In-silico, evolutionary, and functional analysis of CHUP1 and its related proteins in Bienertia sinuspersici—a comparative study across C3, C4, CAM, and SCC4 model plants
- Published
- Accepted
- Received
- Academic Editor
- Sapna Langyan
- Subject Areas
- Agricultural Science, Bioinformatics, Genomics, Plant Science, Taxonomy
- Keywords
- Bienertia sinuspersici, CHUP1 protein, CHUP1-like proteins, Single-Cell C4 plants, Phylogenetic analysis, Subcellular compartmentalization
- Copyright
- © 2023 Won et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits using, remixing, and building upon the work non-commercially, as long as it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. In-silico, evolutionary, and functional analysis of CHUP1 and its related proteins in Bienertia sinuspersici—a comparative study across C3, C4, CAM, and SCC4 model plants. PeerJ 11:e15696 https://doi.org/10.7717/peerj.15696
Abstract
Single-cell C4 (SCC4) plants with bienertioid anatomy carry out photosynthesis in a single cell. Chloroplast movement is the underlying phenomenon, where chloroplast unusual positioning 1 (CHUP1) plays a key role. This study aimed to characterize CHUP1 and CHUP1-like proteins in an SCC4 photosynthetic plant, Bienertia sinuspersici. Also, a comparative analysis of SCC4 CHUP1 was made with C3, C4, and CAM model plants including an extant basal angiosperm, Amborella. The CHUP1 gene exists as a single copy from the basal angiosperms to SCC4 plants. Our analysis identified that Chenopodium quinoa, a recently duplicated allotetraploid, has two copies of CHUP1. In addition, the numbers of CHUP1-like and its associated proteins such as CHUP1-like_a, CHUP1-like_b, HPR, TPR, and ABP varied between the species. Hidden Markov Model analysis showed that the gene size of CHUP1-like_a and CHUP1-like_b of SCC4 species, Bienertia, and Suaeda were enlarged than other plants. Also, we identified that CHUP1-like_a and CHUP1-like_b are absent in Arabidopsis and Amborella, respectively. Motif analysis identified several conserved and variable motifs based on the orders (monocot and dicot) as well as photosynthetic pathways. For instance, CAM plants such as pineapple and cactus shared certain motifs of CHUP1-like_a irrespective of their distant phylogenetic relationship. The free ratio model showed that CHUP1 maintained purifying selection, whereas CHUP1-like_a and CHUP1-like_b have adaptive functions between SCC4 plants and quinoa. Similarly, rice and maize branches displayed functional diversification on CHUP1-like_b. Relative gene expression data showed that during the subcellular compartmentalization process of Bienertia, CHUP1 and actin-binding proteins (ABP) genes showed a similar pattern of expression. Altogether, the results of this study provide insight into the evolutionary and functional details of CHUP1 and its associated proteins in the development of the SCC4 system in comparison with other C3, C4, and CAM model plants.
Introduction
Plants acquired chloroplasts from a cyanobacterial endosymbiont about 1,000 million years ago (MYA) (Jensen & Leister, 2014). Plants exploit well-developed chloroplasts for fixing atmospheric CO2 into organic compounds (Jarvis & López, 2013) using C3, C4, and crassulacean acid metabolism (CAM) photosynthetic mechanisms (Sage, 2004; Sage, Way & Kubien, 2008; Yang et al., 2015). Approximately 85% of terrestrial plants have adapted the C3 photosynthetic pathway. It was estimated that C3 plants could have originated from the Paleozoic and Mesozoic eras preceding C4 plants (Sage, Christin & Edwards, 2011). Exposure of C3 plants to adverse conditions led to higher photorespiration. To adapt to drought and high-temperature conditions, C4 pathways have evolved at least 60 times independently in approximately 19 families of angiosperms. Approximately 3% of plants have adapted the C4 photosynthesis (Sage, 2004; Sage, Christin & Edwards, 2011). It can be classified into three types based on the type of four carbon acids, such as NAD-dependent malic enzyme (NAD-ME), NADP-dependent malic enzyme (NADP-ME), and PEP carboxykinase (PEPCK) (Sage, 2004; Offermann, Okita & Edwards, 2011). Apart from the C3 and C4 pathways, 10% of plants growing in extreme conditions (deserts and epiphytes) adopted the CAM pathway, which evolved independently in 400 distinct genera in 36 families (Sage, Christin & Edwards, 2011; Ming et al., 2015; Yang et al., 2015). In this pathway, during the night, stomata remain open and fix malate via a phosphoenolpyruvate (PEP) reaction. In the daytime, the C3 pathway takes place with closed stomata (Ming et al., 2015; Yang et al., 2015). In C3 plants, photosynthesis occurs only in the mesophyll (MS) cells, whereas almost all C4 plants require Kranz anatomy, where both MS and bundle sheath (BS) cells participate in primary and secondary carbon fixation, respectively (Sage, 2004; Koteyeva et al., 2016). In CAM plants, both the C3 and C4 photosynthesis processes take place in the same MS cells (Ghannoum et al., 2013; Yang et al., 2015). Recent findings indicate that plants have developed an efficient single-cell C4 (SCC4) photosynthetic mechanism which can perform C4 photosynthesis in a single cell in the absence of C4 Kranz anatomy (Sage, 2004). This type was demonstrated in four terrestrial species within the Chenopodiaceae family. SCC4 plants employ dimorphic chloroplasts to carry out C4 photosynthesis (Mai et al., 2019). In Suaeda aralocaspica (hereafter, Suaeda), two types of chloroplasts are arranged at proximal and distal positions in elongated chlorenchyma cells. In contrast, Bienertia sinuspersici (hereafter, Bienertia, SCC4 model plant), B. kavirense, and B. cycloptera possess bienertioid anatomy, wherein two types of chloroplasts, namely the central chloroplast (CCp) and the peripheral chloroplast (PCp), are localized and compartmentalized in central and peripheral cytoplasmic regions within a single cell, respectively (Mai et al., 2019). PCp generates a C4 organic acid from atmospheric CO2, which is decarboxylated in the CCp by the Calvin-Benson-Bassham cycle (Offermann et al., 2015). Young Bienertia chlorenchyma cells have a uniform distribution of chloroplasts operating in C3 photosynthesis without partition of the cytoplasm (Koteyeva et al., 2016). The partition of two chloroplasts is observed only in mature chlorenchyma cells. As the chlorenchyma cells mature, the vacuoles fuse to create the unusually large central vacuole, which is distinct from most plant cells, and these cells operate C4 photosynthesis (Park et al., 2009). The distribution of chloroplasts is achieved by moving chloroplasts to the peripheral cytoplasmic compartments and the central cytoplasmic compartments, and yet they are connected by cytoplasmic channels, which limit the gas diffusion between the two compartments. The function of peripheral and central compartments is similar to that of MS cells and BS cells in C4-type plants with Kranz anatomy, respectively (Offermann, Okita & Edwards, 2011).
Chloroplast movement is one of the mechanisms that sessile plants have developed to receive more light in low-light conditions and to reduce photodamage in high-light conditions. Under low light, chloroplasts have a “periclinal” face position to utilize maximum light, whereas, during high light, an “anticlinal” position is taken to minimize photodamage (Wada, 2016). Chloroplast movement is carried out by many factors, such as phototropins (PHTs), Chloroplast Unusual Positioning 1(CHUP1), Plastid Movement Impaired 1 (PMI1), Kinesin-like Protein for Actin-based Chloroplast Movement (KAC), and THRUMIN 1, a glutaredoxin-like protein (Dwyer & Hangarter, 2022; Shi et al., 2022; Gao et al., 2023). Among these factors, CHUP1 is crucial for generating and maintaining chloroplast actin (cp-actin) filaments (Oikawa et al., 2003; Oikawa et al., 2008). In the Arabidopsis model plant (C3 plant), CHUP1 protein has been reported to be involved in chloroplast movement (Oikawa et al., 2003; Lehmann, Bohnsack & Schleiff, 2011). It is localized in the chloroplast outer membrane, and it triggers chloroplast movement by polymerizing the cp-actin filaments using profilactins. CHUP1 belongs to the hydroxyproline-rich (HPR) glycoprotein family and is a multi-domain protein consisting of an N-terminal hydrophobic domain (HD), a coiled-coil domain (CCD), an actin-binding domain (ABD), two leucine zipper (LZ) domains, and a proline-rich motif (PRM) (Von Braun & Schleiff, 2008). The N-terminal of the CHUP1 protein anchors to the outer envelope membranes of chloroplasts in MS cells, and the CCD of CHUP1 facilitates the anchorage of chloroplasts to the plasma membrane (Oikawa et al., 2003; Oikawa et al., 2008; Lehmann, Bohnsack & Schleiff, 2011). The Arabidopsis chup1 mutant showed defects in both the chloroplast movement and its anchorage to the plasma membrane (Von Braun & Schleiff, 2008; Kadota et al., 2009; Suetsugu et al., 2010; Ichikawa et al., 2011; Lehmann, Bohnsack & Schleiff, 2011; Lehmann et al., 2011; Manandhar-Shrestha et al., 2013; Suetsugu et al., 2015). Therefore, CHUP1 is considered a key binding factor between the chloroplast and cytoskeleton filaments. Similarly, in Zea mays (maize) and Eleusine coracana (finger millet) plants (NAD-malic enzyme-type C4 plant), it is reported that CHUP1 links the chloroplasts to actin filaments in MS cells (Kobayashi et al., 2009). In Z. mays, a dramatic increase in phosphorylation of CHUP1 during midday was demonstrated (Gao et al., 2023).
The movement of chloroplasts into the two internal cytoplasmic compartments separated into proximal-distal and central-peripheral locations in SCC4-type plants is fascinating and is a recent trending topic in chloroplast research. Though studies were carried out on the investigation of the response of chloroplasts under different light conditions, such as low light and high light, where they move to “periclinal” and “anticlinal” for maximum utilization and avoidance responses, respectively (Wada, 2016), no research was carried out on the involvement of CHUP1 in the chloroplast movement in SCC4-type plants during the development process. This work was carried out to identify the copy numbers, conserved and diverged structural patterns, and natural selection between the different plant systems such as C3, C4, CAM, and SCC4 in both monocot and dicot plants for CHUP1 and its related genes. The genome analysis mentioned that Amborella is the single living plant that could be the sister lineage to all extant flowering plants (Albert et al., 2013). Therefore, we have included Amborella as an extant lineage for both monocots and dicots. Our in-silico analysis using AtCHUP1 as a query showed that CHUP1-like and other proteins shared functional domains such as tetratricopeptide repeat (TPR) and actin-binding protein (ABP). In addition, hydroxyproline-rich glycoprotein (HRGP) was identified only in specific species. We considered that these proteins could be an interactive partner with CHUP1. Therefore, we included CHUP1-like proteins and their associated proteins in this genome-wide analysis. Also, the studies on CHUP1 genes and isoforms at the genome level are sparse. Hence, in this study, we attempt to characterize CHUP1 and its paralogs among the lineages of C3, C4, and CAM in monocot and dicot orders, with the main focus on the SCC4 model plant. The results of this study could be very useful in understanding the evolutionary and functional adaptation of CHUP1 and its homologs among plant lineages, particularly the SCC4 species Bienertia.
Materials and Methods
Plant material
Initial identification of the sample was done by Prof. Gerald Edwards(School of Biological Sciences and Center for Integrated Biotechnology, Washington State University, USA) (Akhani et al., 2005). Bienertia sinsuspersici (accession PRJNA273351) was received from one of our collaborators, Prof. Sascha Offermann (Institute for Botany, Leibniz University Hannover, Germany) as seeds. Seeds were germinated, and tissue culture was performed in vitro for propagation and maintenance at the National Institute of Agricultural Sciences, Rural Development Administration, Jeonju, Republic of Korea.
Identification of CHUP1 proteins
A well-characterized Arabidopsis CHUP1 protein (NCBI ID: NP_189197.2) (Oikawa et al., 2003; Lehmann, Bohnsack & Schleiff, 2011) was used as a query to BLAST against the Arabidopsis whole proteome (Lamesch et al., 2012). After the selection of proteins, according to a previous method (Lamesch et al., 2012), CHUP1-like and other associated proteins of Arabidopsis were taken as queries to BLAST against Amborella (Amborella trichopoda) (Albert et al., 2013), maize (Zea mays) (Schnable et al., 2009), sorghum (Sorghum bicolor) (McCormick et al., 2018), rice (Oryza sativa) (Ouyang et al., 2007), pineapple (Anana cosmosus) (Ming et al., 2015), Cactus (Cactus gigantea) (Copetti et al., 2017), quinoa (Chenopodium quinoa) (Jarvis et al., 2017), Amaranthus (Amaranthus hypochondriacis) (Sunil et al., 2014), Suaeda (Suaeda aralocaspica) (Wang et al., 2019), and Bienertia (Bienertia sinuspersici). For Bienertia, the genome project was launched in 2016, and genome assembly has since been completed with about 97.5% coverage of the anticipated genome size of 3.7 Gb (Soundararajan et al., 2019). For the identification of CHUP1 and its associated genes in Bienertia, whole genome protein sequences have been used. Recently, we published a comparative analysis of the YABBY gene in Bienertia with our existing data (Soundararajan et al., 2019). As the first step in genome sequencing, cytogenetic analysis of Bienertia has been published (Soundararajan et al., 2019; Sevilleno et al., 2020). All genome links have been given in Table S3.
Phylogenetic tree, functional domain, motif, gene structural analysis, and subcellular localization
Phylogenetic trees were constructed using MEGA 6.0 with ClustalW using the maximum likelihood method. The nearest neighbor-interchange and partial deletion (95) were chosen with a 1,000 bootstrap value (Tamura et al., 2013). CHUP1 is a multifunctional domain-containing protein. Therefore, functional domains have been mapped using different databases and tools. CCD has been identified using the COILS program (https://bio.tools/coils), with Windows 28 showing more than a 0.95 value and other settings defaulted (Lupas, Van Dyke & Stock, 1991). LZ domain position was determined with the 2ZIP server (http://2zip.molgen.mpg.de/index.html) (Lupas, Van Dyke & Stock, 1991; Bornberg-Bauer, Rivals & Vingron, 1998). The ABD position was recognized with a motif in CHUP1 and a pattern in CHUP1_like proteins. PRM is mapped based on the UniProt (https://www.uniprot.org/) curation of homologous sequences (Table S4). Residues involved in interaction with other proteins on the N-terminal of CHUP1 proteins (HD) were predicted based on linear interacting peptides (LIPs) analysis (https://mobidb.bio.unipd.it/) (Piovesan et al., 2018). For CHUP1-like homologs, N-terminal sequence homology with CHUP1 was also considered to predict HD.
Conserved protein motifs were predicted using the MEME (v. 4.9.1) online tool with options such as zero or one occurrence and 20 motifs with 6 to 200 widths (Bailey et al., 2009). Coding sequences (CDS) of Bienertia, Suaeda, and cactus were obtained by tBLASTn of proteins against their genomes (Soundararajan et al., 2019). CHUP1-like and its associated proteins were identified using BLASTp with a 10−5 e-value of at least 30% homology against their CHUP1 proteins in all genomes. Proteins with the same chromosomal positions, truncation, ambiguity, and highly diverged sequences were removed. Complete CDS and genome regions were extracted using the FGENESH+ Hidden Markov Model (HMM) profile and BEDtools, respectively. For other species, the genomic region of CHUP1 and its associated proteins were obtained from the respective databases. Exon-intron arrangements were constructed with Gene Structure Display Server 2.0 (GSDS) (Hu et al., 2015). The subcellular localization of CHUP1 and its related proteins was analyzed using the DeepLoc 2.0 Prediction of eukaryotic protein subcellular localization tool (https://services.healthtech.dtu.dk/services/DeepLoc-2.0/#: :text=The%20DeepLoc%202.0%20server%20predicts,localizations%20inside%20the%20eukaryotic%20cel). The amino acid sequences in FASTA format were used as input (Almagro Armenteros et al., 2017).
Positive Selection-Selectivity pressure analysis
The codeml program of a phylogenetic analysis by maximum likelihood(PAML) was used for identifying the dN/dS ratio of CHUP1 and CHUP1-like proteins. The PAL2NAL program was used to generate codon alignment (Suyama, Torrents & Bork, 2006). A free-ratio model (NSsites = 0, model = 1), which allows different dN/dS ratios for each branch of the tree, was used to detect adaptive selection (Yang, 2007). The unrooted tree has been constructed using Phylip.
RNA isolation, cDNA synthesis, and qRT-PCR analysis
RNA was isolated using the CTAB method from three stages of Bienertia leaves: young (1∼3 mm) with yellowish leaves, intermediate(3∼6 mm) containing green tips with yellowish leaves, and mature upper one cm sections of whole green leaves. Finely ground samples (100 mg) in liquid nitrogen were used for RNA isolation by following the CTAB protocol.
For cDNA synthesis, 2000 ng of RNA was used (amfiRivert II cDNA Synthesis Master Mix, GenDEOPT, USA). Ten-fold diluted cDNA was used for quantitative real-time PCR (qRT-PCR) analysis (iQ™SYBR Green®Supermix; Bio-Rad, Hercules, CA, USA) with the primers mentioned in Table S5. The two-step SYBR method with 58 °C amplification was used in the thermocycler (CFX96 Touch™ Real-Time PCR Detection System; Bio-Rad). Denaturation was performed at 95 °C for 3 min in 40 cycles with 95 °C/15s, 58 °C/30s, and a melting curve with 95 °C/10s, 65 °C/5s, and 60 °C/50s. All samples were analyzed with three individual biological replications. The expression of CHUP1, CHUP1-like, and its associated genes was calculated by their relative expression to internal control (GAPDH). The analysis of qRT-PCR was done with three biological and two technical repeats. Statistical analysis was done using one-way ANOVA followed by Duncan’s multiple range tests (p ≤ 0.05) in SAS software (Statistical Analysis System, V. 6.12; SAS, Inc, Cary, NC, USA).
Microscopy
Protoplasts were prepared from young (0.3–0.5 cm long), intermediate (0.5–1.0 cm long), and mature (between 1.0–1.5 cm long) stage leaf tissues according to the protocol published previously (Wimmer et al., 2017). Images were captured under the 20X objective lens of a Zeiss (Jena, Germany) Axioplan fluorescence microscope with a cooled CCD camera and the final images were processed using Photoshop CS6.
Results
Identification of CHUP1 and CHUP1-like proteins in C3, C4, CAM, and SCC4 model plants
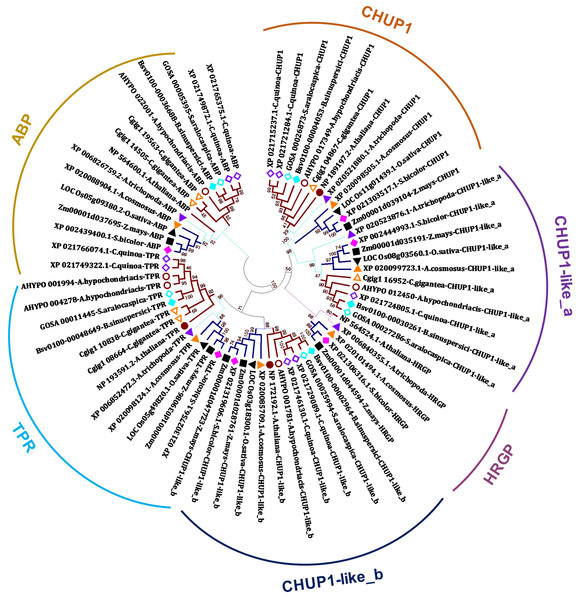
In the present study, we identified CHUP1, CHUP1-like proteins, and their associated proteins, including actin-binding proteins (ABP), tetratricopeptide repeat (TPR), and other HPR glycoproteins, across the model plants from C3, C4 and CAM of both monocot and dicot taken for the study using BLAST with AtCHUP1 as reference (Table 1). Phylogenetic trees showed that all CHUP1, CHUP1-like, and its associated proteins other than ABP of Arabidopsis were present as monophyletic between monocots and dicots. Similarly, CHUP1-like_a was recognized as monophyletic. The recently duplicated Chenopodium quinoa (hereafter, quinoa) has two copies of CHUP1 and CHUP1-like_b, whereas CHUP1-like_a was a single-copy gene. Contrasting to the C3 monocot rice and its close C4 relatives’ sorghum, maize possessed two copies of CHUP1-like_b. One is located on chromosome 1, and the other is on chromosome 9 (Fig. 1, Table 1, Table S1).
| Plant type | Type | Species | Gene ID | Gene Name | Genome location | Genomic position | Strand | CDS (bp) | Amino acids |
|---|---|---|---|---|---|---|---|---|---|
| Dicot | SCC4 | Bienertia | Bsv0100-00004053 | CHUP1 | 000130F | 1118154-1124532 | + | 2949 | 982 |
| Bsv0100-00030261 | CHUP1-like_a | 003553F | 178764-201267 | + | 2295 | 765 | |||
| Bsv0100-00002964 | CHUP1-like_b | 000073F | 919011-919485 | − | 1110 | 370 | |||
| Suaeda | GOSA_00026873 | CHUP1 | Contig 295 | 64362- 58799 | − | 2958 | 985 | ||
| GOSA_00027286 | CHUP1-like_a | Contig 751 | 381394-398100 | + | 2466 | 821 | |||
| GOSA_00025994 | CHUP1-like_b | Contig 607 | 272990-279085 | + | 1197 | 398 | |||
| C4 | Amaranthus | AHYPO_017349 | CHUP1 | Scaffold 380 | 54205-62742 | − | 2880 | 959 | |
| AHYPO_012450 | CHUP1-like_a | Scaffold 149 | 314501-316325 | + | 1215 | 404 | |||
| AHYPO_001781 | CHUP1-like_b | Scaffold 9 | 174263-178053 | + | 975 | 324 | |||
| C3 | Quinoa | XP_021721284.1 | CHUP1 | US | 1550610-1544417 | + | 2943 | 980 | |
| XP_021715237.1 | US | 974091-967433 | + | 2943 | 980 | ||||
| XP_021724805.1 | CHUP1-like_a | US | 1539562-1546012 | − | 2301 | 766 | |||
| XP_021729089.1 | CHUP1-like_b | US | 3186783-3189139 | − | 1242 | 413 | |||
| XP_021746130.1 | US | 3209412-3211923 | + | 1242 | 413 | ||||
| Arabidopsis | NP_189197.2 | CHUP1 | Chr 3 | 9352444-9357953 | + | 3015 | 1004 | ||
| NP_172192.1 | CHUP1-like_b | Chr 1 | 2186627-2184759 | + | 1179 | 392 | |||
| CAM | Cactus | Cgig1_04867 | CHUP1 | Scaffold 1884 | 35922-42874 | − | 2958 | 985 | |
| Cgig1_16952 | CHUP1-like_a | Scaffold 5088 | 52262-57326 | − | 2319 | 772 | |||
| Monocot | CAM | Pineapple | XP_020098505.1 | CHUP1 | Chr 11 | 10132256-10137677 | + | 2973 | 990 |
| XP_020099723.1 | CHUP1-like_a | Chr 12 | 12107512-12111995 | − | 2343 | 780 | |||
| XP_020085709.1 | CHUP1-like_b | Chr 4 | 13960552-13957835 | + | 1266 | 421 | |||
| C3 | Rice | LOC_Os11g01439.1 | CHUP1 | Chr 11 | 262011-256827 | − | 2790 | 930 | |
| LOC Os08g03560.1 | CHUP1-like_a | Chr 8 | 1658274-1653274 | − | 2397 | 798 | |||
| LOC Os03g18300.1 | CHUP1-like_b | Chr 3 | 10255348-10257715 | + | 1251 | 416 | |||
| C4 | Sorghum | XP_021303517.1 | CHUP1 | Chr 9 | 57875196-57870434 | + | 3195 | 1064 | |
| XP_002444993.1 | CHUP1-like_a | Chr 7 | 2531140-2526115 | + | 2394 | 797 | |||
| XP_021319606.1 | CHUP1-like_b | Chr 1 | 69005221-69001827 | − | 1257 | 418 | |||
| Maize | Zm00001d039104.1 | CHUP1 | Chr 6 | 170108921-170113150 | − | 2826 | 941 | ||
| Zm00001d0351491.1 | CHUP1-like_a | Chr 6 | 8696067-8700892 | − | 2394 | 797 | |||
| Zm00001d028761.1 | CHUP1-like_b | Chr 1 | 45864595-45867995 | + | 1272 | 423 | |||
| Zm00001d047723.1 | Chr 9 | 140000414-140003136 | + | 1293 | 430 | ||||
| Outgroup | C3 | Amborella | XP_020521880.1 | CHUP1 | US | 751159-742747 | − | 3042 | 1013 |
| XP_020523876.1 | CHUP1-like_a | US | 133663-141544 | − | 2577 | 858 |
Notes:
- US
-
Unplaced Scaffold
Figure 1: Phylogenetic tree of CHUP1, CHUP1-like, and its associated proteins.
Am.t (Amborella trichopoda, purple up triangle); Zm (Zea mays, dark green square); Sb (Sorghum bicolor, pink diamond); Os (Oryza sativa, light green down triangle); Ac (Ananas cosmosus, orange up triangle); At (Arabidopsis thaliana, brown circle); Cg (Carnegiea gigantea, orange-outlined up triangle); Cq (Chenopodium quinoa, purple-outline diamond); Ah (Amaranthus hypochondriacis, brown-outlined circle); Sa (Suaeda aralocaspica, aqua-outlined diamond); and Bs (Bienertia sinuspersici, aqua diamond). The percentage of 1,000 bootstrap replicates was represented in branches. ABP, actin-binding protein; HPR, hydroxyproline-rich glycoproteins; TPR, tetratricopeptide repeat. Dicots, monocots, and Amborella are highlighted in bold dark orange-colored branches, bold dark blue-colored branches, and thin light blue-colored branches, respectively.Functional domain prediction
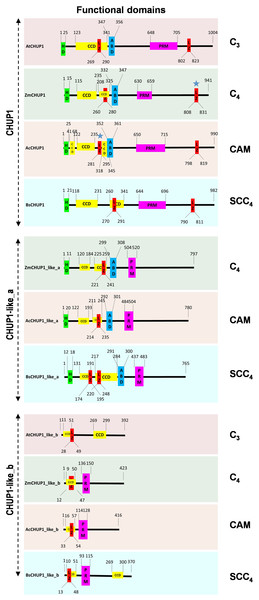
To identify the functional domains in CHUP1 and CHUP-like proteins, we used the COILs program for CCD, the 2bZIP server for LZ, UniProt for PRM, and LIP analysis for HD predictions. The position of functional domains such as HD, CCD, LZs, ABD, and PRM of CHUP1 and CHUP1-like proteins was mapped in the C3, C4, CAM, and SCC4 model plants (Fig. 2). Functional domain results showed that, unlike CHUP1, CHUP1-like_a was deficient in the C-terminal LZ domain. Except for the C3 photosynthetic plant Arabidopsis, all other model plants had PRM along with CCD and LZ domains in the CHUP1-like_b protein.
Figure 2: Functional domain prediction of CHUP 1 and CHUP 1_like proteins from the C3, C4, CAM, and SCC4 model plants.
HD, hydrophobic domain; CCD, coiled-coil domain; LZ, leucine zipper region; ABD, actin-binding domain; PRM, proline-rich motif. At, A. thaliana; Zm, Z. mays; Ac, A. cosmosus; and Bs, B. sinuspersici. The blue star represents the position of the LZ region that has been manually curated based on the motif pattern.Gene structure and motif analysis of CHUP1 and CHUP1-like genes in C3, C4, CAM, and SCC4 plants
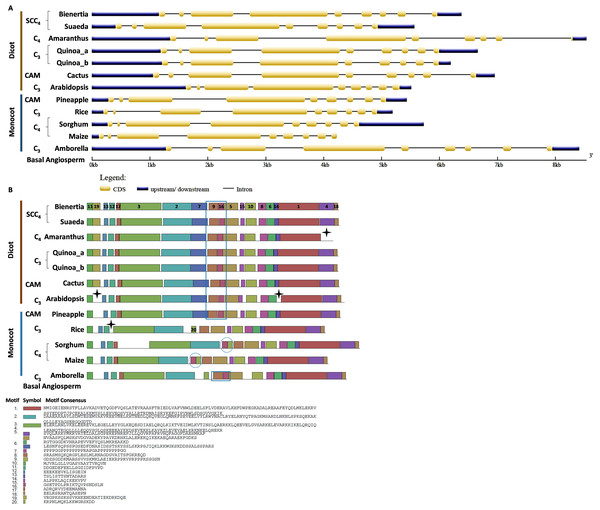
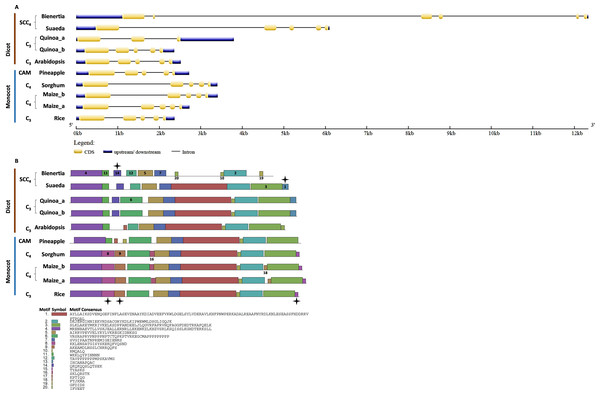
We investigated the intron/exon pattern and the presence and positions of motifs in all CHUP1 and CHUP1-like genes across the model plants under study. Gene structure analysis revealed that despite the variation in gene size, the CHUP1 gene of all species had nine exons (Fig. 3A). A total of 20 motifs were identified, of which motif 19 was observed only in the Caryophyllales and motif 20 was detected as Poales-specific. Interestingly, Amborella shared motifs 9 and 16 with all dicot species and pineapple, and motif 20 with Poales. The pineapple shared motif 7, which was identified only in dicots. All species contained motifs 4 and 18 in their C-terminal region except Amaranthus. As observed in sorghum and maize, C4 plants had rearrangement displays on motifs 16 and 20 (Fig. 3B).
Figure 3: Gene structure and motif of CHUP1-like_a from C3, C4, CAM, and SCC4 plants.
(A) The gene structure of CHUP1 was analyzed using GENE STRUCTURE DISPLAY SERVER (GSDS) v. 2.0 (http://gsds.gao-lab.org/index.php). The CDS and gene sequences were used as input. (B) The motif of the CHUP1 was identified using the MULTIPLE EM for MOTIF ELICITATION (MEME) server v. 4.9.1 (https://meme-suite.org/meme/tools/meme). The protein sequence was used as input. The motif numbers are labeled in order. The black stars denote the differential motif either between species, lineages, types, or clades. The square box indicates the shared motifs among lineages. The circle (blue) around the motifs specifies the unique motifs present in monocots. Note: The sequences of each motif are provided at the bottom of the figure in a black square box.For the comparative analysis of CHUP1-like_a and CHUP1-like_b, Amaranthus was excluded from gene structure and motif analysis because of its smaller size (Table 1). Several key differences between C3, C4, CAM, and SCC4 species were found in the gene structure and motif of CHUP1-like_a (Fig. 4). Apart from Cactus and Suaeda, all species had seven exons. In the cactus, splits in the first and second exons were observed. Suaeda possessed an extra exon compared to the other species. The CHUP1-like_a gene size of all species was between 5kb and 8kb, whereas in SCC4 species, Bienertia and Suaeda had about 22kb and 17kb, respectively (Fig. 4A). Distinct motifs between the compared species were found in the CHUP1-like_a protein (Fig. 4B). The motifs 17 and 19 were Amaranthaceae-specific whereas motifs 5 and 6 were Poaceae-specific. Interestingly, Amborella and the monocot CAM plant pineapple shared the motifs 8, 9, 10, 11, 12, and 15 with other dicot plants. Additionally, motif 7, which was present only in dicots, was shared with Amborella. Motif 14 was dicot-specific. The difference between C3 and C4 monocots was the absence of motif 20 in rice.
Figure 4: Gene structure and motif of CHUP1-like_a from C3, C4, CAM, and SCC4 plants.
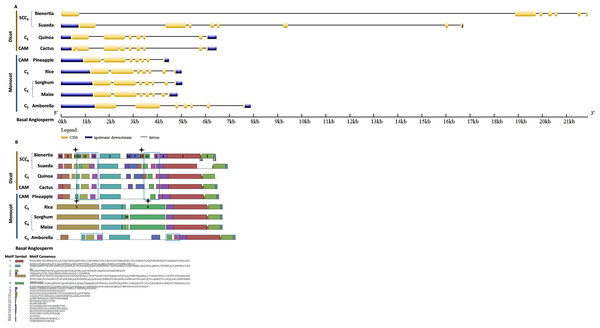
(A) The gene structure of CHUP1-like_a was analyzed using GENE STRUCTURE DISPLAY SERVER (GSDS) v. 2.0 (http://gsds.gao-lab.org/index.php). The CDS and gene sequences were used as input. (B) The motif of the CHUP1 was identified using the MULTIPLE EM for MOTIF ELICITATION (MEME) server v. 4.9.1 (https://meme-suite.org/meme/tools/meme). The protein sequence was used as input. The motif numbers are labeled in order. The black stars denote the differential motif either between species, lineages, types, or clades. The square box indicates the shared motifs among lineages. Note: The sequences of each motif are provided at the bottom of the figure in a black square box.Bienertia had an extra exon insertion between the first and third exons compared to other plants. The CHUP1-like_b gene size of all species was approximately between 2.2 and 3.6kb, whereas Bienertia and Suaeda had 12kb and 6kb, respectively (Fig. 5A). From the identified motifs, 13 and 14 were Amaranthaceae-specific and 8, 9, and 15 were Poaceae-specific (Fig. 5B). Interestingly, two motifs 1 and 3, commonly present in all species, were absent in Bienertia. Similarly, Amaranthaceae-specific motif 13 was also not detected in Bienertia. Among monocots, motif 16 existed only in sorghum and maize, both C4 plants. We also identified the gene structure and motifs of CHUP1-associated proteins such as ABP (Fig. S1), HPR (Fig. S2), and TPR (Fig. S3).
Figure 5: Gene structure and motif of CHUP1-like_b from C3, C4, CAM, and SCC4 plants.
(A) The gene structure of CHUP1-like_b was analyzed using GENE STRUCTURE DISPLAY SERVER (GSDS) v. 2.0 (http://gsds.gao-lab.org/index.php). The CDS and gene sequences were used as input. (B) The motif of the CHUP1 was identified using the MULTIPLE EM for MOTIF ELICITATION (MEME) server v. 4.9.1 (https://meme-suite.org/meme/tools/meme). The protein sequence was used as input. The motif numbers are labeled in order. The black star denotes the differential motif either between species, lineages, types, or clades. Note: The sequences of each motif are provided at the bottom of the figure in a black square box.Adaptive evolution
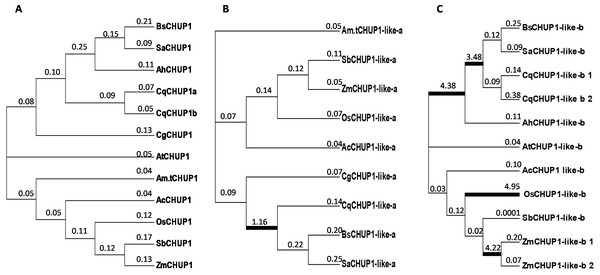
We calculated the non-synonymous substitution rate (dN) and synonymous substitution rate (dS) to evaluate the divergence and selection pressure in the CHUP1 gene among the species under study. The analysis showed that the ratio of non-synonymous to synonymous substitutions (dN/dS, ω) was less than 1. So CHUP1 underwent purifying selection along the lineages. However, adaptive evolution was detected between the Caryophyllales lineages of C3 (Quinoa) and SCC4 plants on the CHUP1-like_a protein. Similarly, adaptive selection was observed in C3-C4-SCC4 branches of Caryophyllales and also in monocots such as rice and maize (Fig. 6).
Figure 6: Phylogenetic analysis by maximum likelihood.
The unrooted images show the dN/dS ratio of (A) CHUP1, (B) CHUP1-like_a, and (C) CHUP1-like_b proteins from plant species included in the study. A free-ratio model was used to calculate independent ω for each branch of C3, C4, CAM, and SCC4 plants. The ratio of dN/dS was mentioned on the branches and lineages. Branches with an estimated ω ratio > 1 were emphasized in thick black color lines.Stage-specific morphological development and quantitative expression of CHUP1 and its associated genes
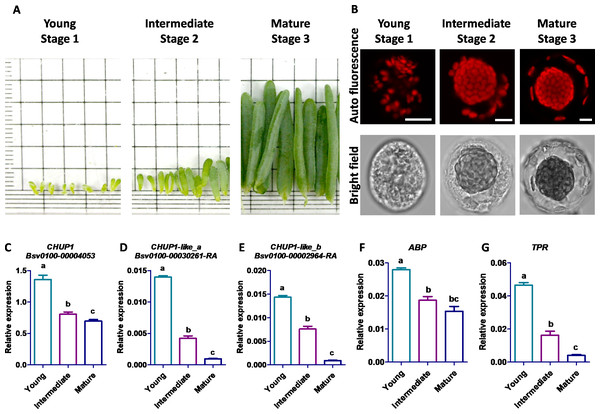
The young stage cells were sampled from Bienertia leaves less than three mm long, the intermediate stage cells from leaves 4–6 mm long, and the mature stage cells from the tip region of leaves three cm long (Fig. 7A). The scattered starch-like granules with pre-CCps appeared in the youngest stage, and no proper differentiation was observed between CCp and PCp. The progressive transition of PCp was observed in the intermediate stage. The complete subcellular localization and compartmentalization of PCp and CCp occurred in the mature stage (Fig. 7B). The transcript expression analysis of five genes in Bienertia displayed two different patterns during leaf developmental stages. Though the CHUP1 gene expression was decreased in the intermediate stage compared with that in the young stage, it was almost constitutively expressed in the mature stage, and its expression level was highest among the homologous genes, as shown by the normalized expression value against the internal control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Expression of ABP was in a similar pattern toCHUP1. The CHUP1-like and TPR genes showed decreased expression patterns from the young to intermediate to mature stages (Fig. 7C). Subcellular localization analysis showed that BsCHUP1, SaCHUP1, BsABP, and SaABP are predicted to localize at the chloroplast outer membrane, whereas BsCHUP1-like_a, BsCHUP1-like_b, BsTPR, SaCHUP1-like_a, SaCHUP1-like_b, and SaTPR are predicted to be localized in the cytoplasm. This result shows that CHUP1 and CHUP1-like proteins are involved in central and peripheral chloroplast cytoplasmic compartmentation (Table S2).
Figure 7: Eexpression studies of CHUP1, CHUP1-like, and its associated genes in B. sinuspersici.
(A) Leaf samples of B. sinuspersici from young (Stage 1), intermediate (Stage 2), and mature (Stage 3) stage plants (details in materials and methods) (B) Development of central chloroplasts and peripheral chloroplasts in the different stages of leaves. Chloroplast autofluorescence (top panels) and bright-field (bottom panels) images, scale bar = 10 µm. Relative expression of (C) CHUP1, (D) CHUP1-like_a, (E) CHUP1-like_b, (F) ABP, and (G) TPR was analyzed using qRT-PCR in three different developmental stages. ABP, actin-binding protein; TPR, tetratricopeptide repeat. Different letters indicate that treatments are significantly different at p ≤ 0.05. The error bar indicates the standard error of the mean.Discussion
CHUP1 is involved in the chloroplast accumulation and avoidance response to facilitate photosynthetic efficiency. CHUP1 was extensively studied in the Arabidopsis model plant (C3), and its orthologs were studied in millet and maize (C4) (Kobayashi et al., 2009). In Arabidopsis, it has been shown that CHUP1 is involved in the light avoidance mechanism, which was demonstrated in chup1 mutants. At the chloroplast outer envelope, CHUP1 links chloroplasts with actin (Oikawa et al., 2003; Von Braun & Schleiff, 2008; Lehmann, Bohnsack & Schleiff, 2011). In the case of Bienertia, the role of CHUP1 is not known. Since Bienertia is an SCC4 plant with bienertioid anatomy, we hypothesized that it may have paralogs of CHUP1 and its associated evolved proteins. Therefore, this study found CHUP1 and its associated novel proteins such as CHUP1-like_a, CHUP1-like_b, HPR, TPR, and ABP in the genomes included in the study. In this study, genome-wide identification analysis showed the presence of one copy of CHUP1, CHUP1-like_a, and CHUP1-like_b in SCC4 species, which is similar to the other plants. However, variation in gene size and amino acid number was observed (Fig. 1, Table 1). Variations in CHUP1, CHUP1-like_a, and CHUP1-like_b gene numbers are correlated with gene duplication and deletion of gene architecture between the species (Table 1, Table S1) (Long & Deutsch, 1999; Wang et al., 2018). In phylogenetic analysis, it was found that CHUP1, CHUP1-like_a, and CHUP1-like_b were placed in different clades. However, CHUP1 was found to share the monophyletic group with the CHUP1-like_a protein (Fig. 1). In addition, Suaeda and Bienertia were observed with an additional exon on CHUP1-like_a (Fig. 4A) and CHUP1-like_b (Fig. 5A), respectively. Moreover, exon splits were observed in the 5′ end of CHUP1-like_a of cactus (Fig. 5A). These molecular changes in the intron or exon can be attributed to splicing events or elements and genome evolution (Bondarenko & Gelfand, 2016). Studies have demonstrated that ND, CCD, ABD, and PRM domains are significant for chloroplast movement by characterizing the mutants and their complements in plants (Oikawa et al., 2003; Oikawa et al., 2008; Yamada et al., 2009; Li et al., 2010; Chotewutmontri & Barkan, 2016; Wang et al., 2018). The absence of the C-terminal LZ domain in CHUP1-like_a proteins was observed in all species in our analysis (Fig. 2), and it showed it might not undergo a dimerization process (Wang et al., 2018), unlike CHUP1. In CHUP1, dimerization is one of the key processes for chloroplast anchoring (Lehmann et al., 2011). Similarly, unlike the CHUP1 protein, functional domains such as the HD region, ABD, and C-terminal LZ are absent in the CHUP1-like_b protein (Fig. 2). Though the significance of the absence of conserved motifs in BsCHUP1-like_b is unclear, the presence of a higher number of transposons in its genome may be attributed to the loss of motifs (Offermann, Okita & Edwards, 2011; Panchy, Lehti-Shiu & Shiu, 2016).
The presence of functional domains was clearly distinguished between CHUP1, CHUP1-like_a, and CHUP1-like_b proteins (Fig. 3). The absence of HD in CHUP1-like_b showed that it is not involved in the chloroplast movement. No chloroplast relocation movement was observed in an Arabidopsis mutant with 300 aa in the N-terminal region of CHUP1 (Oikawa et al., 2008). This indicates the importance of ABD and PRM in chloroplast relocation. The CCD and LZ (also called short CCD) regions of CHUP1-like_b showed their involvement in the signaling networks, interaction with filaments, and organization of cellular processes like cell division (Yamada et al., 2009). Though the C-terminal LZ role remains elusive in CHUP1, it could have been involved in intra-molecular interaction (Lehmann, Bohnsack & Schleiff, 2011). The presence of ABD and PRM (Fig. 6) showed the involvement of CHUP1-like_a in binding with G-actin and profilin (Von Braun & Schleiff, 2008). Further, in vivo studies are needed to know the role of CHUP1-like_a in chloroplast movement. Well-characterized functional domains for homo-dimerization, anchoring the chloroplast, and the photo-relocation process (Von Braun & Schleiff, 2008) are absent in the CHUP1-like_b protein (Fig. 6). This concurs with the fact that BS CHUP1-like_b is not engaged in chloroplast relocation movements (Li et al., 2010; Chotewutmontri & Barkan, 2016). The existence of differential regulatory motifs between CHUP1 (Fig. 4B) and CHUP1-like_b (Fig. 5B) was enriched with the different composites of the protein-protein interactive network. Intriguingly, no closely related homologs for NP_564524.1, HPR, were identified in other dicots (Fig. 1). Oikawa et al. (2008) reported that the CCD of CHUP1 is apparent for oligomerization followed by firm anchorage of the chloroplast on the plasma membrane (Chotewutmontri & Barkan, 2016). The MS chloroplasts of maize are structurally similar to those of C3 plants (Oikawa et al., 2008). Usually, the BS chloroplast is not influenced by light intensity. The structure of the BS chloroplast is neither affected when plants are grown under tropical sunlight nor in low-light conditions (Drozak & Romanowska, 2006).
Compared with CHUP1, CHUP1-like proteins possessed different motifs (Figs. 3B, 4B, and 5B). For chloroplast movement, the N-terminal has been considered the key translocation component (Li & Chen, 1996; Oikawa et al., 2003). However, we have found that CHUP1-like_a had a conserved C-terminal region (Fig. 4B). Interestingly, pineapple, a monocot, shared comparable functional motifs of CHUP1-like_a with cactus, a distant CAM-relative, rather than other close grass family members (Fig. 4B). It may be a grass-specific change showing the similarity between pineapple and the dicots (Kondo et al., 2004). Since Amborella shared motifs with the dicots and CAM plants (Fig. 4B), an adaptive response could be a possible correlation for the CHUP1-like_a function.
Though CHUP1 proteins exhibited purifying selection, CHUP1-like_a protein showed strong positive selection in Caryophyllales, which contains the SCC4 plant lineage and C3 (quinoa). This represents the functional adaptation of CHUP1-like_a in the SCC4 species, and it could be involved in the unique chloroplast localization mechanisms within the single cell (Fig. 6B). However, positive selection was found commonly in the Caryophyllales clade. More combinations of evolutionary models need to be conducted to find the possibility of CHUP1-like_a on SCC4 adaptation. CHUP1-like_b possessing positive selection signifies differential function on BS chloroplast movement in maize and also its functional adaptation in lineages of C3 (rice) (Fig. 6C). The existence of positiveselection on CHUP1-like_a and CHUP1-like_b denotes the neo-functionalization/sub-functionalization of modified photosynthetic or other regulatory mechanisms as well as independent evolution in certain plant lineages, including SCC4 plants (Panchy, Lehti-Shiu & Shiu, 2016; Wang et al., 2018).
Previous studies by Offermann et al. (2015) showed that Bienertia exhibits no chloroplast differentiation in the early stages. Organelle position occurs during the developmental stages of leaves (Offermann, Okita & Edwards, 2011; Offermann et al., 2015). The constitutive expression of CHUP1 in the intermediate and mature stages showed its involvement in the partitioning of MS chloroplasts towards the plasma membrane (Li & Chen, 1996; Kondo et al., 2004; Slewinski, 2013; Miyake, 2016). Increased CHUP1 gene expression in MS was observed during the developmental changes in maize (Drozak & Romanowska, 2006; Li et al., 2010). Our study shows a similar pattern of BsCHUP1 expression(Fig. 7C). Similar expression patterns of CHUP1 and ABP from young to mature stage leaf development (Fig. 7C) showed their significance in the involvement of compartmentalization of PCp in Bienertia. Suetsugu & Wada (2016) stated that chloroplast movement depends on specialized actin filaments, namely cp-actin filaments (Suetsugu & Wada, 2016). Lesser expression of CHUP1-like genes is correlated with their localization in the central cytoplasmic compartment. CHUP1-like gene expression patterns were found to be similar to those of TPR gene (Fig. 7C). Additionally, the TPR protein has been reported to facilitate the interaction between proteins. It is intricate in chloroplast gene expression and essential for signal transduction pathways (Hu et al., 2014).
A transcriptomic study on maize leaf detected that 64% and 21% of genes are differentially expressed along with the developmental gradient of BS and MS chloroplasts, respectively (Li et al., 2010). Accordingly, ZmCHUP1 homologs to AtCHUP1 and OsCHUP1 are expressed three-fold higher in MS than in BS, and higher phosphorylation of CHUP1 in MS cells was demonstrated in maize at mid-day with high light intensity (Gao et al., 2023). In finger millet and maize, BS chloroplasts migrate toward vascular bundles and form the centripetal position during cell maturation (Li et al., 2010). The establishment of positioning is related to tissue development and cytoskeletal changes (Koteyeva et al., 2016). In maize, extensive partitioning of the photosynthetic process occurs between MS and BS during the leaf developmental changes from the basal zone, transitional zone, maturing zone, and mature zone (tip, +1 cm below the leaf tip), with an active expression of the ZmCHUP1 gene in MS cells (Kondo et al., 2004; Drozak & Romanowska, 2006). In most cases, organelle movements are dependent on actin filaments. Similar expression patterns of CHUP1 and ABP from young to mature leaf development (Fig. 7C) showed their significance in the involvement of compartmentalization of PCp in Bienertia. Suetsugu & Wada (2016) stated that chloroplast movement depends on specialized actin filaments, namely cp-actin filaments. CHUP1-like gene expression patterns are similar to those of TPR gene (Fig. 7C). Additionally, the TPR protein has been reported to be involved in facilitating structural interactions between proteins. It is involved in chloroplast gene expression and is essential for signal transduction pathways. A mutant of tpr in Arabidopsis displayed a slow greening phenotype (Yang et al., 2011). Therefore, TPR has been considered an important gene involved in chlorophyll biosynthesis and photosynthetic signaling.
Conclusions
Overall, CHUP1 and its associated proteins were identified and characterized in SCC4 with reference to existing C3, C4, and CAM models, including the extant plant Amborella. In summary, the phylogeny-based analysis showed CHUP1-like_a shared monophyly with CHUP1 since it might have the property of MS-specific function. Monophyletic clade BsCHUP1-like_b without HD domain showed that it could be expressed in CCp. In contrast, decreased expression of both BsCHUP-like genes in the well-developed leaves was observed. CHUP1 contains domains that anchor the chloroplast to the plasma membrane (Oikawa et al., 2008). In MS cells, chloroplasts are anchored to the plasma membrane, whereas in BS cells, chloroplasts are located at the center surrounding the vascular bundle (Oikawa et al., 2003). High expression and activation of CHUP1 were reported in MS cells to facilitate the anchorage (Li et al., 2010; Gao et al., 2023). Similarly, in Bieneritia, higher CHUP1 expression was observed in the younger stage as chloroplasts are evenly distributed. But in the matured stage, chloroplasts are released from the plasma membrane and aggregated at the center. This phenomenon supports the low expression of CHUP1 in the mature stage. In addition, the subcellular localization prediction tool has predicted CHUP1 to be localized in the chloroplast and CHUP1-like proteins in the cytoplasm. Amplification of genes was observed only in SCC4 species. The CHUP1-like_a and CHUP1-like_b genes showed that these could have undergone evolutionary changes. Finally, positive selection on SCC4 gives the primary clue about the independent evolution in the Caryophyllales clade. Similarly, positive selection in CHUP1-like_b in rice and maize showed that this species could also vary in the MS-specific chloroplast mechanism.Because these genes are crucial for chloroplast mobility, their involvement in SCC4 photosynthesis does not necessitate further positively selected modifications, at least not at the level of amino acid sequence. Positive selection was observed in gene phylogenies, although it was not always associated with SSC4 function. However, non-coding regulatory sequences could have undergone positive selection. However, establishing this would call for a population genetics-style method, such as looking for indications of a “selective sweep” around a specific locus. The results of this study shed light on the functional and evolutionary details of the CHUP1 protein and its related proteins. Detailed experiments on the CHUP1-regulatory networks are essential to reveal the evolution of chloroplast compartmentalization networks, positive selection, and interacting networks in the SCC4 system.