abundant in the early stage (1.92-fold); meanwhile, the extracellular region (GO:0005576) remained without changes in both developmental stages of the eggs (Fig. 1B). The main components of the cell were in the cytoplasm (GO:0005737) in both stages of development. Mitochondrion (GO:0005739) had the most considerable number of G.O. terms associated, followed by cytosol (GO:0005829), ribosome (GO:0005840), nucleus (GO:0005634) and chromosome (GO:0005654); E.R. (GO:0005783), nucleoplasm (GO:0005654), endosome (GO:0005768) and Golgi (GO:0005794) were only present in the early stage (Fig. 3).
Figure 3: Molecular GO terms for the intracellular component protein classification from M. tenellum eggs.
Analysis was performed using the Slim2 classification system on February 17th, 2018. All proteins identified in the late and early stages are red and blue, respectively.Biological process
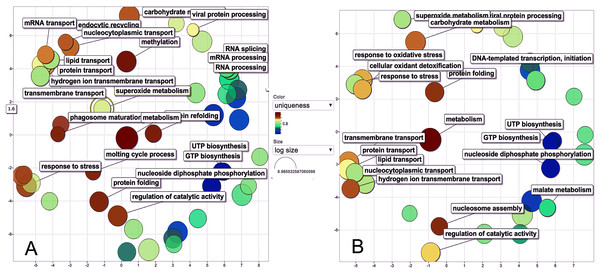
The biological process was the most complex classification in M. tenellum eggs; according to the GO-Slim2 analysis, the scatter plot of the main biological processes during ontogeny showed a significant complexity during the early stage (Fig. 4A) than during the late stage (Fig. 4B). We found eight primary generic biological processes; Metabolism (GO:0008152) was the most active on the eggs, followed by Transport (GO:0006810), Response to Stress (GO:0006950), Cell Organization and Biogenesis (GO:0016043), with the same activity were Cell Communication (GO:0007987), Cell Homeostasis (GO:0018725), Viral Cell Life (GO:0016032), and Signal Transduction (GO:0007165) (Fig. 1C).
Figure 4: Scatter plot of GO Terms of biological process for proteins from M. tenellum eggs.
(A) Early and (B) late stage of development. Analysis was performed by using the REVIGO web-based software.For the main biological process during ontogeny, we found more GO terms (1.32-fold more) in the early stage than in the late stage (Fig. 1C). Metabolism-related terms were more abundant, but the response to stress and transport was 2.33 and 1.71-fold more active, respectively, in the early than in the late stage. The metabolic processes were more active in the early than in the late stage; the most important metabolic processes in the early stage were nucleotide-related metabolism (GO:0006139), followed by biosynthesis (GO:0009058), metabolite and energy precursor and protein metabolism (GO:0006091 and GO:0019538), carbohydrate metabolism and catabolism (GO:0005975 and GO:0009056) and protein modifications and protein biosynthesis (GO:0006464 and GO:0006412); in late stages, protein metabolism was more active than precursor metabolites and energy metabolism, and protein biosynthesis was more involved than protein modifications.
Discussion
The study of the ontogeny of crustaceans has been an essential factor in the development of the aquaculture of the principal commercial species. Molecular-level studies on marine shrimps have provided necessary information about their energetic metabolism and embryonic development for breeding and reproduction. There are limited studies on the ontogeny of aquatic prawns, mainly concerning the proximate composition of metabolites. For those species, the primary metabolites during the ontogeny are carbohydrates and lipids as a source of energy and precursors (Guerrero & Sandoval, 2012). At the same time, proteins are the main structural components of embryonic development. Some reports suggest a higher protein concentration in the early development stage than in the late stage because of their use as structural materials (Bliss, 1982; Zhao, Zhao & Zeng, 2007; Guerrero & Sandoval, 2012; Varadharajan, 2012).
The prawn M. tenellum is a species with regional commercial importance. The low quantity of proteins identified compared to the amount present in electrophoretic gels may be related to the limited information contained in the Macrobrachium databases associated with only seven species (M. rosenbergii, M. nipponense, M. amazonicum, M. aff. Pilimanus, M. acanthurus, M. lanchesteri, and M. bullatum); this problem is broader, since in the only similar report -to our knowledge- in Pacifastacus leniusculus (crayfish) eggs and spermatophore, were 41 proteins identified; the most diverse being respiratory and cytoskeleton related, while no proteins were related to DNA or cell defense (Niksirat et al., 2014).
The most abundant protein found in eggs, according to our data, was Vitellogenin (Vg), the most studied and well-known protein among crustacean eggs that disappears during embryogenesis after cleavage as vitellin, which explains the lower concentration of total protein in the late-stage of development (Chang & Bradley, 1983; Byrne, Gruber & Ab, 1989; Raikhel & Dhadialla, 1992). Vg is the yolk protein precursor from eggs synthesized in the hepatopancreas and released to the hemolymph, which yields several subunits (Okuno et al., 2002; Wilder, Okumura & Tsutsui, 2010). In M. nipponense, Bai et al. (2015) reported a fluctuation of Vg expression during embryonic development, with a peak at the beginning of the larval development (Bai et al., 2015); Vg seems to be an essential protein during embryogenesis, especially during the first stages of life of the organism, providing critical metabolites and energy; any deficiency on its synthesis or accumulation could carry developmental defects. In insects, the amino acids from diet induce Vg synthesis and egg development via the juvenile hormone biosynthesis (Lu et al., 2016); in M. tenellum, we found 1.54 fold more Vg concentration during the initial stage of embryogenesis, and according to the GO Term (GO:0005319), and as reported by Wyatt & Davey (1996), it enables and participates in the lipid transport activity (Wyatt & Davey, 1996); moreover, this suggests that it could be related to juvenile hormone feedback, where farnesoic acid O-methyltransferase (Prot. Id. H6UXP2) and juvenile hormone epoxide oxidase (Prot. Id. A0A0H3WF09) catalyze the conversion of farnesoic acid to juvenile hormone III (White, 1972; Defelipe et al., 2011; Duan et al., 2014). This hormone is related to general protein synthesis and the molt cycle control (Homola & Chang, 1997); both processes are more active during the initial stages of embryonic development.
According to the relative content of protein, Vitellogenin is associated with lipid mobilization in the initial developmental stages; meanwhile, the use of carbohydrates as energy supply is more active during the late stages of embryonic development since the relative content of proteins related to glycolysis is lower (RA > 1); Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is the second more abundant protein; this is a cytosolic enzyme of eukaryotic cells, and is involved in the regulation of the energetic metabolism of the eggs; it plays a central role on glycolysis and gluconeogenesis and catalyzes the conversion of D-glyceraldehyde-3-phosphate (G3P) to 1,3 diphosphoglycerate (Harding et al., 1975; Ou et al., 2006), it has also reported having a role in cytoskeleton and apoptosis (Dugaiczyk et al., 1983; Huang et al., 1989; Berry & Boulton, 2000). From the glycolytic pathway, we also identified phosphofructokinase, which converts fructose-6-phosphate to fructose-1,6-biphosphate; enolase, which catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate (PEP); and the pyruvate kinase, which converts PEP to pyruvate. For gluconeogenesis, besides GAPH and enolase, we found pyruvate carboxylase, an enzyme needed to convert pyruvate to oxaloacetate, and malate dehydrogenase, an enzyme that catalyzes the interconversion of oxaloacetate to malate (an intermediary of the citric acid cycle); downstream the metabolic process we found proteins related to the electron transport chain and the oxidative-phosphorylation for ATP synthesis (Cytochrome B, Cytochrome C, and ATP-synthase).
H4 histone is a protein involved in eukaryotic cells’ chromatin structure and functions as a component of the nucleosome, in a central role in regulating transcription, DNA repair, DNA replication, and chromosomal stability. Is mainly recognized as a DNA binding and a hetero dimerization activity protein implicated in the biological process of DNA-template transcription, initiation, and nucleosome assembly due to its ability to undergo several post-translational modifications, is expressed in all tissues, mainly in gills, and it is upregulated after viral and bacterial injection; In addition, peptides derived from its N- and C-terminus present antibacterial activity (Chaurasia et al., 2015).
Beta-actin is a ubiquitous, conserved protein with multi-diverse functions, which participates in several processes such as cell motility, cell division, vesicle movement, apoptosis, gene expression, and cell signaling (Doherty & McMahon, 2008). Its molecular function is ATP binding. In Macrobrachium rosenbergii, it is a cytoplasmic protein distributed widely in all tissues: it is highly expressed in muscle, being upregulated during embryonic development; interestingly, beta-actin is downregulated in M. tenellum in the same period (Zhu et al., 2005).
Hemocyanin is a conserved extracellular, multi-subunit respiratory protein among arthropods and mollusks (van Holde & Miller, 1995; Adachi et al., 2005). It is a copper-containing protein that typically constitutes up to 85% of the total proteins in the hemolymph. Other functions reported for this protein are mainly in the defense mechanism as phenol oxidase, antiviral and antibacterial protein (García-Carreño, Cota & Del Toro, 2008; Lei et al., 2008; Fan et al., 2009; Zhang et al., 2009; Somboonwiwat et al., 2010). Its molecular functions related to M. nipponense are metal ion binding and oxidoreductase activity; it belongs to the hemocyanin and tyrosine kinases family and is distributed in most tissues, more expressed in hepatopancreas. After bacterial infection, hemocytes’ expression is downregulated at 3 h and upregulated after 6 h (Kong et al., 2016).
From the less abundant proteins identified, we identified two embryonic development-related proteins and two immune-related functions: the retinoid X receptor (Prot. Id: V9PNQ4), a nuclear receptor related to retinoic acid (vitamin A metabolite) and an essential protein that guides the development of the posterior side of the vertebrate embryo; and the dorsal protein (Prot. Id: A0A173G7X5), which is required for the generation of embryonic dorsoventral polarity and probably at later developmental stages for an innate immune response in Drosophila. In addition, Prophenoloxidase (Prot. Id: Q58HZ8), a crucial inactive protein of melanization in crustaceans, the very large inducible GTPase 1 (VLG1; Prot. Id: A0A173GTS1) and VLG2 (Prot. Id: A0A173GTR2) are members of a protein family related to multiple biological processes.
The study of gene ontology is crucial for an integrated vision of the identified proteins. Our data suggest that during the ontogeny of this species, proteins are more abundant and participate more actively in cellular and biological processes and molecular functions (according to the GO terms found) in the early stage, revealing a more active metabolism and decreasing at the final stage.
Our data shows that the majority and more complex relation of the GO terms identified in M. tenellum eggs were related to metabolic processes (Fig. 5); during ontogeny, the primary molecular functions are catalytic and binding proteins (Fig. 1A) associated with the intracellular space in the cytoplasm (Fig. 1B). The metabolism of the embryo was focused on nucleotides (Fig. 5), and it encompasses a complex interaction between biosynthesis and the generation of metabolites and energy amino acids; protein and carbohydrate metabolism decrease in the late stage; moreover, the generation of precursor metabolites and energy decreases their abundance, as protein metabolism becomes more critical.
Figure 5: Interactive graph of GO biological processes from M. tenellum egg proteins.
(A) early stage of development, (B) late stage of development. Analysis was performed by the use of the REVIGO web-based software.Conclusions
To our knowledge, this is the first report on quantitative proteomic profiling of ontogeny of crustacea. This study found that embryogenesis in crustaceans exhibits a typical pattern in both morphological and protein profiles. Our findings on the protein concentration during the early stage of embryonic development in Macrobrachium tenellum could suggest that females carrying early-stage development eggs may require a high protein nutrient supply to ensure proper nutrition and growth; this can be optimized by providing a suitable protein-rich diet for these females. Moreover, our findings on the proteomic analysis, about 89 proteins within M. tenellum eggs, none of which were previously reported for this species, 27 proteins found exclusively in the early stages of development, and three proteins exclusively presented in the late stages, all with diverse functions, contributes to the understanding of the intricate molecular mechanisms driving the Ontogenic development of this species. Understanding the role of these proteins can help develop more effective feeding strategies and optimize the species’ development in aquaculture settings. For example, proteins involved in metabolic processes could drive effective feeding strategies to use specific micronutrients, which could rely on a better metabolism. Our research contributes to the understanding of the ontogenic development of M. tenellum. It can be used to create a more suitable growth and development environment for crustaceans, leading to improved aquaculture practices.