Genome-wide analysis of the WRKY genes and their important roles during cold stress in white clover
- Published
- Accepted
- Received
- Academic Editor
- Lin Zhang
- Subject Areas
- Agricultural Science, Bioinformatics, Genomics, Molecular Biology, Plant Science
- Keywords
- White clover, WRKY, Genetic regulation network, Cold stress
- Copyright
- © 2023 Li et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Genome-wide analysis of the WRKY genes and their important roles during cold stress in white clover. PeerJ 11:e15610 https://doi.org/10.7717/peerj.15610
Abstract
Background
White clover (Trifolium repens L) is a high-quality forage grass with a high protein content, but it is vulnerable to cold stress, which can negatively affect its growth and development. WRKY transcription factor is a family of plant transcription factors found mainly in higher plants and plays an important role in plant growth, development, and stress response. Although WRKY transcription factors have been studied extensively in other plants, it has been less studied in white clover.
Methods and Results
In the present research, we have performed a genome-wide analysis of the WRKY gene family of white clover, in total, there were 145 members of WRKY transcription factors identified in white clover. The characterization of the TrWRKY genes was detailed, including conserved motif analysis, phylogenetic analysis, and gene duplication analysis, which have provided a better understanding of the structure and evolution of the TrWRKY genes in white clover. Meanwhile, the genetic regulation network (GRN) containing TrWRKY genes was reconstructed, and Gene Ontology (GO) annotation analysis of these function genes showed they contributed to regulation of transcription process, response to wounding, and phosphorylay signal transduction system, all of which were important processes in response to abiotic stress. To determine the TrWRKY genes function under cold stress, the RNA-seq dataset was analyzed; most of TrWRKY genes were highly upregulated in response to cold stress, particularly in the early stages of cold stress. These results were validated by qRT-PCR experiment, implying they are involved in various gene regulation pathways in response to cold stress.
Conclusion
The results of this study provide insights that will be useful for further functional analyses of TrWRKY genes in response to biotic or abiotic stresses in white clover. These findings are likely to be useful for further research on the functions of TrWRKY genes and their role in response to cold stress, which is important to understand the molecular mechanism of cold tolerance in white clover and improve its cold tolerance.
Introduction
During the life cycle of plants on Earth, they often encounter various stresses, seriously hindering their growth and development (Chinnusamy, Schumaker & Zhu, 2004; Katagiri, 2004). Among these stresses, most of them are abiotic stresses, such as drought, salt, and cold, etc. In long-term evolution, plants have gradually developed numerous molecular regulatory mechanisms to confer various stresses. These regulatory mechanisms have employed many genes involved in complex regulatory processes, which adjust plants’ physiological and biochemical processes in adapting to adverse environments. Among these genes, transcription factors (TF) are richly distributed in plants, which play very important roles in response to stress (Singh, Foley & Onate-Sanchez, 2002; Yamaguchi-Shinozaki & Shinozaki, 2006). The TF genes can bind to DNA regions, which are named as cis-acting elements, and regulate downstream gene expression. According to kinds of DNA regions, TF genes are divided into many classes, such as AP2/ERF, bZIP, WRKY, MYB, bHLH, etc., (Singh, Foley & Onate-Sanchez, 2002).
The WRKY gene family is a transcription factor family that has been specifically and widely identified in plants, mainly in higher plants, but rarely in lower organisms (Eulgem et al., 2000). These WRKY TFs can bind to cis-acting element (named as W-box, (T)(T)TGAC(C/T)), and regulate expression of downstream target genes containing W-box in promotors. These genes are characterized with critical roles in response to biotic, abiotic, and hormonal signaling processes (Ülker & Somssich, 2004). The WRKY gene was first isolated from sweet potato and named Sweet Potato Factor 1 (SPF1) (Ishiguro & Nakamura, 1994), it has since been widely identified and characterized from a lot of plants, and has become one of the largest TF families in plants, such as 72 WRKY TFs in Arabidopsis thaliana and 102 WRKY members in rice (Oryza sativa) (Abdullah-Zawawi et al., 2021). With the development of genome sequencing projects, many WRKY TFs have been well identified in dozens of plants with completed genome sequencing, such as cucumber (Cucumis sativus L) (Chen et al., 2020b), poplar (Populus tremula), grape (Vitis vinifera) (Wang et al., 2014), alfalfa (Medicago sativa L) (Mao et al., 2020), tea tree (Camellia sinensis), etc., (Chen et al., 2015, 2016; Rinerson et al., 2015; Singh et al., 2019; Xie et al., 2018). The roles of WRKY TFs have been widely documented in regulating plant growth, development process, and especially in response to abiotic stresses (Li et al., 2020). Yoo et al. (2014) have found that multiple WRKY genes could modulate response to abiotic stress, while over-expression of OsWRKY genes would enhance its tolerance to abiotic stress, and many WRKY TFs have also been characterized with a high capability to improve abiotic stress tolerance in various plants (Ülker & Somssich, 2004).
White clover (Trifolium repens L) is a perennial legume plant which is widely distributed in temperate and cool-temperate regions due to its strong root development, prostrate growth, and rapid regeneration (Wu et al., 2021). It also has a high yield, good quality, and adaptability, making it an important economic crop and an ideal forage for reforestation and improvement of natural grassland in some regions, which is also widely used as a landscape plant in gardens. However, during its growth process, white clover inevitably encounters various abiotic stresses, such as high salt, drought, and cold stress, especially in hard-winter of high-latitude regions, which causes extremely abnormal death of white clover and severely affects their production and promotion. Therefore, improving the cold tolerance of white clover has become an incisive problem in white clover production, while its genetic knowledge is still poor (Bao et al., 2020; Inostroza et al., 2018). Until 2019, the genome sequences of white clover were released, and researchers have been able to investigate gene function at the genome-level, which greatly promoted genetic improvement works in white clover (Griffiths et al., 2019). For example, Ma et al. (2022) have identified 37 SPL transcription factors from the white clover genome and characterized their key functions in inflorescence development.
Based on these results, our study has used bioinformatics methods to identify WRKY TFs from white clover at the genome-wide level, and systematically characterized their structure constitutions, cis-acting elements analysis, chromosome distributions, and genetic regulation network by integrating various datasets. Furthermore, RNA-seq was adopted to investigate WRKY TFs in response to cold stress, and qRT-PCR also confirmed their expression profiles. These findings would provide valuable insights into the exploration of white clover WRKY functions in response to cold stress.
Materials and Methods
Identification and classification of the TrWRKY gene family in white clover
The white clover genome resource information was released from the previous study, and all files were provided by Stig Uggerhøj Andersen from Aarhus University (Griffiths et al., 2019). DNA, CDS, and protein sequences were retrieved from the white clover genome. The sequences of Arabidopsis WRKY family proteins were collected from the TAIR database and used as BLAST (version 2.9.0+) query sequences to search the genome of white clover (Altschul et al., 1997), with an evaluation setting of 1E-05 and the coverages were set as 80%. The HMM file (version 3.3) (PF03106) was downloaded from the Pfam database (Punta et al., 2011), and the HMMER (evalue: 0.01) was used to identify and confirm WRKY DNA-binding domain, which was characterized as candidate WRKY proteins (Finn, Clements & Eddy, 2011). Annotated information for all candidate WRKY genes was retrieved from the white clover genome, including genome position, protein length, intron numbers, and these WRKY genes were classified into groups based on similar WRKY genes in Arabidopsis.
Phylogenetic analysis of the TrWRKY genes in white clover
Multiple sequence alignment analysis of Arabidopsis thaliana and white clover WRKY proteins using MUSCLE (version 5.1.0) with default parameters (Edgar, 2004). The phylogenetic tree of the WRKY gene family was constructed using the MEGA (version 11; Tamura, Stecher & Kumar, 2021) method with the following parameters: (1) Neighbor-joining (NJ); (2) Poisson correction; (3) genetic distance; (4) pair-wise deletion; (5) bootstrap: 1,000 replications. The TrWRKY genes were classified into different groups and subgroups based on the phylogenetic tree of AtWRKY and TrWRKY sequences.
Motif composition distribution analysis of TrWRKY genes in white clover
The conserved motifs of white clover WRKY protein sequences were identified using MEME (Multiple EM for motif Elicitation, Version 4.8.1) with the following parameters: (1) minimum and maximum width: 10 and 50, respectively; (2) the maximum number of motifs, 10; (3) number of appearances of a single pattern distributed in the sequences with model: 0 or 1 per sequence (-modzoops) (Bailey et al., 2009). All results were displayed with TBtools (version 1.098; Chen et al., 2020a).
Cis-acting elements analysis in the promoters of members of the TrWRKYs gene family
The 1,000 bp genomic sequence upstream of the transcription start site of the WRKYs gene family members was obtained from the white clover genome and the cis-acting elements in the promoter region of TrWRKYs gene family members were predicted by the PlantCARE online tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Chromosomal location, gene duplication and Ka/Ks analysis of WRKY genes in white clover
All white clover proteins were compared to each other using the software BLASTP (version 2.9.0+), and the gene duplications were identified and characterized based on BLAST results by software MCSanX (version Python) with the default parameter (Wang et al., 2012). Based on the positional information of WRKY genes in the white clover genome and the duplication between genes, the software CIRCOS (version 0.69-8) was used to display the distribution of the WRKY gene family in the white clover genome (Krzywinski et al., 2009). Based on the phylogenetic tree and gene duplications results, a molecular evolutionary analysis of the TrWRKY genes was performed by calculating the nonsynonymous (Ka) to synonymous (Ks) substitution ratio of the duplicated gene pairs in S. tuberosum using the KaKs_Calculator in TBtools (Chen et al., 2020a).
Gene regulation network analysis of white clover WRKY gene family
The information on the Arabidopsis gene regulatory network (GRN) was extracted from the AraNet database (V2) (Lee et al., 2014), including 22,894 Arabidopsis genes and 895,000 interactions (links). All white clover proteins were subjected to BLAST searched with Arabidopsis proteins, with an e-value cut-off 1e-05, and the highest scoring hits were confirmed as homologous genes of the white clover gene. Meanwhile, all Arabidopsis proteins were also BLAST searched against white clover proteins with the same set, and hits with the highest scores were identified as homologous genes for Arabidopsis genes; the two genes were identified as homologous pairs with two BLAST results. Then, the GRN of white clover was constructed using the GRN of Arabidopsis based on homologous pairs. The sub-networks that contained the TrWRKY gene of white clover were searched and evaluated; the results were visualized using Cytoscape software (version 3.9.1), as well as the genes were annotated using GO information based on white clover genome annotation information (Shannon et al., 2003). Then, GO enrichment analysis of the sub-networks was performed using topGO software (version 2.50.0; Alexa & Rahnenfuhrer, 2019), the threshold level was set to 0.05 to show the most significant terms, and terms with high enrichment were assigned as GRN functions as described in the software protocol.
Expression analysis of white clover TrWRKY genes in response to cold stress
The RNA-seq data have been reported, the data included eight time points in response to cold stress; our previous work has described in detail, which could be assessed with accession numbers: PRJNA781064 (Zhang et al., 2022). All of the RNA-seq reads were mapped to the transcript sequences of white clover genomes using the Salmon software (version 0.12.0), and the expression level of each gene (FPKM value) was estimated using the subroutine quant of software Salmon (Patro et al., 2017). These expressional data were transformed using the “log2” function and they were centered using the “scale” function of the R program (version 4.2.1; R Core Team, 2022); then, all expression data were clustered and plotted using the “heatmap.2” function of the ggplots package (version 3.1.3).
Plant growth and qRT-PCR analysis
Seeds of white clover cv. Haifa were purchased from Barenbrug China Ltd. Com. (Beijing, China). All seeds were germinated and transferred to a mixture of perlite and sand each with a volume of 3:1, as our previously described (Zhang et al., 2022). In briefly, all seeds were growing in the pots, about 10–15 plants per pot. The growth temperature was 24 °C in light and 18 °C in darkness per day and irrigated with half-strength Hoagland solution once every 2 days. After 4 weeks, they were randomly divided into four groups for cold stress treatment. We collected at 0 min (control), 30 min, 1 h, and 3 h (4 time points in total) at a setting of 4 °C. For each group, three samples were randomly chosen of five seedlings were pooled to form a biological replicate. All samples were frozen in liquid nitrogen and stored at −80 °C. Total RNA was extracted from white clover seedlings at different time points of cold stress at 4 °C using the Total Plant RNA Extraction Kit (Tiangen, Beijing, China), divided into four time points (0 min (control), 30 min, 1 h, 3 h), and used Prime Script RT kit (Toyobo, Shanghai, China) was reverse transcribed into cDNA as a template for quantitative reverse transcription PCR. Primers were designed using Primer3 based on the nucleotide sequences of WRKY family genes (Table S1) (Untergasser et al., 2012). qRT-PCR was performed using a Light Cycler® 96 system (Roche, Rotkreuz, Switzerland) and SYBR Premix Ex TaqTMII (Toyobo, Shanghai, China). Three replicates of each experiment were performed. The PCR conditions were set as follows. 95 °C for 2 min, 40 cycles, 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min. Fold change values were calculated using expression abundance, which is based on the 2−ΔΔCT method.
Results and analysis
Identification of WRKY genes in white clover
A total of 145 TrWRKY genes were successfully identified from the white clover genome by multiple sequence alignment based on the amino acid sequence of the white clover WRKY gene family. These genes were proven to contain WRKY domains according to domain analysis and they were named TrWRKY1 to TrWRKY145 according to their chromosome locus and structure. All genomic information of these TrWRKY genes, including names, gene locus, chromosomal locations, group, introns and protein length (aa) were retrieved and summarized in Table 1. For these 145 TrWRKY genes, the largest protein was TrWRKY145, comprising 1,063 amino acids (aa), while the smallest one was TrWRKY65 (137aa). There are 16 chromosomes in the white clover genome; the TrWRKY genes were not evenly distributed across all chromosomes. The intron distribution was 1—11, of which the TrWRKY121 had the most introns (11 introns), while fifteen members contained only one intron; the results showed TrWRKY genes highly diverged.
| Name | Locus | Chromosomal locations | Group | Intron | Length (aa) |
|---|---|---|---|---|---|
| TrWRKY001 | chr1.jg4946 | Tr1O:35253397–35255542 | I | 4 | 155 |
| TrWRKY002 | chr5.jg5422 | Tr5O:36224162–36225107 | I | 1 | 195 |
| TrWRKY003 | chr1.jg4948 | Tr1O:35276740–35279067 | I | 2 | 206 |
| TrWRKY004 | chr5.jg4677 | Tr5O:31183372–31185712 | I | 3 | 290 |
| TrWRKY005 | chr7.jg5842 | Tr7O:36085774–36087462 | I | 2 | 340 |
| TrWRKY006 | chr12.jg3166 | Tr4P:21046044–21048271 | I | 3 | 378 |
| TrWRKY007 | chr2.jg5856 | Tr2O:39405643–39408558 | I | 3 | 379 |
| TrWRKY008 | chr3.jg9578 | Tr3O:62913893–62916736 | I | 3 | 425 |
| TrWRKY009 | chr15.jg2764 | Tr7P:18230475–18233187 | I | 4 | 436 |
| TrWRKY010 | chr9.jg5404 | Tr1P:36641010–36643525 | I | 4 | 441 |
| TrWRKY011 | chr4.jg377 | Tr4O:2750219–2753428 | I | 4 | 462 |
| TrWRKY012 | chr4.jg517 | Tr4O:3682810–3686140 | I | 4 | 462 |
| TrWRKY013 | chr13.jg5881 | Tr5P:38784060–38785660 | I | 2 | 470 |
| TrWRKY014 | chr9.jg4026 | Tr1P:27139671–27144775 | I | 4 | 487 |
| TrWRKY015 | chr16.jg6482 | Tr8P:46034663–46039091 | I | 4 | 493 |
| TrWRKY016 | chr3.jg1725 | Tr3O:11498359–11502499 | I | 4 | 507 |
| TrWRKY017 | chr3.jg810 | Tr3O:5276114–5280219 | I | 4 | 511 |
| TrWRKY018 | chr7.jg4527 | Tr7O:28275075–28279600 | I | 3 | 517 |
| TrWRKY019 | chr16.jg6484 | Tr8P:46056816–46061810 | I | 5 | 522 |
| TrWRKY020 | chr13.jg5948 | Tr5P:39165179–39168802 | I | 5 | 536 |
| TrWRKY021 | chr7.jg2559 | Tr7O:16132786–16135642 | I | 4 | 542 |
| TrWRKY022 | chr15.jg1220 | Tr7P:8126860–8129794 | I | 4 | 546 |
| TrWRKY023 | chr11.jg2218 | Tr3P:14403172–14409407 | I | 5 | 550 |
| TrWRKY024 | chr5.jg6632 | Tr5O:44854411–44858046 | I | 5 | 550 |
| TrWRKY025 | chr13.jg5557 | Tr5P:36394815–36398400 | I | 4 | 566 |
| TrWRKY026 | chr3.jg3718 | Tr3O:24080424–24083938 | I | 4 | 568 |
| TrWRKY027 | chr3.jg9591 | Tr3O:62992430–62995677 | I | 4 | 576 |
| TrWRKY028 | chr13.jg5883 | Tr5P:38801923–38805881 | I | 4 | 611 |
| TrWRKY029 | chr7.jg4480 | Tr7O:27919168–27923981 | I | 7 | 625 |
| TrWRKY030 | chr7.jg4830 | Tr7O:30168002–30171661 | I | 4 | 659 |
| TrWRKY031 | chr3.jg54 | Tr3O:364289–368532 | I | 4 | 661 |
| TrWRKY032 | chr13.jg5616 | Tr5P:36858900–36863941 | I | 4 | 667 |
| TrWRKY033 | chr9.jg1959 | Tr1P:13473819–13478644 | I | 4 | 668 |
| TrWRKY034 | chr1.jg2939 | Tr1O:21500495–21505105 | I | 4 | 669 |
| TrWRKY035 | chr14.jg4072 | Tr6P:26714269–26720795 | I | 6 | 897 |
| TrWRKY036 | chr1.jg1973 | Tr1O:14366719–14368228 | IIa | 3 | 249 |
| TrWRKY037 | chr9.jg1140 | Tr1P:7918363–7919974 | IIa | 3 | 249 |
| TrWRKY038 | chr1.jg1972 | Tr1O:14356694–14358566 | IIa | 3 | 284 |
| TrWRKY039 | chr9.jg1139 | Tr1P:7914343–7916022 | IIa | 3 | 285 |
| TrWRKY040 | chr1.jg2956 | Tr1O:21601386–21603577 | IIa | 3 | 288 |
| TrWRKY041 | chr10.jg540 | Tr2P:3685109–3687534 | IIa | 3 | 288 |
| TrWRKY042 | chr12.jg1297 | Tr4P:8797774–8800048 | IIa | 4 | 314 |
| TrWRKY043 | chr4.jg283 | Tr4O:2101646–2103890 | IIa | 4 | 314 |
| TrWRKY044 | chr14.jg4718 | Tr6P:31206787–31208142 | IIa | 3 | 321 |
| TrWRKY045 | chr6.jg5830 | Tr6O:39546655–39548025 | IIa | 3 | 322 |
| TrWRKY046 | chr3.jg2104 | Tr3O:14203754–14206210 | IIb | 4 | 367 |
| TrWRKY047 | chr6.jg3356 | Tr6O:22856414–22858929 | IIb | 3 | 388 |
| TrWRKY048 | chr3.jg2106 | Tr3O:14209147–14217688 | IIb | 7 | 407 |
| TrWRKY049 | chr11.jg4323 | Tr3P:28623299–28628203 | IIb | 5 | 420 |
| TrWRKY050 | chr13.jg6040 | Tr5P:39734181–39741546 | IIb | 5 | 445 |
| TrWRKY051 | chr5.jg5942 | Tr5O:39784978–39789205 | IIb | 5 | 450 |
| TrWRKY052 | chr4.jg9125 | Tr4O:64569075–64575507 | IIb | 6 | 473 |
| TrWRKY053 | chr3.jg4106 | Tr3O:26548251–26551282 | IIb | 3 | 494 |
| TrWRKY054 | chr8.jg8224 | Tr8O:57583206–57585522 | IIb | 5 | 496 |
| TrWRKY055 | chr8.jg8226 | Tr8O:57595609–57597977 | IIb | 5 | 496 |
| TrWRKY056 | chr11.jg6061 | Tr3P:40558111–40560701 | IIb | 4 | 497 |
| TrWRKY057 | chr9.jg3992 | Tr1P:26841184–26844303 | IIb | 3 | 501 |
| TrWRKY058 | chr14.jg619 | Tr6P:4268713–4272199 | IIb | 3 | 521 |
| TrWRKY059 | chr4.jg1879 | Tr4O:13724492–13727623 | IIb | 4 | 556 |
| TrWRKY060 | chr8.jg7568 | Tr8O:52902921–52906615 | IIb | 4 | 556 |
| TrWRKY061 | chr10.jg1880 | Tr2P:13055576–13058377 | IIb | 5 | 564 |
| TrWRKY062 | chr10.jg1240 | Tr2P:8565304–8567908 | IIb | 5 | 565 |
| TrWRKY063 | chr6.jg3259 | Tr6O:22013991–22017015 | IIb | 3 | 573 |
| TrWRKY064 | chr4.jg5478 | Tr4O:39794266–39798649 | IIb | 4 | 611 |
| TrWRKY065 | chr14.jg1701 | Tr6P:11603027–11604397 | IIc | 2 | 137 |
| TrWRKY066 | chr9.jg1339 | Tr1P:9315424–9316217 | IIc | 2 | 152 |
| TrWRKY067 | chr16.jg6071 | Tr8P:43479876–43480502 | IIc | 1 | 172 |
| TrWRKY068 | chr9.jg4957 | Tr1P:33609237–33616647 | IIc | 1 | 188 |
| TrWRKY069 | chr3.jg7827 | Tr3O:50998463–50999731 | IIc | 1 | 191 |
| TrWRKY070 | chr12.jg4938 | Tr4P:32185999–32188206 | IIc | 1 | 215 |
| TrWRKY071 | chr3.jg7996 | Tr3O:52089540–52090962 | IIc | 1 | 229 |
| TrWRKY072 | chr8.jg6405 | Tr8O:44781392–44782814 | IIc | 1 | 229 |
| TrWRKY073 | chr7.jg896 | Tr7O:5551407–5554787 | IIc | 2 | 241 |
| TrWRKY074 | chr16.jg4775 | Tr8P:33960705–33961961 | IIc | 3 | 242 |
| TrWRKY075 | chr11.jg7038 | Tr3P:46907394–46909934 | IIc | 2 | 246 |
| TrWRKY076 | chr8.jg6410 | Tr8O:44809752–44812152 | IIc | 2 | 250 |
| TrWRKY077 | chr14.jg1115 | Tr6P:7734201–7741598 | IIc | 4 | 266 |
| TrWRKY078 | chr2.jg886 | Tr2O:6217545–6220827 | IIc | 2 | 273 |
| TrWRKY079 | chr7.jg7388 | Tr7O:46299748–46301293 | IIc | 2 | 289 |
| TrWRKY080 | chr3.jg7829 | Tr3O:51008820–51011087 | IIc | 2 | 313 |
| TrWRKY081 | chr4.jg7787 | Tr4O:55224476–55225976 | IIc | 2 | 318 |
| TrWRKY082 | chr1.jg12558 | Tr1O:86758945–86760457 | IIc | 2 | 323 |
| TrWRKY083 | chr12.jg7469 | Tr4P:49027733–49032235 | IIc | 6 | 343 |
| TrWRKY084 | chr1.jg10510 | Tr1O:72595887–72597616 | IIc | 2 | 358 |
| TrWRKY085 | chr10.jg2272 | Tr2P:15800686–15803310 | IIc | 2 | 368 |
| TrWRKY086 | chr16.jg5753 | Tr8P:41361956–41363810 | IIc | 2 | 404 |
| TrWRKY087 | chr8.jg8112 | Tr8O:56771672–56773457 | IIc | 2 | 406 |
| TrWRKY088 | chr14.jg1447 | Tr6P:9967786–9968685 | IId | 1 | 149 |
| TrWRKY089 | chr16.jg297 | Tr8P:1981047–1981864 | IId | 1 | 153 |
| TrWRKY090 | chr5.jg6489 | Tr5O:43735385–43739232 | IId | 1 | 165 |
| TrWRKY091 | chr7.jg1916 | Tr7O:11673947–11677464 | IId | 1 | 165 |
| TrWRKY092 | chr3.jg10600 | Tr3O:70128861–70129969 | IId | 2 | 223 |
| TrWRKY093 | chr3.jg376 | Tr3O:2419836–2420993 | IId | 2 | 245 |
| TrWRKY094 | chr5.jg633 | Tr5O:4407752–4409399 | IId | 2 | 250 |
| TrWRKY095 | chr3.jg418 | Tr3O:2696699–2704267 | IId | 6 | 258 |
| TrWRKY096 | chr11.jg9390 | Tr3P:63448611–63450828 | IId | 3 | 260 |
| TrWRKY097 | chr8.jg8664 | Tr8O:60972016–60973116 | IId | 2 | 290 |
| TrWRKY098 | chr12.jg3044 | Tr4P:20303684–20304867 | IId | 2 | 314 |
| TrWRKY099 | chr4.jg8047 | Tr4O:57107571–57109224 | IId | 2 | 327 |
| TrWRKY100 | chr9.jg1028 | Tr1P:7138533–7141172 | IId | 2 | 345 |
| TrWRKY101 | chr9.jg954 | Tr1P:6662343–6664904 | IId | 2 | 349 |
| TrWRKY102 | chr9.jg5416 | Tr1P:36708761–36713161 | IId | 3 | 350 |
| TrWRKY103 | chr8.jg7125 | Tr8O:49744559–49746242 | IIe | 1 | 154 |
| TrWRKY104 | chr10.jg758 | Tr2P:5125509–5126589 | IIe | 2 | 215 |
| TrWRKY105 | chr8.jg6926 | Tr8O:48516893–48517971 | IIe | 2 | 215 |
| TrWRKY106 | chr4.jg10192 | Tr4O:72095432–72096468 | IIe | 2 | 237 |
| TrWRKY107 | chr7.jg7373 | Tr7O:46208011–46209900 | IIe | 2 | 257 |
| TrWRKY108 | chr15.jg4042 | Tr7P:26578475–26580507 | IIe | 2 | 259 |
| TrWRKY109 | chr6.jg179 | Tr6O:1356330–1358362 | IIe | 2 | 259 |
| TrWRKY110 | chr13.jg4575 | Tr5P:30183713–30187166 | IIe | 2 | 262 |
| TrWRKY111 | chr4.jg3667 | Tr4O:26832093–26835148 | IIe | 2 | 262 |
| TrWRKY112 | chr9.jg5891 | Tr1P:39840694–39844509 | IIe | 5 | 277 |
| TrWRKY113 | chr4.jg8395 | Tr4O:59509470–59511001 | IIe | 1 | 296 |
| TrWRKY114 | chr1.jg12229 | Tr1O:84484169–84486864 | IIe | 2 | 307 |
| TrWRKY115 | chr9.jg7902 | Tr1P:53219706–53221002 | IIe | 2 | 317 |
| TrWRKY116 | chr16.jg1080 | Tr8P:7667356–7669114 | IIe | 2 | 324 |
| TrWRKY117 | chr16.jg3334 | Tr8P:23935985–23937743 | IIe | 2 | 324 |
| TrWRKY118 | chr8.jg542 | Tr8O:4059377–4061115 | IIe | 2 | 329 |
| TrWRKY119 | chr1.jg7088 | Tr1O:49874567–49875820 | IIe | 2 | 337 |
| TrWRKY120 | chr7.jg3290 | Tr7O:20634234–20635490 | IIe | 2 | 337 |
| TrWRKY121 | chr1.jg8379 | Tr1O:58443647–58446096 | IIe | 2 | 370 |
| TrWRKY122 | chr6.jg3054 | Tr6O:20633120–20636355 | IIe | 2 | 483 |
| TrWRKY123 | chr10.jg1494 | Tr2P:10329830–10331679 | IIe | 2 | 517 |
| TrWRKY124 | chr12.jg6707 | Tr4P:43890308–43904513 | IIe | 11 | 919 |
| TrWRKY125 | chr14.jg2719 | Tr6P:18441055–18442214 | III | 1 | 182 |
| TrWRKY126 | chr3.jg6078 | Tr3O:40310959–40312118 | III | 1 | 182 |
| TrWRKY127 | chr16.jg69 | Tr8P:454817–456279 | III | 2 | 242 |
| TrWRKY128 | chr3.jg8340 | Tr3O:54563576–54565215 | III | 2 | 263 |
| TrWRKY129 | chr1.jg1680 | Tr1O:12254543–12257169 | III | 2 | 302 |
| TrWRKY130 | chr9.jg3115 | Tr1P:20932379–20934360 | III | 2 | 305 |
| TrWRKY131 | chr9.jg6983 | Tr1P:47150282–47152529 | III | 3 | 307 |
| TrWRKY132 | chr16.jg1458 | Tr8P:10549490–10551175 | III | 2 | 309 |
| TrWRKY133 | chr2.jg4287 | Tr2O:28961889–28963324 | III | 2 | 321 |
| TrWRKY134 | chr4.jg11883 | Tr4O:83642647–83644458 | III | 2 | 321 |
| TrWRKY135 | chr2.jg6049 | Tr2O:40646492–40650667 | III | 2 | 331 |
| TrWRKY136 | chr5.jg4951 | Tr5O:33015650–33019304 | III | 2 | 335 |
| TrWRKY137 | chr13.jg1501 | Tr5P:10308563–10310061 | III | 2 | 338 |
| TrWRKY138 | chr7.jg7162 | Tr7O:44915496–44917119 | III | 2 | 338 |
| TrWRKY139 | chr16.jg1332 | Tr8P:9621061–9623396 | III | 2 | 345 |
| TrWRKY140 | chr12.jg6924 | Tr4P:45351376–45352661 | III | 2 | 357 |
| TrWRKY141 | chr12.jg6940 | Tr4P:45450209–45451499 | III | 2 | 357 |
| TrWRKY142 | chr6.jg2768 | Tr6O:18717189–18720732 | III | 3 | 807 |
| TrWRKY143 | chr5.jg5407 | Tr5O:36181101–36185875 | III | 3 | 835 |
| TrWRKY144 | chr5.jg5428 | Tr5O:36242747–36247552 | III | 4 | 913 |
| TrWRKY145 | chr5.jg5405 | Tr5O:36173904–36179186 | III | 4 | 1,063 |
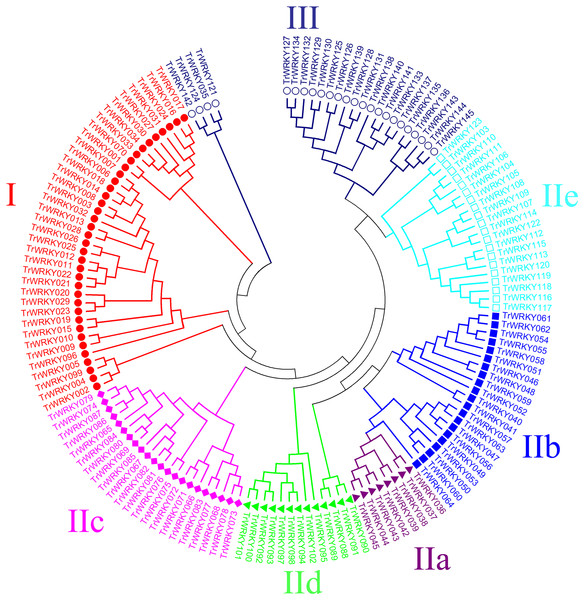
Classification and phylogenetic analysis in white clover
An unrooted phylogenetic tree with 145 TrWRKY genes using neighbor joining methods (Fig. 1) was constructed to further explore the phylogenetic relationship of the WRKY transcription factor family in the white clover. This unrooted tree intuitively reflected the evolutionary status and grouping attribution of 145 members of the WRKY family. As shown in Fig. 1, the white clover WRKY proteins could be classified into three large groups (Group I-II) on the basis of the classifications of WRKYs in Arabidopsis. Specifically, the largest number of WRKY members in Group II was 84, while Group I and Group III had 37 and 24 members, respectively. In addition, Group II was further classified into five subgroups (SubGroup IIa-IIe). The most numerous subgroups were subgroup IIc, with 22 members. Next are IIb and IIe, with 21 and 20 members, respectively. Subgroup IId has 13 members. The subgroup with the lowest number of members in Group II is IIa, with only eight members. At the same time, the unrooted phylogenetic tree showed that the distribution of WRKY genes in white clover and Arabidopsis was highly consistent.
Figure 1: Phylogenetic analysis of white clover WRKY proteins.
The NJ tree was constructed from the amino acid sequences of TrWRKY using MEGA4 with 1000 bootstrap replicates. The white clover WRKY proteins were grouped into three groups (Group I, labeled with red solid circle, II, and III, labeled with blue black hollow circle), and the Group II was further divided into five subgroups (IIa labeled with purple solid triangle, IIb labeled with blue solid square, IIc labeled with pink solid diamond, IId labeled with green solid triangle, and IIe labeled with cyan hollow square).Motif composition distribution analysis of TrWRKY proteins in white clover
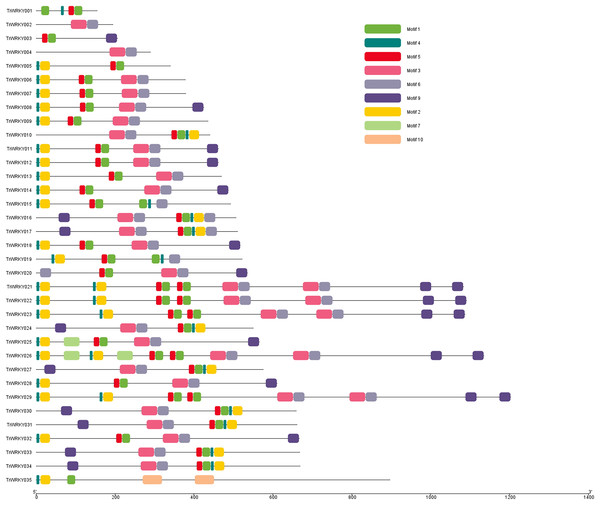
To further our understanding of the molecular structure and function of the TrWRKY gene family in white clover, we have analyzed the conserved motifs of WRKY gene family members and found the same subgroup had similar motif composition. Ten individual motifs were identified by the MEME tool, revealing the distinct regions of TrWRKYs (Fig. 2, and Figs. S1–S4). As Fig. 2 shows, the TrWRKY genes in Group I have contained nine motifs in total, most of them (except TrWRKY001, 003, 005, 015, 035) contained motif 3 and motif 6, while motif 3 has “WRKY” residues (see Fig. S1), which confirmed WRKY domain in these TrWRKY genes. Motif 1 is present in TrWRKY genes without motif 3, which also contained “WRKY” residues (see Fig. S1). The results showed WRKY domain has diverged in white clover. In addition, there are double WRKY domains identified in Group I members, even three copies of WRKY domains, which is also supported by domain search results. Group II and III have shown similar results, each TrWRKY gene contained motif 1 or motif 3, even two motifs, which consisted of BLAST and domain search results (Figs. S2 and S3). Meanwhile, the results of conservation motif composition also supported the results of sequence similarity and phylogenetic tree analysis, demonstrating clear structural motif differences between the three group. For example, most members of Group I contain motif 3, while members of Group III contain motif 1 (see Fig. S4), similar appearance was also discovered in Group II, each subgroup has diff motif composition patterns, see Fig. S2. In each subgroup, the proteins harbor a similar number and type of motif, which suggested the functional similarities of these TrWRKYs.
Figure 2: Distribution of conserved motifs of TrWRKY genes Group I in white clover.
Cis-acting elements analysis of the TrWRKYs promoter
Promoter cis-elements influence the initiation of gene transcription. We performed a bioinformatics analysis to identify possible cis-elements in the promoter sequences of TrWRKYs. PlantCARE was used to identify putative cis-acting elements in the 1,000 bp upstream sequence of each TrWRKY gene promoter. A total of 15 stress response elements, consisting of TC-rich repeats (the cis-regulatory element for defense along with stress response), ACE (cis-regulatory element that engages in light response), LTR (cis-regulatory element that plays a role in low-temperature response), TCA-element (cis-regulatory element with a role in salicylic acid response), SARE (cis-regulatory element with a role in salicylic acid response), ABRE (cis-regulatory element associated with the abscisic acid response), AuxRR-core (cis-regulatory element with a role in auxin response), G-box (cis-regulatory element with a role in light response), CGTCA-motif (cis-regulatory element with a role in the MeJA-response), TGACG-motif (cis-regulatory element associated with MeJA-response), P-box (gibberellin-responsive element), GARE-motif (gibberellin-responsive element), WUN-motif (wound-responsive element), MBS (MYB binding site associated with drought-inducibility), and MRE (MYB binding site associated with light response), were identified (Fig. S5). All TrWRKYs had at least one stress response-linked cis-regulatory element. The cis-regulatory elements for hormone modulation consisting of CGTCA motifs, ABREs, AuxRR cores, P-boxes, TCA elements and TGA elements were also uncovered in numerous TrWRKY promoter regions. Overall, 64 TrWRKYs (44%) had more than one ABRE motif, which indicated the prospective abscisic acid response under stress conditions. Approximately 66 TrWRKYs (46%) had one or more CGTCA motifs that demonstrated the MeJA response potential, and the TCA element, TGACG motif, P-box, and AuxRR core were found in 30, 60, 12 and 8 TrWRKYs, respectively (Fig. S5). 68 G-box, 23 LTR, 29 MBS, and 25 TC-rich repeats were also found in TrWRKY promoter regions, which illustrated that these genes might play a role in cold, drought inducibility and defense responses.
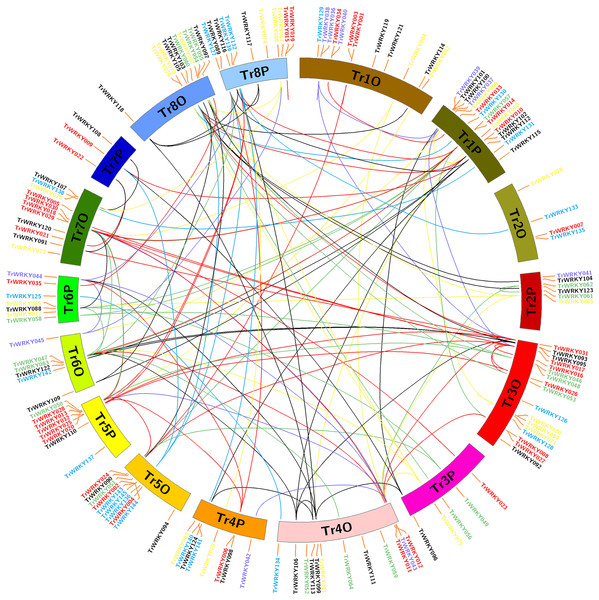
Chromosome localization, gene duplication and Ka/Ks analysis of TrWRKY genes in white clover
To determine the evolution and expansion of WRKY genes, we used the MCScanX and Circos softwares to construct the distribution of WRKY genes across chromosomes. All TrWRKY genes were distributed across 16 chromosomes, but they were not uniformly located on these chromosomes, see Fig. 3. For example, the chromosomes TrChr1O, TrChr1P, TrChr8O and TrChr8P harbor more TrWRKY genes than other chromosomes, such as TrChr2O and TrChr2P. Based on BLAST results, there were 124 gene duplication events identified by MCScanX software, including 118 segment duplications (SD) and six tandem duplications (TD). The results have suggested chromosome doubling helps to bring about WRKY expansion in white clover, and distributions of TrWRKY were similar between some doubling chromosomes, for example, chromosomes TrChr6O and TrChr6P, chromosome TrChr5O and TrChr5P. However, there are some divergences between doubling chromosomes, such as chromosome TrChr3O and TrChr3P, chromosome TrChr7O and TrChr7P. The results suggested TrWRKY genes have undergone deep diversion in sub-genome evolution, some chromosomes have expanded TrWRKY members by gene duplication events, while some duplications have been purged, which caused some TrWRKY genes hots with numerous members clustering.
Figure 3: Chromosome distribution and expansion analysis of WRKY transcription factors in white clover.
Red lines show duplications between members of the Group I, purple lines show duplications between members of the subgroup IIa, green lines show duplications between members of the subgroup IIb, yellow lines show duplications between members of the subgroup IIc, greys lines show duplications between members of the subgroup IId, cyan lines show duplications between members of the subgroup IIe, and blue lines show duplications between members of Group III.To estimate the divergence time of white clover WRKYs, synonymous (Ks) and nonsynonymous (Ka) substitutions between gene duplication pairs were calculated using the KaKs_Calculator in TBtool. When Ka/Ks <1, the genes experience purifying selection, which means the selection process could neutralize mutation to maintain the stability of the protein; in contrast, when Ka/Ks >1, the genes experience positive selection, which means great mutation happens in genes and eventually leads to a change in coded proteins. Our identified WRKY gene pairs had Ka/Ks values ranging from 0.06 to 0.89, proving that all of these genes experienced a purification selection process in white clover (Table S2).
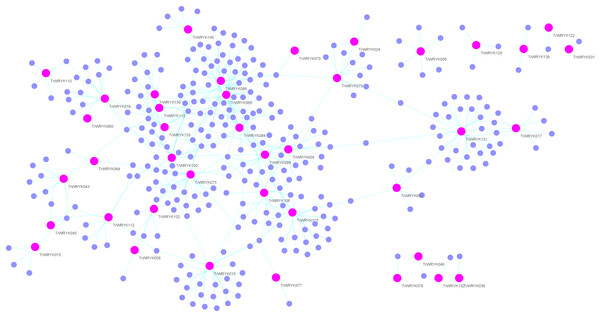
Genetic regulation network analysis of white clover WRKY genes
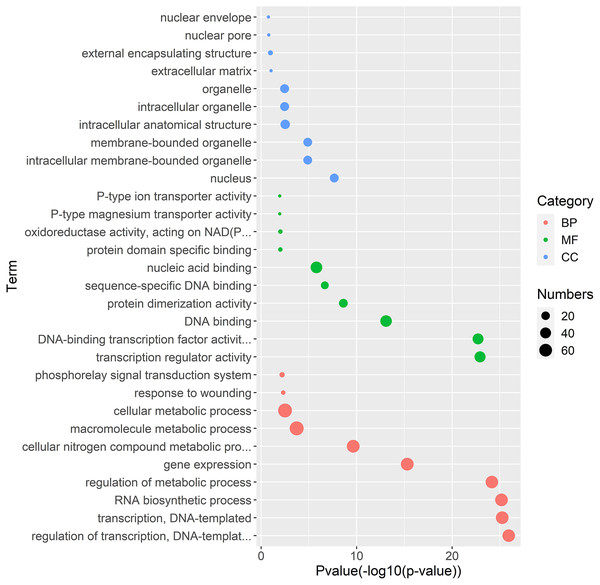
Gene regulation networks (GRN) are increasingly used to explore the system-level functions of genes, we have reconstructed GRNs of TrWRKY and their interacting genes based public interaction database. The GRNs consisted of 349 genes and 463 interactions, as Fig. 4 shown. From GRNs, we have found most of TrWRKY have interacted with dozens of function genes, consisting of TrWRKY function on the transcription regulation process. For example, TrWRKY084 interacted with 37 genes, TrWRKY131 with 32 genes, and TrWRKY100 with 30 genes, the results indicated these TrWRKY genes played important roles in white clover lifespan. Gene Ontology (GO) annotation of these interacting genes was retrieved, and GO enrichment analysis was performed using the topGO package on the R platform. The results showed they were mainly distributed in the nucleus, see Fig. 5, which supported TrWRKY genes also functioned in the nucleus. In addition, molecular functions of these function genes were highly focused on transcription regulator activity, while they were mainly participating in the regulation of the transcription process, the results have confirmed the function of TrWRKY genes. It was notable that these genes were also enriched on terms “response to wounding” and “phosphorylay signal transduction system”, which are plant popular descriptions in response to abiotic stress, these results suggested TrWRKY genes probable function in response to abiotic stress.
Figure 4: Gene regulatory network analysis of TrWRKY genes and their interactions in white clover.
Gene regulatory network (GRN) of TrWRKY genes and their interactions were generated based Arabidopsis interactions, which was displayed with Cytoscape. Pink nodes correspond TrWRKY genes, while violet nodes correspond the genes interacted with TrWRKY genes, the cyan lines represented interactions in white clover.Figure 5: Gene Ontology enrichment analysis of interaction genes with TrWRKY genes.
The GO enrichment analysis showed the involvement of interaction genes with TrWRKY genes in biological processes, molecular functions, and cellular components. Red dots represent GO terms from biological process (BP), green dots represent GO terms from molecular function (MF), while blue dots represent GO terms from cellular component (CC). Dot size represents the number of genes involving in the GO term, the X-axis is p-value of topGO enrichment analysis, with -log10 transformation, -log10 (p), while the Y-axis is GO terms.Expression analysis of TrWRKY genes in response to the cold stress
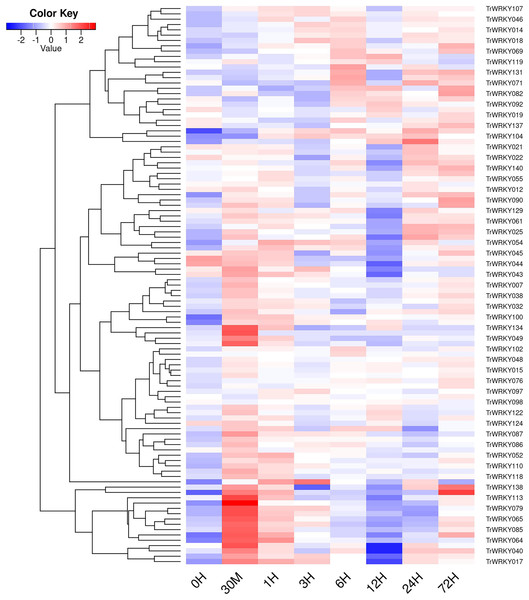
In order to investigate TrWRKY genes function in response to abiotic stress, we have adopted our previous RNA-seq data under cold stress to assess their expression profiles. The white clover was treated with cold stress at 4 °C, and RNA-seq was analyzed at eight time points, including 0 h, 30 min, 1 h, 3 h, 6 h, 12 h, 24 h, and 72 h. All expressing TrWRKY genes (with FPKM value larger than 1) were collected, and their expressional value (FPKM) was grouped with a violin plot, and results showed TrWRKY genes were increased in response to cold stress, see Fig. S6. Especially, TrWRKY genes were rapidly activated at 30 min, and keeping a high expressional level in the following stages, the results suggested TrWRKY gene played critical roles in the early stage under cold stress. Expression profiles of these TrWRKY genes were clustered and displayed using the heatmap function, the results showed most of the TrWRKY genes were highly expressed at 30 min in response to cold stress, see Fig. 6. The finding consisted of violin plot analysis, which confirmed their rapid response to cold stress. Among these TrWRKY members, some were expressly high-expressing at 30 min, for example, TrWRKY017, 040, 049, 064, 065, 079, 085, 100, 102, 113, 138. Combining their hub-function in previous GRNs, such as TrWRKY100 and TrWRKY 113, they would be assigned rapid and critical regulation function in response to cold stress.
Figure 6: The expressional profiles of TrWRKY genes in response to cold stress.
The expressional profiles of TrWRKY genes were retrieved from RNA-seq database with accession numbers: PRJNA781064. There were eight time points, including 0 h, 30 min, 1, 3, 6, 12, 24, and 72 h, each time point has three biological replications. Mean expression levels (FPKM values) were measured by Salmon software (version 0.12.0), and they were displayed using ggplots package of R platform.qRT-PCR validation of TrWRKY genes expression in response to cold stress
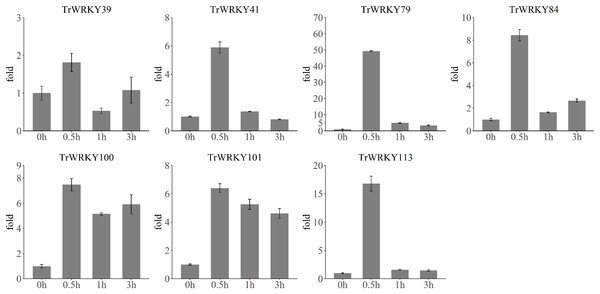
To validate the rapid response to cold stress, we have performed qRT-PCR analysis of seven TrWRKY genes with four time points, including 0 h, 30 min (0.5 h), 1 h, and 3 h. The qRT-RCR analysis results have confirmed their rapid and highly expressed in response to cold stress, see Fig. 7. All TrWRKY genes have shown a dramatic increase followed by a mild decrease in expression, the qRT-PCR and RNA-seq results are concordant, showing a similar pattern of TrWRKY genes expression in response to cold stress. These findings demonstrate that the TrWRKY genes are actively involved in the early response to cold stress and play critical roles in regulating gene expression in white clover.
Figure 7: qRT-PCR analysis of TrWRKY genes in response to cold stress.
The X-axis represent time points in response to cold stress, and Y-axis represent relative expression level of TrWRKY genes, which set expression level at “0 h” time point as 1. The expression level were calculated using the 2−ΔΔCT method as method section described.Discussion
WRKY TFs are well indicated to regulate various physiological processes in plants, from plant growth, development, to respond to abiotic stress (Eulgem et al., 2000; Ülker & Somssich, 2004). With more and more plant genome sequencing accomplished, numerous WRKY TFs have been identified from many plants, with important roles in various physiological processes, especially their critical function in response to biotic or abiotic stress (Chen et al., 2015; Chen et al., 2016; Dong et al., 2019; Li et al., 2014; Rinerson et al., 2015; Singh et al., 2019). However, there is no report on the white clover, which is widely distributed on global earth.
In the present research, there are 145 WRKY TFs identified in white clover, whose assembly genome size is 841 Mb, the number is about two folds of Arabidopsis (72 members) (Abdullah-Zawawi et al., 2021). Based on previous research, the numbers of WRKY TFs are variations between different plants, which are not straightly related to genome sizes. For example, Arabidopsis contains 72 members in 119 Mb, cucumber contains 61 members in 225 Mb (Chen et al., 2020b), rice contains 128 members in 466 Mb (Abdullah-Zawawi et al., 2021), Medicago truncatula contains 93 members in 420 Mb (Song & Nan, 2014), and soybean contains 188 members in 994 Mb (Yu et al., 2016), etc. In these plants, rice and soybean have undergone whole genome duplication, which brought about WRKY TFs double in the genome. Therefore, white clover with the tetraploid genome, contained more WRKY TFs than plants with the diploid genome, such as Arabidopsis, cucumber. Besides, gene duplication is also considered with important role in the expansion of gene families, which are mainly classified as tandem duplication and segmental duplication based on duplication patterns. There were 118 segmental duplication events and six tandem duplication events in white clover, the results suggested segmental duplication played critical roles in the expansion of TrWRKY TFs. Meanwhile, the tandem duplication also has made minor contributions in the expansion of TrWRKY TFs compared to those with less WRKY members in the genome, such as cucumber (Chen et al., 2020b). Hence, gene duplication is the primary means of expansion and evolution of the white clover WRKY gene family.
Increasing reports have shown WRKY genes have played important roles in the regulation of signaling transduction, and transcription, especially in response to biotic or abiotic stress (Li et al., 2020; Waqas et al., 2019; Wei et al., 2021; Xie et al., 2018). In the present research, the genetic regulation network was reconstructed with 463 interactions. Most of the TrWRKY genes have more interacting genes, for example, TrWRKY065 interacts with 46 genes, TrWRKY086 with 42 genes, TrWRKY084 with 37 genes, TrWRKY131 with 32 genes, and TrWRKY018 with 31 genes, etc. According to GO enrichment analysis results, most genes had focused on the transcription process, regulation of the metabolic process, gene expression, etc. These results also confirmed that WRKY TFs functions were highly conservative in white clover. Meanwhile, molecular function results were also supported these findings, most interaction genes were annotated with DNA binding activity, implying that they are regulated by transcription factors. It is worth noting that the GO term “protein dimerization activity” was also enriched in GRN genes, which intensely suggested some TrWRKY genes interacted with each other to perform their function, for example, TrWRKY065 interacted with TrWRKY100, TrWRKY084 interacted with TrWRKY086, TrWRKY135 interacted with TrWRKY113, etc. Similar opinions were also reported in other plants, for example, the interaction of homoeologous WRKY18 and WRKY40 in Arabidopsis was confirmed with important roles in response to biotic stress (Abeysinghe, Lam & Ng, 2019); JrWRKY2 and JrWRKY7 were interacting and formatting into homodimers in response to abiotic stress and ABA treatment from Juglans (Yang et al., 2017); and WRKY genes were also found to form dimerization in rice (Cheng et al., 2019). These findings indicated function types of TrWRKY genes were consistent with other plants, which is needed more molecular experiments to validate. In addition, TrWRKY genes were characterized to participate in response to wounding, this result was also consisted with previous reports (Srivastava et al., 2022). In Arabidopsis, WRKY8 expression was wounding-induced and confirms involvement in basal defense (Chen, Zhang & Yu, 2010); Preferential expression of PsWRKY and its interaction with downstream genes in benzylisoquinoline alkaloids (BIAs) were possible involvement in response to wounding in Papaver somniferum (Mishra et al., 2013); OsWRKY53 was characterized in response to wounding stress, which was regulated by OsMKK4-OsMPK1 cascade, implying WRKY was involved in phosphorelay signal transduction pathway (Yoo et al., 2014). This conclusion was also testified in the present research, we have found that interactions of TrWRKY genes were focused on the phosphorelay signal transduction system, it has consisted of WRKY regulated by MAPK cascade in rice. The above analysis and discussion showed that the WRKY gene family could widely participate in various signaling pathways in response to biotic or abiotic stresses.
WRKY TFs have well documented in plant growth and various stress processes. Our previous RNA-seq has shown most of TrWRKY TFs were remarkably upregulated in response to cold stress (Zhang et al., 2022). All cold treatment samples were more highly expressed than the control sample, except the 12H sample, see Fig. S6. These results showed genes were down-regulated expressions at night, while TrWRKY genes were up-regulated by cold stress, and the final expressions were slightly upregulated. Other time points showed the expressions of most TrWRKY genes were sharply up-regulated under cold stress, especially at 30 min, there were 43 TrWRKY genes up-regulated at this time point, implying that these TrWRKY genes quickly response to cold stress. This finding was consistent with other plants, for example, in grapevine, Wang et al. (2014) have demonstrated 36 VvWRKYs were changed following cold exposure and identified 15 VvWRKYs in two or more cold expression datasets, intensely suggesting their key functions in response to cold stress; similar research has reported 10 WRKYs were strongly expressing during cold stress in tomato (Chen et al., 2015). These genes represented candidate genes for future functional analysis of WRKYs involved in the cold related signal pathways. By transgenic analysis, VvWRKY28 has greatly improved the tolerance of Arabidopsis to cold stress, bound to promote downstream genes, including (RAB18, COR15A, ERD10, PIF4, COR47, and ICS1), and promoted their expressions (Liu et al., 2022). PmWRKY57 was also identified to improve cold tolerance, and induce the expression levels of cold-response genes in Arabidopsis transgenic lines, including AtCOR6.6, AtCOR47, AtKIN1, and AtRCI2A (Wang et al., 2022). In addition, KoWRKY40 was demonstrated to involved in ICE-CBF-COR signaling pathway, functioned as an important regulator under cold stress (Fei et al., 2022). Meanwhile, CdWRKY2 from bermudagrass was also revealed as a positive regulator in cold stress by targeting CdCBF1 promoters and activating its expression, improving cold tolerance by opening the CBF-signaling pathway (Huang et al., 2022). In white clover, there were 19 target genes of TrWRKY genes identified with the AP2 domain, see Table S3, which was the conservative domain of CBF genes, implying TrWRKY genes probably conferred cold tolerance by regulated ICE-CBF-COR signaling pathway. However, the molecular mechanism of their regulation is still unclear, there are more experiments needed to explore their function in detail.
Conclusion
In summary, we have identified 145 WRKY genes in white clover, and the following analysis was performed: gene identification and classification, phylogenetic and motif composition distribution analysis, cis-acting elements analysis, chromosomal mapping and gene duplication analysis, gene expression analysis, Ka/Ks analysis, genetic regulation network analysis and Gene Ontology annotation, and quantitative real-time reverse transcription PCR (qRT-PCR) analysis. We found that the evolution and expansion of the WRKY gene family may be closely related to the replication of segment and tandem replication within the WRKY genes. Meanwhile, we identified WRKY genes that may be involved in response to cold stress. The results of RNA-seq demonstrated that WRKY gene expression was partially up-regulated in response to cold stress. qRT-PCR results directly revealed that the WRKY gene plays an important role in response to early cold stress in white clover. The results of this study suggest a foundation for further studies on the function of the WRKY gene family in response to biotic or abiotic stresses in white clover.