Genome-wide identification and analysis of the evolution and expression pattern of the HVA22 gene family in three wild species of tomatoes
- Published
- Accepted
- Received
- Academic Editor
- Héctor Mora-Montes
- Subject Areas
- Agricultural Science, Bioinformatics, Biotechnology, Cell Biology, Plant Science
- Keywords
- HVA22, Bioinformatics, Phylogenetic analysis, Abiotic stress, Wild tomato
- Copyright
- © 2023 Zhao et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Genome-wide identification and analysis of the evolution and expression pattern of the HVA22 gene family in three wild species of tomatoes. PeerJ 11:e14844 https://doi.org/10.7717/peerj.14844
Abstract
Wild tomato germplasm is a valuable resource for improving biotic and abiotic stresses in tomato breeding. The HVA22 is widely present in eukaryotes and involved in growth and development as well as stress response, such as cold, salt, drought, and biotic stress. In the present study, we identified 45 HVA22 genes in three wild species of tomatoes. The phylogenetic relationships, gene localization to chromosomes, gene structure, gene collinearity, protein interactions, and cis-acting element prediction of all 45 HVA22 genes (14 in Solanum pennellii, 15 in S. pimpinellifolium, and 16 in S. lycopersicoides) were analyzed. The phylogenetic analysis showed that the all HVA22 proteins from the family Solanaceae were divided into three branches. The identified 45 HVA22 genes were grouped into four subfamilies, which displayed similar number of exons and expanded in a fragmentary replication manner. The distribution of HVA22 genes on the chromosomes of the three wild tomato species was also highly similar. RNA-seq and qRT-PCR revealed that HVA22 genes were expressed in different tissues and induced by drought, salt, and phytohormone treatments. These results might be useful for explaining the evolution, expression patterns, and functional divergence of HVA22 genes in Lycopersicon.
Introduction
The HVA22 gene was first isolated from the dextrin layer of barley (Hordeum vulgare) in 1993 (Shen, Uknes & Ho, 1993). Homologs to the HVA22 gene have been identified in eukaryotes, such as yeast, cereals, Arabidopsis thaliana, nematodes, mice, and humans. Approximately 367 HVA22 homologs have been found in eukaryotes, which present a conserved TB2/DP1 (deleted-in-polyposis) domain (PF03134) (Guo & David Ho, 2008; Sharon & Suvarna, 2017; Gomes Ferreira et al., 2019). Notably, no homologs have been described in prokaryotes to date, suggesting that HVA22 is likely involved in eukaryote-specific functions.
In plants, the expression patterns of HVA22 during development are well characterized. The accumulated transcripts of HVA22 homologs in leaves are highly induced by abscisic acid (ABA), drought, cold, and salt stresses in both barley and A. thaliana (Shen, Uknes & Ho, 1993; Shen et al., 2001; Chen et al., 2002). The heterologous expression of the barley HVA22 protein improves the salt tolerance and the survival rate of E. coli under cold stress by delaying its proliferation (Lu, 2013). Additionally, a recent study on barley discovered that the amount of the transcription product of the HVA22 gene was significantly higher in barley hvabi5d mutant strains than in wild-type plants under drought conditions (Collin et al., 2020). Subsequently, the HVA22 gene was also shown to be induced by drought and salt stresses in different plants. For example, the accumulation of transcript of HVA22 homologs in common wheat roots under drought stress was reported (Grzesiak et al., 2019). The expression of the HVA22 gene was significantly higher in drought-tolerant bermudagrass subjected to external simulated drought (Liu et al., 2014). The HVA22-like protein encoded by BG598159 was found to play an important role in salt and drought stress in potato by interacting with the StPDI1 protein and participating in the sucrose transport pathway (Eggert et al., 2016). The expression of the tomato HVA22 gene was also significantly induced by salt and drought stresses (Wai et al., 2022). Also, the heterologous expression of the Citrus clementina CcHVA22d gene in tobacco enhanced dehydration tolerance and significantly reduced the H2O2 content in a short-term dehydration environment (Gomes Ferreira et al., 2019).
Studies have shown that the HVA22 gene also plays an important role in endoplasmic reticulum–related pathways. The Yop1p gene, a homolog of HVA22 in yeast, appears to be involved in the translocation of substances from the endoplasmic reticulum to the Golgi apparatus during cellular activity (De Antoni et al., 2002). Previous studies on the yeast Yop1p protein revealed that the Yop1p/DP1 protein interacted with the Rtn4/NogoA protein, thereby co-regulating the interactions between other proteins in vivo as well as endoplasmic reticulum function (Hu et al., 2008). The HVA22 gene in the dextrin layer of barley seeds also has a similar function as yeast Yop1p. The accumulation of the HVA22 gene in the dextrin layer after induction by abscisic acid (ABA) inhibits vesicle transport in cells, thereby delaying the incorporation of storage protein vesicles, which is a process thought to play a role in regulating seed germination and seedling growth (Guo & David Ho, 2008). More recent studies in rice have shown that the rice HVA22 family gene OsHLP1 promotes disease resistance mechanisms in plants by maintaining endoplasmic reticulum homeostasis (Meng et al., 2022). To date, studies on the involvement of HVA22 homologs in the cellular vesicle transport pathway are scarce.
The supply of food and vegetable production have become major issues with the continuous rise in the world population. This is compounded by the potential impact of an increasingly changing climate on crop productivity. Extreme temperatures, drought, and soil salinization are the main adverse environments often encountered by plants (Gong et al., 2020). Tomatoes are a favorite vegetable worldwide. However, adverse environments such as salt, drought, and cold severely affect tomato growth and development (Chaudhary et al., 2019a; Chaudhary et al., 2019b). Wild tomatoes belonging to the genus Lycopersicon have higher tolerance to salt, drought, and cold than cultivated tomatoes (Szymański et al., 2020). Thus, wild tomatoes were an important genetic resource for our study on tomato response to adversity. It has been demonstrated that HVA22 genes are significantly upregulated in rice (Zhao et al., 2021), A. thaliana (Chen et al., 2002), barley (Shen et al., 2001), and tomatoes (Wai et al., 2022) in response to salt and drought stresses. However, systematic studies on HVA22 family genes in wild tomatoes have not been reported. In this study, we used the bioinformatics methods to comprehensively identify HVA22 family genes in three species of wild tomatoes (S. pimpinellifolium, S. pennellii, and S. lycopersicoides). This study might provide a theoretical reference for elucidating HVA22 family gene members and mining tomato genes for resistance to abiotic stresses.
Materials & Methods
Identification of HVA22 family genes in the family Solanaceae
The protein sequence of the Arabidopsis thaliana HVA22 family gene was downloaded from the Ensembl database (http://plants.ensembl.org/index.html) (Yates et al., 2022). Protein sequence files for three species of wild tomatoes (S. pimpinellifolium, S. pennellii, and S. lycopersicoides), tobacco (Nicotiana benthamiana), pepper (Capsicum annuum), eggplant (Solanum melongena), and potato (Solanum tuberosum), as well as genome files from the Solanaceae genome database (https://Solgenomics.net/), were downloaded (Fernandez-Pozo et al., 2015). The hidden Markov model of the structural domain of the HVA22-like protein TB2/DP was obtained from the Pfam (PF03134) (http://pfam.xfam.org/) and PANTHER (PTHR12300) (http://www.pantherdb.org/) database (Mistry et al., 2021; Thomas et al., 2022). The screened HVA22 protein sequences were validated using the online protein structural domain prediction tool HMM search (http://hmmer.org/) (Finn, Clements & Eddy, 2011), and genes that did not contain the TB2/DP structural domain were removed. The physicochemical properties of the screened tomato HVA22 family of proteins were predicted on the ExPASy website (https://www.expasy.org/protparam/) (Artimo et al., 2012). The subcellular localization prediction of HVA22 family genes in three species of wild tomatoes was performed on the WoLF PSORT online tool (https://wolfpsort.hgc.jp/) (Horton et al., 2007).
Construction of conserved motifs, cis-acting elements, and phylogenetic tree of HVA22 gene in three species of wild tomatoes
The HVA22 family gene motifs (Grundy et al., 1997) in the three species of wild tomatoes were searched using the MEME online tool (https://meme-suite.org/meme/tools/meme). The number of search base sequences was set to 20, and the minimum and maximum widths were set to 6 and 50, respectively. The results from the MEME search were used to map the conserved modal motifs and gene structures using TBtools. Multiple sequence comparisons were performed using MEGA 11 software, and a phylogenetic tree was constructed using the maximum likelihood method (ML) (Tamura, Stecher & Kumar, 2021). The constructed phylogenetic trees were embellished using the online tools ITOOL (https://itol.embl.de/) (Letunic & Bork, 2021). The 2,000-bp promoter sequence upstream of the HVA22 family gene in the three species of wild tomatoes was extracted, and the cis-acting element of the HVA22 family gene was predicted using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al., 1999) and visualized using TBtools (Chen et al., 2020).
Interaction network and expression analysis of HVA22 family homologous genes
MCScanX was used to analyze the HVA22 gene in four species of Lycopersicon (S. lycopersicum, S. lycopersicoides, S. pennellii, and S. pimpinellifolium) and Solanaceae [tobacco (N. benthamiana), pepper (C. annuum), potato (S. tuberosum) and eggplant (S. melongena)] interspecies as well as the intraspecific collinearity in four species of Lycopersicon. The substitution rate of paralogous homologous genes was calculated using Ka/Ks_Calculator 2.0 (Wang et al., 2010). The direct homologous genes between species and the paralogous homologous gene collinearity within species were visualized using TBtools. The STRING online website was used to predict protein–protein interaction relationships (Szklarczyk et al., 2019), interactions, after which the relationship data given by the predictions were visualized using Cytoscape 3.9.1. The expression matrices for different tissues and developmental stages of S. pimpinellifolium were downloaded from the Tomato Function Genomics database (http://ted.bti.cornell.edu/), which included expression data for root, stem, leaves, young flower buds, anthesis flowers, 10 days post anthesis, 20 days post anthesis, 30 days post anthesis, and ruptured fruit (Fei et al., 2010). From these, HVA22 family genes were selected and the expression profiles were heat-mapped using TBtools.
Total RNA extraction and reverse transcription
The extraction of plant leaf RNA was accomplished using a Tiangen plant polyphenol polysaccharide total RNA extraction kit (Beijing, China). The cDNA synthesis of extracted total RNA was performed using a 5 × All-ln-one RTMasterMix (AccuRT Genomic DNA Removal Kit; G492, ABM, Vancouver, Canada) reverse transcription kit. qRT-PCR-specific primers were designed using the NCBI online primer tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome), and the designed qRT-PCR primers were sent to Biotech Biologicals (Shanghai, China) for synthesis (Table S1). Quantitative PCR (qPCR) analysis was subsequently performed on a LightCycler machine using ChamQ Universal SYBR qPCR Master Mix (Q711, Vazyme, Nanjing, China) with Slactin as the internal reference gene. Three replicates of each treatment were performed. The relative expression was calculated by the 2−ΔΔCt Method (Livak & Schmittgen, 2001).
Plant material and growing conditions
The plant material used in this study was a wild tomato variety (S. pimpinellifolium, LA1589). For stress treatment, the seeds were sown in nutrient soil and vermiculite (v/v = 2:1) in a growth room at 24 ± 2 °C under 16-h light /8-h dark cycle. The seedlings were treated with Hoagland every week until use. Then, the plants were treated with different stresses, including 100 mM abscisic acid (ABA) or methyl jasmonate (MeJA), 200 mM NaCl, and 15% PEG6000. The leaves were collected and stored in liquid nitrogen quickly for RNA extraction at different time points (0, 2, 6, 12, and 24 h). Three independent biological replicates were included for each sample in the experiment.
Results
Identification of physicochemical properties and prediction of subcellular localization of HVA22 family proteins from the three species of wild tomatoes
In the present study, we used the Simple HMM Search function of the TBtools tool to search the HVA22 gene in S. lycopersicoides, S. pennellii, and S. pimpinellifolium. The retrieved genes were then subjected to structural domain validation using the HMM search (http://hmmer.org/) (Finn, Clements & Eddy, 2011) and InterPro (https://www.ebi.ac.uk/interpro/result/InterProScan/#table) online tool, and those without TB2/DP1 structural domains were discarded. The 45 HVA22 family genes were finally determined in three species of wild tomatoes (15 in S. pimpinellifolium, 16 in S. lycopersicoides, and 14 in S. pennellii) and named as HVA22a–HVA22p according to their positions on chromosomes. Subsequently, we characterized the physicochemical properties and predicted the subcellular localization of the HVA22 gene family member proteins from these three species of wild tomatoes. All HVA22 family proteins in three species of wild tomatoes (15 in S. pimpinellifolium 14 in S. pennellii, and 16 in S. lycopersicoides) had amino acid lengths between 88 and 603 and protein molecular masses between 9989.65 to 68493.8 Da, with an isoelectric point of 5.53–10.09 and aliphatic index of 51.68–123.66. In terms of hydrophilicity, most of the proteins exhibited hydrophilic proteins (GRAVY <0) and a few exhibited hydrophobic proteins (GRAVY >0). The subcellular localization predictions showed that HVA22-like was localized to multiple organelles in three species of wild tomatoes; most HVA22 family genes were localized to the endoplasmic reticulum, chloroplasts, cytoplasm, and nucleus; only the SpiHVA22k gene was localized to the vesicle (Table 1).
| Species | Gene id | Gene name | Length | MW(Da) | pI | Aliphatic index | GRAVY | Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| S.pimpinellifolium | Spim05g006320.1.1 | SpiHVA22h | 171 | 20248.41 | 7.67 | 98.71 | −0.101 | E.R |
| Spim03g025310.1.1 | SpiHVA22c | 182 | 20623.89 | 6.65 | 102.86 | 0.066 | Chlo | |
| Spim06g022990.1.1 | SpiHVA22i | 133 | 15554.63 | 9.49 | 115.11 | 0.249 | Extr | |
| Spim06g026920.1.1 | SpiHVA22j | 135 | 15665.57 | 8.8 | 104.74 | 0.21 | Chlo | |
| Spim11g010400.1.1 | SpiHVA22o | 167 | 19963.73 | 9.32 | 113.23 | 0.34 | Chlo | |
| Spim03g033180.1.1 | SpiHVA22d | 176 | 20507.66 | 6.41 | 96.93 | 0.005 | Extr | |
| Spim04g011050.1.1 | SpiHVA22e | 105 | 12446.7 | 8.66 | 104.86 | 0.13 | Cyto | |
| Spim10g007900.1.1 | SpiHVA22l | 156 | 18030.22 | 9.26 | 95.64 | 0.026 | Chlo | |
| Spim04g032680.1.1 | SpiHVA22g | 323 | 35618.35 | 8.81 | 71.05 | −0.346 | Chlo | |
| Spim01g007550.1.1 | SpiHVA22a | 180 | 21270.52 | 5.95 | 92.11 | −0.116 | Cyto | |
| Spim09g019680.1.1 | SpiHVA22k | 241 | 28172.67 | 8.93 | 84.94 | −0.026 | Vacu | |
| Spim10g015050.1.1 | SpiHVA22m | 603 | 68493.8 | 8.68 | 81.51 | −0.408 | Nucl | |
| Spim04g027770.1.1 | SpiHVA22f | 494 | 56507.39 | 9.34 | 89.82 | −0.226 | Nucl | |
| Spim01g044830.1.1 | SpiHVA22b | 255 | 29350.56 | 8.68 | 108.94 | 0.308 | Nucl | |
| Spim10g025730.1.1 | SpiHVA22n | 150 | 17597.28 | 9.1 | 80.73 | −0.339 | Cyto | |
| S.pennellii | Sopen05g003250.1 | SpHVA22g | 171 | 20234.38 | 7.67 | 98.13 | −0.102 | E.R. |
| Sopen11g005670.1 | SpHVA22m | 142 | 16848.74 | 8.58 | 95.42 | 0.063 | Cyto | |
| Sopen03g028180.1 | SpHVA22b | 136 | 15845.88 | 9.37 | 115.44 | 0.21 | Extr | |
| Sopen04g006560.1 | SpHVA22e | 131 | 15267.96 | 5.53 | 106.41 | 0.186 | Extr | |
| Sopen03g030110.1 | SpHVA22c | 180 | 20364.57 | 6.65 | 104 | 0.076 | Nucl | |
| Sopen06g029050.1 | SpHVA22h | 135 | 15652.57 | 8.8 | 104.74 | 0.231 | Chlo | |
| Sopen10g022980.1 | SpHVA22k | 187 | 21851.19 | 7.02 | 83.9 | −0.07 | E.R. | |
| Sopen03g035280.1 | SpHVA22d | 176 | 20507.66 | 6.41 | 96.93 | 0.005 | Extr | |
| Sopen10g003720.1 | SpHVA22i | 156 | 18020.24 | 9.24 | 93.14 | 0.013 | Chlo | |
| Sopen12g031160.1 | SpHVA22n | 302 | 33844.67 | 9.5 | 68.15 | −0.447 | Chlo | |
| Sopen04g034960.1 | SpHVA22f | 304 | 33623.99 | 8.95 | 68.12 | −0.379 | Chlo | |
| Sopen10g032550.1 | SpHVA22l | 188 | 22108.6 | 8.63 | 86.7 | −0.227 | Extr | |
| Sopen10g018090.1 | SpHVA22j | 559 | 63615.22 | 8.69 | 81.82 | −0.444 | Nucl | |
| Sopen01g003320.1 | SpHVA22a | 155 | 18113.25 | 10.09 | 51.68 | −0.977 | Nucl | |
| S.lycopersicoides | Solyd05g052440.1 | SlydHVA22h | 171 | 20176.3 | 7.67 | 97.6 | −0.113 | E.R. |
| Solyd11g055150.1 | SlydHVA22n | 123 | 14697.04 | 7.9 | 91.14 | −0.112 | Cyto | |
| Solyd11g055100.1 | SlydHVA22m | 145 | 17473.82 | 8.63 | 123.66 | 0.675 | Chlo | |
| Solyd03g072250.1 | SlydHVA22d | 190 | 21617.78 | 6.16 | 90.37 | −0.049 | Cyto | |
| Solyd06g073510.1 | SlydHVA22i | 135 | 15689.63 | 8.8 | 104.74 | 0.205 | Chlo | |
| Solyd03g077950.1 | SlydHVA22e | 176 | 20507.66 | 6.41 | 96.93 | 0.005 | Extr | |
| Solyd12g070530.1 | SlydHVA22o | 229 | 26296.16 | 9.05 | 65.15 | −0.345 | Chlo | |
| Solyd12g070590.1 | SlydHVA22p | 302 | 33831.62 | 9.28 | 68.15 | −0.456 | Chlo | |
| Solyd04g078410.1 | SlydHVA22g | 313 | 34770.38 | 8.87 | 68.02 | −0.35 | Chlo | |
| Solyd01g052910.1 | SlydHVA22a | 182 | 21641.02 | 5.91 | 95.88 | −0.036 | Nucl | |
| Solyd10g061800.1 | SlydHVA22l | 241 | 27758.68 | 7.05 | 78.92 | −0.22 | Chlo | |
| Solyd10g059860.1 | SlydHVA22k | 547 | 62120.36 | 8.28 | 79.85 | −0.452 | Nucl | |
| Solyd10g052770.1 | SlydHVA22j | 119 | 14080.39 | 9.14 | 85.21 | −0.242 | Chlo | |
| Solyd01g086740.1 | SlydHVA22b | 306 | 34201.23 | 8.87 | 110.56 | 0.206 | Nucl | |
| Solyd03g070260.1 | SlydHVA23c | 88 | 9989.65 | 9.64 | 84.2 | −0.172 | Cyto |
Phylogenetic tree analysis of HVA22 family genes
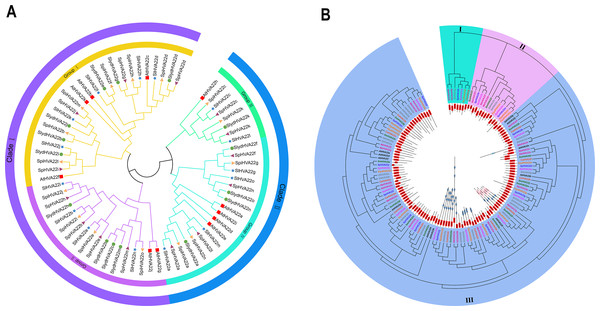
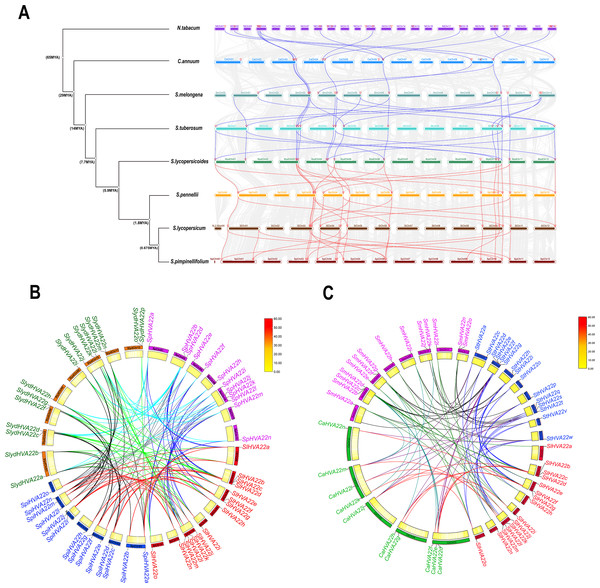
We combined 45 HVA22 genes from three species of wild tomatoes (16 in S. lycopersicoides, 14 in S. pennellii, and 15 in S. pimpinellifolium) with HVA22 genes from A. thaliana and cultivated tomatoes (S. lycopersicum) to construct a complete phylogenetic tree (Table S3). The phylogenetic tree showed that HVA22 genes in the cultivated tomatoes, three species of wild tomatoes, and A. thaliana were divided into two clades: I and II (Fig. 1A). The subgroups I and II were included in clade I, and the subgroups III and IV were included in clade II. The HVA22 family genes in the three species of wild tomatoes in groups I, II, III, and IV clustered on a subgroup with the A.thaliana and cultivated HVA22 genes. To enable a comprehensive understanding of the quantitative distribution of HVA22 family gene members in tobacco (N. benthamiana), potato (S. tuberosum), eggplant (S. melongena), pepper (C. annuum), and three kinds of wild tomatoes (S. pimpinellifolium, S. pennellii, and S. lycopersicoides). We identified 120 HVA22 family genes in selected Solanaceae species, constructed a phylogenetic tree, and designated these genes based on their chromosomal locations (Table S4). All HVA22 genes were classified into three groups (Fig. 2B). Large differences were found in amino acid length and structural domains of HVA22 family genes. HVA22 family proteins in group III had the most pronounced differences in length and structural domains. However, the amino acid lengths were essentially similar in each cluster in group III. The HVA22 family proteins in group III also possessed the Zf-met, RVT-3, and LRR_8 structural domains besides the TB2/DP structural domain unique to HVA22 proteins. In addition, the Solanaceous HVA22 family proteins with Zf-met (SpiHVA22m, SpHVA22j, SlydHVA22k, StHVA22v, CaHVA22i, SmHVA22o, NbHVA22c, NbHVA22u, SpiHVA22b, SlydHVA22b, StHVA22 h, and NbHVA22n) and RVT-3 (SpiHVA22f, SlydHVA22f, StHVA22g, SmHVA22f, CaHVA22g, and NbHVA22g) structural domains clustered in group III. On the contrary, NbHVA22r containing the LRR_8 structural domain was independently classified into a distinct cluster, which might be related to the fact that only NbHVA22r contained the LRR_8 structural domain among the numerous HVA22 family proteins in species belonging to Solanaceae. Moreover, the distribution position of the TB2/DP structural domain in the amino acid sequence of the HVA22 family in Solanaceae was at the N-terminal, except for SlydHVA22m, NbHVA22a, CaHVA22l, and SmHVA22d, which were at the C-terminal.
Figure 1: Phylogenetic tree analysis of the HVA22 gene.
(A) Phylogenetic trees were constructed for 60 HVA22 genes form Lycopersicon using the ML method with 1,000 bootstrap repetitions. A species abbreviation was provided prior to each HVA22 protein name: Sl, Solanum lycopersicum; Sp, Solaunm pennellii; At, Arabidopsis thaliana; Spi, Solaunm pimpinellifolium; and Slyd, Solaunm lycopersicoides. (B) Phylogenetic tree of the HVA22 family in Solanaceae. The phylogenetic tree was constructed using the ML method with 1,000 bootstrap repetitions. The different coloured HVA22 genes were derived from different Solanaceae species, and the conserved structural domains of the corresponding HVA22 genes are shown inside the evolutionary tree, with the TB2/DP1 structural domain in red, the Zf-met structural domain in blue, and the RVT-3 structural domain in pink.Figure 2: Genetic structure analysis.
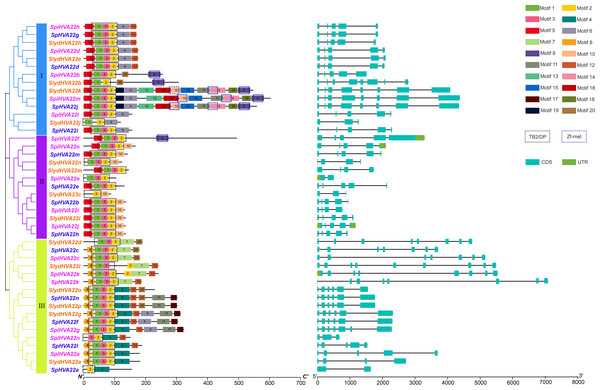
Phylogenetic relationships, Structure and conserved motifs of HVA22 genes in three types of wild tomatoes. The different coloured HVA22 genes were derived from different wild tomato species. Cyan boxes indicate exons, green boxes indicate UTR and black lines indicate introns. The numbers 1–20 and the different colored boxes indicate motifs.Conserved motif and gene structure analyses of the HVA22 gene family
The conserved motifs of the HVA22 family genes of three wild tomato species were predicted using the online tool MEME to understand the specific distribution of conserved motifs in HVA22 genes in the three species of wild tomatoes, and a total of 20 conserved motifs were identified (Fig. 2). Motifs 1, 2, and 3 formed the TB2/DP structural domain, which was distributed in HVA22 family genes in the three species of wild tomatoes. Additionally, the Zf-met structural domain consisting of Motifs 9, 10, and 14 was also distributed in some wild tomato HVA22 family of proteins. The HVA22 protein possessing the Zf-met structural domain was mainly concentrated on a small cluster in group I. The analysis indicated that the amino acid motif composition of the HVA22 family in the same group was approximately similar. HVA22 genes containing a Zf-met structural domain exhibited more motifs, with the exception of SpiHVA22f, SpiHVA22b, and SlydHVA22b.
The structural differences in exon–intron arrangement are an important source of gene family variation and plant diversity. Different structures lead to differences in gene expression and function (Xu et al., 2012). Our results showed that the HVA22 family genes in the three species of wild tomatoes were divided into three major groups by the phylogenetic tree, with a large degree of similarity in the exon–intron arrangement in most of the same clusters (Fig. 2). However, large differences existed in the arrangements in some of the clusters. In groups I and II, the number of exons was mostly 5, and the number of exons in a few individual HVA22 family genes was 2 (SpiHVA22e), 3 (SlydHVA22c, and SlydHVA22j), 4 (SlydHVA22n, SlydHVA22m, and SpiHVA22i), and 8 (SlydHVA22b, SlydHVA22k, SpiHVA22m, and SpHVA22j). The number of group III exons was highly variable, ranging from 2 to 9. Despite the large variation in the number of exons in group III, the HVA22 genes in each subgroup in group III exhibited similar gene structures.
HVA22 gene promoter analysis in the three species of wild tomatoes
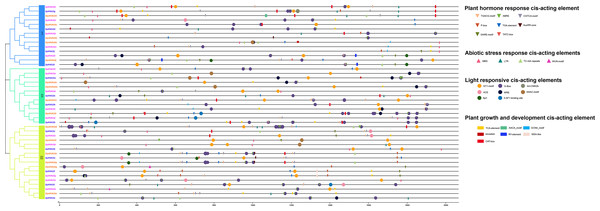
We performed a cis-acting element analysis of the 2,000-bp promoter sequence upstream of the HVA22 gene in three species of wild tomatoes (S. lycopersicoides, S. pennellii, and S. pimpinellifolium) (Fig. 3). The analysis showed that the cis-acting elements in the HVA22 gene were divided into four categories: light-responsive cis-acting elements, phytohormone-responsive cis-acting elements, biotic/abiotic stress cis-acting elements, and growth and development cis-acting elements. Cis-acting elements involved in light response, phytohormone response, and development were distributed in HVA22 family genes in all three species of wild tomatoes, while cis-acting elements involved in plant growth and abiotic stress were only present in the promoters of some HVA22 family genes. In the present study, five hormone response elements were identified to be involved in the transcriptional initiation of the HVA22 gene: abscisic acid response element (ABRE), salicylic acid response element (TCA-element and SARE), gibberellin response element (TATC-box, GARE-motif and P-box), auxin-responsive element (TGA-element, AuxRR-core, and TGA-box), and methyl jasmonate response element (TGACG-motif and CGTCA-motif). Four response plant biotic/abiotic stress elements were found, namely, cis-acting elements involved in defense and stress response (TC-rich repeats, and WUN-motif), cis-acting elements involved in low-temperature response (LTR), and MYB-binding sites involved in drought-inducing elements (MBS). Six species (CAT-box, AACA_motif, GCN4-motif, circadian, RY-element, and MSA-like) were involved in plant growth and developmental response elements. The largest number of cis-acting element types were involved in light response, with eight light response elements identified in the HVA22 family of genes; among these, except for SpHVA22j, the promoter sequences of the remaining HVA22 family genes were distributed with cis-acting elements associated with light response. Among the promoters of the HVA22 family genes in the three species of wild tomatoes, apart from SlydHVA22b, SlydHVA22c, SlydHVA22e, SlydHVA22f, SlydHVA22g, SlydHVA22j, SlydHVA22n, SlydHVA22o, SlydHVA22p, SpHVA22b, SpHVA22b, SpHVA22f, SpHVA22 h, SpHVA22i, SpHVA22n, SpiHVA22a, SpiHVA22g, and SpiHVA22l, all other HVA22 family genes contained cis-acting elements in response to low temperature or drought. Four cis-acting elements involved in biotic/abiotic stress were not as widely distributed in the promoters of HVA22 family genes in the three species of wild tomatoes as were light-responsive elements and phytohormone-responsive elements. This also suggested that some HVA22 family genes were involved in responding to abiotic stresses.
Figure 3: Distribution of CREs of HVA22 genes in three species form wild tomato.
Different colored squares indicate different branches of HVA22 family genes in the phylogenetic tree. Different CREs were indicated by different shapes, inverted triangles indicate hormone response elements, circles indicate light response elements, boxes indicate growth and development related elements, triangles indicate stress response related elements, and different elements were indicated by different colors.Gene localization to chromosomes and collinearity analysis of HVA22 in wild tomatoes
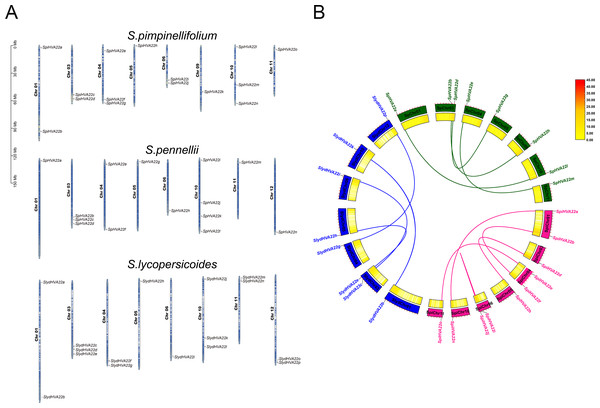
The HVA22 family genes in three species of wild tomatoes were mainly distributed on chromosomes Chr 01, Chr 03, Chr 04, Chr 05, Chr 06, Chr 09, Chr 10, Chr 11, and Chr 12 (Fig. 4A). Among these, the chromosomes distribution of the HVA22 family genes in S. pennellii and S. lycopersicoides was consistent; they were all distributed on chromosomes Chr 01, Chr 03, Chr 04, Chr 05, Chr 06, Chr 10, Chr 11, and Chr 12. The chromosomal distribution of HVA22 family genes in S. pimpinellifolium differed from that of S. pennellii and S. lycopersicoides. The HVA22 family gene in S. pimpinellifolium was distributed on chromosome Chr 09; however, no HAV22 gene family members were found on chromosome Chr 12. We performed intraspecific MCScanX analysis on three species of wild tomatoes to gain a clear understanding of the linear relationships between HVA22 family genes within species. The results showed five pairs of paralogous homologous genes within the HVA22 family in S. pimpinellifolium: SpiHVA22o/SpiHVA22e, SpiHVA22n/SpiHVA22a, SpiHVA22j/SpiHVA22i, SpiHVA22h/SpiHVA22d, and SpiHVA22f/SpiHVA22b. Four pairs of paralogous homologous genes existed in S. lycopersicoides and S. pennellii (SlydHVA22b/SlydHVA22k, SlydHVA22c/SlydHVA22i, SlydHVA22e/SlydHVA22h, and SlydHVA22g/SlydHVA22p in S. lycopersicoides; SpHVA22a/SpHVA22l, SpHVA22b/SpHVA22n, SpHVA22d/SpHVA22g, and SpHVA22e/SpHVA22m in S. pennellii) (Fig. 4B). We performed a Ka/Ks analysis of the identified paralogous homologs to enable a comprehensive understanding of the HVA22 family genes in three species of tomatoes. The final results showed that all 13 pairs of paralogous homologous genes in the HVA22 families had Ka/Ks greater than 1 (Table 2). This also suggested that the paralogous homologous gene pairs were subjected to stronger environmental stresses, and the gene evolution and protein function were stabilized.
Figure 4: Gene localization to chromosomes and collinearity analysis of HVA22 within species.
(A) Chromosome localization of three species form wild tomato HVA22 genes. (B) Collinearity analysis of HVA22 within species. Pink, green, and blue lines, which indicate the colinearity of the HVA22 gene between Solaunm pimpinellifolium and Solaunm pimpinellifolium, Solaunm pennellii and Solaunm pennellii, and Solaunm lycopersicoides and Solaunm lycopersicoides, respectively.| Species | Duplicated gene pairs | Ka | Ks | Ka/Ks | Selective pressure | Type |
|---|---|---|---|---|---|---|
| S.lycopersicoides | SlydHVA2b/SlydHVA22k | 0.80688 | 1.83965 | 0.43861 | Purify selection | Segmental |
| SlydHVA2c/SlydHVA22i | 0.43342 | 1.57164 | 0.27577 | Purify selection | Segmental | |
| SlydHVA22e/SlydHVA22h | 0.22811 | 3.48793 | 0.0654 | Purify selection | Segmental | |
| SlydHVA22g/SlydHVA22p | 0.18797 | 0.6627 | 0.28364 | Purify selection | Segmental | |
| S.pimpinellifolium | SpiHVA22a/SpiHVA22n | 0.34191 | 2.79239 | 0.12244 | Purify selection | Segmental |
| SpiHVA22b/SpiHVA22 f | 1.13033 | 3.13699 | 0.36032 | Purify selection | Segmental | |
| SpiHVA22d/SpiHVA22h | 0.24031 | 3.49311 | 0.0688 | Purify selection | Segmental | |
| SpiHVA22e/SpiHVA22o | 0.20661 | 0.78885 | 0.26192 | Purify selection | Segmental | |
| SpiHVA22i/SpiHVA22j | 0.17878 | 0.9293 | 0.19239 | Purify selection | Segmental | |
| S.pennellii | SpHVA22a/SpHVA22l | 0.46107 | 3.2625 | 0.14132 | Purify selection | Segmental |
| SpHVA22b/SpHVA22h | 0.18824 | 1.15914 | 0.16239 | Purify selection | Segmental | |
| SpHVA22d/SpHVA22g | 0.22977 | 3.50697 | 0.06552 | Purify selection | Segmental | |
| SpHVA22e/SpHVA22m | 0.18263 | 0.65296 | 0.27969 | Purify selection | Segmental |
Evolution and collinearity analysis of the HVA22 gene family in the three species of wild tomatoes
We performed an interspecific collinearity analysis of HVA22 family genes in eight Solanaceae species, tobacco, pepper, potato, eggplant, and four species of Lycopersicon (S. lycopersicum, S. lycopersicoides, S. pennellii and S. pimpinellifolium), based on their divergence times (Wu & Tanksley, 2010), to explore the homology of HVA22 family genes in Solanaceae plants. Our results showed a significant increase in HVA22 family homologous genes and a significant acceleration in the rate of evolution from pepper onward. The distribution of HVA22 homologs was similar on the chromosomes of the remaining Solanaceae members except for tobacco. The HVA22 genes on chromosomes Chr 03, Chr 04, Chr06, and Chr10 in Solanaceae (excluding tobacco) had high homology. A large similarity and homology were seen in the chromosomal distribution of HVA22 genes in four species of Lycopersicon (Fig. 5A). To gain further insight into the homology of HVA22 family genes in Solanaceae, we performed a collinearity analysis of HVA22 family genes in Solanaceae plants one by one; the analysis was also performed in the four species of Lycopersicon by the same method. The final results showed that most of the HVA22 family genes in the three species wild tomatoes were orthologous to each other and the cultivated tomato HVA22 family genes. Among these, the cultivated tomato HVA22 genes were found to have corresponding homologs in all three species of wild tomatoes, except SlHVA22o, which was not identified as a direct homolog in S. piminellifolium (Fig. 5B). The distribution of the HAV22 family genes on the chromosomes of the four tomato species was also highly similar. The HVA22 genes on chromosome Chr03 were found to be highly homologous to each other in the co-linearity analysis of Solanaceae (tomato, pepper, potato, and eggplant). However, the SlHVA22a and SlHVA22e genes located on chromosomes SlChr01 and SlChr04 were found to be orthologous only in eggplant (SmHVA22a and SmHVA22l), while no orthologous genes were found in pepper and potato. The orthologous genes of SlHVA22e, SlHVA22h, and SlHVA22n located on chromosomes SlChr04, SlChr05, and SlChr11 were present in potato (StHVA22v, StHVA22o, and StHVA22v) and eggplant (SmHVA22l, SmHVA22c, and SmHVA22l); their orthologous genes were not detected in pepper (Fig. 5C).
Figure 5: Homologous genes and evolutionary analysis of the HVA22 family.
(A) Co-lineage map for Solanaceae species, with species genomes arranged in evolutionary order and colored lines representing HVA22 genes with direct homologous relationships within each species. (B) Collinearity of HVA22 genes within Solaunm pimpinellifolium, Solaunm lycopersicoides, Solaunm pennellii, and Solanum lycopersicum, with the outer circle showing the chromosomes of each species, the inner circle showing the gene density, the two ends of the lines representing the direct homologous HVA22 genes, and the different colors indicate comparisons between different Lycopersicon. (C) Co-lineage map of HVA22 genes within Solanaceae (C. annuum, S. melongena, S. tuberosum, and S. lycopersicum), with the outer circle showing the chromosomes of each Solanaceae, the inner circle showing gene density, the ends of the lines representing direct homologous HVA22 genes, and the different colors indicate comparisons between different species.Protein–protein network analysis of HVA22 family genes in tomato
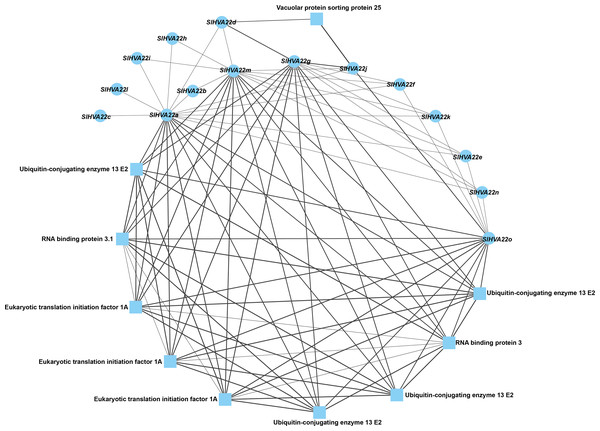
We constructed a protein–protein network expression profile of tomato HVA22 family genes using the STRING database to investigate the interactions between HVA22-like proteins and other proteins. Our results showed interactions between HVA22 family member proteins in tomatoes. Some HVA22 family proteins (SlHVA22o, SlHVA22g, SlHVA22m, and SlHVA22a) interacted with ubiquitin-conjugating enzyme (ubiquitin-conjugating enzyme 13 E2), RNA-binding protein (RNA-binding protein 3.1), and eukaryotic translation initiation factor (ETA). Among these, SlHVA22j and SlHVA22d interacted with the vesicle-sorting protein (vacuolar protein sorting protein 25). The remaining HVA22 family proteins (SlHVA22f, SlHVA22k, SlHVA22e, SlHVA22n, SlHVA22b, SlHVA22i, SlHVA22h, SlHVA22l, and SlHVA22c) interacted with proteins from their own family; interactions between them and with other proteins were not observed (Fig. 6).
Figure 6: Interaction network of the HVA22 family with other proteins.
Each node represents a protein, each connecting line represents the presence of an interaction, and the thickness of the line represents the value of the composite score, with circles representing HVA22 proteins and squares representing other proteins.Expression of tomato HVA22 gene in different tissues
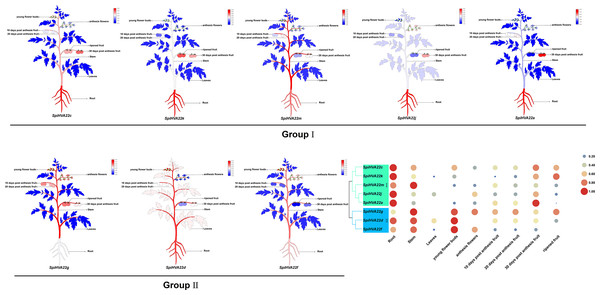
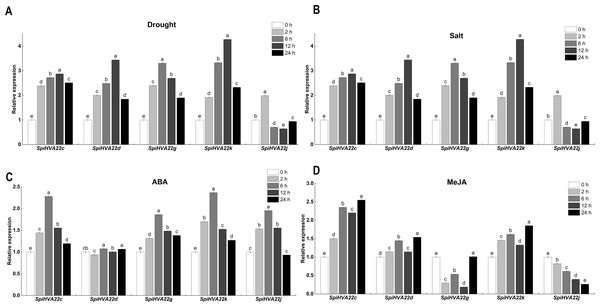
We used published RNA-seq data to map the gene expression heat map of the HVA22 gene in different tissues of S. pimpinellifolium to better understand the role of the HVA22 gene in the growth and development of wild tomatoes. The results showed that most of the HVA22 family members in wild tomatoes were expressed at a lower level in leaves and fruits. In particular, SpiHVA22c, SpiHVA22k, SpiHVA22a, and SpiHVA22g family member genes were highly expressed in ripening fruits. In addition, among the wild tomato HVA22 family genes, SpiHVA22d was slightly more expressed in leaves compared with other HVA22 family members. The roots showed the highest expression of the remaining HVA22 family members, with the exception of SpiHVA22g. The expression of HVA22 family genes in group I was generally low in flower buds. However, the HVA22 family member genes in group II showed higher expression levels in the flower buds. In addition, the HVA22 member genes in group II also showed higher expression levels in the stems. Overall, the expression of tomato HVA22 family genes was concentrated in roots, flowers, and developing fruits (Fig. 7). The high expression of some HVA22 family genes in roots also confirmed the possibility that HVA22 family genes were involved in abiotic stress processes in tomatoes. This result provided important clues for our study of the function of the HVA22 family gene in tomatoes. We treated S. pimpinellifolium seedlings with NaCl, PEG6000, and hormones (ABA and MeJA) to be more confident about how well the wild tomato HVA22 family genes could respond to salt, drought, and hormones. The expression of SpiHVA22c, SpiHVA22d, SpiHVA22g, SpiHVA22k, and SpiHVA22j genes in the leaves of the plants obtained from the treatments was analyzed using qRT-PCR. Our results showed that the expression of SpiHVA22c, SpiHVA22d, SpiHVA22g, and SpiHVA22k was significantly higher under NaCl and PEG6000 treatment than in the control group. The expression of the SpiHVA22j gene was the highest after 2-h treatment and showed lower expression than that in the control group as the treatment time was extended. At the same time, the expression of SpiHVA22k, SpiHVA22g, and SpiHVA22c genes under ABA and MeJA treatment was similar to that under PEG6000 and NaCl treatment. Intriguingly, the SpiHVA22g and SpiHVA22j gene showed negative regulation under MeJA treatment and positive regulation under ABA treatment (Fig. 8). Apart from this, no significant change was found in the expression of the SpiHVA22d gene under ABA treatment, indicating that the SpiHVA22d gene was not regulated by the hormone ABA.
Figure 7: Heat map of tissue-specific expression of HVA22 genes in Solaunm pimpinellifolium.
Root, stem, leaves, young flower buds, anthesis flowers, 10 days post anthesis fruit, 20 days post anthesis fruit, 30 days post anthesis fruit, ripened fruit.Figure 8: QRT-PCR validation of the Solaunm pimpinellifolium HVA22 genes under abiotic stress and hormone induction.
The standard deviations are shown with error bars.Discussion
With advances in gene sequencing technology, next-generation sequencing has improved the accuracy of the genome, thereby avoiding annotation errors in individual gene sequences by whole-gene sequencing. This has also facilitated genetic improvement and directed breeding in tomatoes (Rothan, Diouf & Causse, 2019). The HVA22 gene is commonly found in eukaryotes (Lu, 2013). It is expressed in plants in different tissues such as seeds, stems, and roots and is induced under several environmental stress conditions (e.g., cold, salt, and drought) mediated by ABA (Shen et al., 2001). The HVA22 gene family has been reported in A. thaliana (Chen et al., 2002), C. clementina (Gomes Ferreira et al., 2019), and cultivated tomatoes (Wai et al., 2022), but has not been reported in wild tomatoes (S. lycopersicoides, S. pennellii, and S. pimpinellifolium). The results of the phylogenetic tree showed that the three wild tomato HVA22 family genes were divided into two clades, which were further divided into four groups (Fig. 1A). This result was also consistent with the classification of tomato HVA22 family genes (Wai et al., 2022). Our results showed that the three wild tomato HVA22 family genes in each group were well assigned to the known HVA22 groups of A. thaliana and cultivated tomatoes. This further demonstrated the highly conserved nature of the HVA22 family of genes in Lycopersicon crops. We constructed a phylogenetic tree using HVA22 family genes from seven species of Solanaceae to characterize the HVA22 family genes more comprehensively. The results showed that the HVA22 family genes in seven species of Solanaceae were divided into three major branches (Fig. 1B). Although the number of HVA22 family members varies significantly among Solanaceae, they all carry the TB2/DP1 structural domain specific to HVA22-like. In addition, some of the HVA22 proteins have other structural domains besides TB2/DP1, and HVA22 proteins with other structural domains are clustered together in a phylogenetic tree. This also illustrates the reliability of the phylogenetic structure and the high homology of the HVA22 gene in Solanaceae. This homology may result from the TB2/DP1 structural domain specific to the HVA22 protein. In addition, the analysis of the physicochemical properties of HVA22 family proteins in the three species of wild tomatoes found that the average number of amino acids was 231 (S. pimpinellifolium), 208 (S. pennellii), and 217 (S. lycopersicoides) (Table 1). These were close to the 230 amino acids in tomato HVA22 (Wai et al., 2022), but showed a large difference from the 130 amino acids of barley (Shen et al., 2001) and 151 amino acids of C. clementina (Gomes Ferreira et al., 2019), which might be related to the large evolutionary distance between different species. Interestingly, we found that the chromosomal distribution positions of the HVA22 family genes in the three species of wild tomatoes showed striking similarities (Fig. 4A). At the same time, these HVA22 proteins with similar distributions on the chromosome showed nearly identical physicochemical properties (e.g., theoretical pI, molecular weight, aliphatic index, and grand average of hydropathicity) (Table 1). This also fully corroborated the conserved nature of the HVA22 family genes in tomatoes.
The prediction of subcellular localization of HVA22 helps us understand the function of this gene. Our findings regarding the subcellular localization of HVA22 genes in the three species of wild tomatoes revealed that a majority of the HVA22 genes were predicted to be localized in the endoplasmic reticulum, chloroplast, cytoplasm, and nucleus (Table 1). Evidence showed that after heterologous expression, tomato SlHVA22a, SlHVA22f, and SlHVA22n proteins in rice protoplasts were observed to be localized to the endoplasmic reticulum (Wai et al., 2022). In addition, the rice HVA22 family gene OsHLP1 promoted the mechanism of disease resistance by compromising endoplasmic reticulum homeostasis when plants were infected by pathogens (Meng et al., 2022). In yeast, the HVA22 gene homolog Yop1p played an important role in transporting material between the endoplasmic reticulum and the Golgi apparatus (De Antoni et al., 2002). These findings consistently showed that HVA22 played an important role as an endoplasmic reticulum localization protein in the life activities of eukaryotic cells. The HVA22 protein in the three species of wild tomatoes also seemed to exhibit similar characteristics.
The diversity of exon–intron structures is one of the key factors in the evolution of gene families and underpins the structure of phylogenetic trees (Shiu & Bleecker, 2003; Wang et al., 2014). The structural analysis of the HVA22 family of genes in the three species of wild tomatoes showed that the number of exons was similar in groups I and II; however, a large difference was found in the number of exons in group III (Fig. 2). This also implied that the HVA22 gene in the three species of wild tomatoes could be transcriptionally diversified through processes such as selective splicing to regulate a more complex and broad range of functions. A total of 20 motifs were identified in the structural analysis of the HVA22 family genes in wild tomatoes, among which Motifs 1, 2, and 3 are important motifs that made up the TB2/DP1 structural domain of the HVA22 gene. A membrane protein TB2-like1 exists in animals and belongs to the same TB2/DP1/HVA22 family of proteins. This protein may play an important role in the cell membrane transport of retinal ganglion cells (Sato et al., 2005). The Yop1p gene in yeast (Saccharomyces cerevisiae) is homologous to the barley HVA22 gene, and the proteins it encodes have the TB2/DP2 structural domain (Brands & Ho, 2002). Previous studies showed that yeast Yop1p was an integral membrane protein with a hydrophilic region at its N-terminal end. This region bound specifically to the yeast Yip1p protein, and Yop1p interacted with Yip1p to mediate the biological process of intracellular membrane transport (Calero, Whittaker & Collins, 2001). Also, this membrane protein played a critical physiological role in barley, yeast, and humans (Sato et al., 2005; Voeltz et al., 2006). In a study of the TB2/DP1 structural domain of the HVA22 protein of barley dextrin, the TM1, TM2, and TM3 segments of the HVA22 gene were separately deleted using truncating mutations, so that the separately deleted HVA22 genes would all carry their green fluorescent protein (GFP) tags, which were transferred to barley dextrin cells for cellular sublocalization observation. It was found that the sublocalization of the deletion of TM2 presented a results significant differences compared with HVA22:: GFP (Guo & David Ho, 2008). It further indicated that HVA22 had the properties of a membrane protein. It also showed that the barley HVA22 protein had a transmembrane region in the TB2/DP1 structural domain, and the presence of this transmembrane region provided a theoretical basis for the localization of the HVA22 protein to the organelle membrane. This result was similar to our predicted subcellular localization of the HVA22 gene in the three species of wild tomatoes. This also suggested that the HVA22 gene with the TB2/DP1 structural domain might play an important role in intracellular vesicle transport (Brands & Ho, 2002). However, no relevant studies have been reported on the involvement of the HVA22 gene in intracellular vesicle transport in tomatoes. Moreover, the Zf-met structural domain was found in the amino acid sequence of HVA22 in the three species of wild tomatoes. The Zf-met domain is another zinc-finger domain containing the CxxC(12)Hx(6)H motif, which is associated with RNA binding (Yadav, Fernández-Baca & Cannon, 2020). This is similar to the structure identified in the tomato HVA22 protein. This further indicates that HVA22 family members are conserved in Lycopersicon.
Cis-acting elements are noncoding DNA sequences present in the promoter region of a gene. The distribution of different types of cis-acting elements in the promoter region may determine gene regulation and functional roles (Hernandez-Garcia & Finer, 2014). In this study, we characterized a 2,000-bp promoter sequence upstream of the HVA22 family genes in the three species of wild tomatoes (Fig. 3). The cis-acting elements of HVA22 family genes in the three species of wild tomatoes were divided into four main categories: cis-acting elements involved in light response, cis-acting elements involved in phytohormone response, cis-acting elements involved in biotic/abiotic stress, and cis-acting elements involved in growth and development. Common cis-acting elements associated with light response were ACE, MRE, G-box, GT1-Mofit, Sp1, 4cl-CMA2b, 3-AF1-binding site, and AAAC-Motif. These light-responsive cis-acting elements played an important regulatory role in stress response and growth and development of plants (Kaur et al., 2017). The cis-acting elements responding to phytohormones in the promoter of the HVA22 gene family of three species of wild tomatoes were more widely distributed, including gibberellin, ABA, methyl jasmonate, ethylene, and salicylic acid. This further suggested that the HVA22 gene might be involved in the life activities of tomatoes through a hormone-regulated network, which was consistent with the findings in tomatoes and C. clementina (Gomes Ferreira et al., 2019; Wai et al., 2022). In addition, cis-acting elements involved in multiple stresses were predicted in the HVA22 promoter in the three species of wild tomatoes, such as MBS (drought-inducible), LTR (low temperature responsive), and TC-rich repeats (defense and stress responsive). The HVA22 gene response to low temperature and drought stress has been reported in A. thaliana (Chen et al., 2002), barley (Shen et al., 2001), and rice (Zhao et al., 2021). Six cis-acting elements associated with growth and development were identified in the promoters of HVA22 family genes in the three species of wild tomatoes, including RY-element (involved in seed-specific regulatory element), MSA-like (involved in cell cycle regulatory element), GAT-box (involved in meristematic tissue expression element), circadian (involved in circadian control regulatory element), AACA_motif (involved in the endosperm-specific negative expression), and GCN4_motif (involved in endosperm expression element) (Zhang et al., 2013). The HVA22 gene promoter contained cis-acting elements associated with plant development, particularly involved in seed-specific regulatory elements, which might be relevant to the function of the HVA22 gene in seed maturation and dormancy in barley (Guo & David Ho, 2008). However, further experimental evidence is needed by cloning the upstream promoter of the HVA22 gene in the three species of wild tomatoes to obtain the corresponding experimental evidence and provide direction for the next study of the HVA22 gene function.
Gene duplication provides the material basis for plant evolution and the generation of new functions (Huang et al., 2022). It occurs in several different modes, such as whole-genome duplication, single-gene duplication, and segmental duplication (De Bodt, Maere & Van de Peer, 2005; Paterson et al., 2010). The chromosomal localization and collinearity analysis of the HVA22 gene in the three species of wild tomatoes showed spacer regions in the physical location of the HVA22-encoding gene on the chromosome in the three species of wild tomatoes (Fig. 4A). Thus, it was tentatively determined that the HVA22 gene was amplified in the three species of wild tomatoes mainly by large-scale segmental duplication or whole-genome duplication of the gene family. Subsequently, the intraspecific collinearity analysis of the HVA22 family genes of the three species of wild tomato showed that the HVA22 genes were amplified in the three species of wild tomatoes in an all-segmental duplication manner, which was consistent with our previous speculation on the distribution of HVA22 genes on chromosomes (Fig. 4B). At the same time, the same results were obtained in cultivated tomatoes (Wai et al., 2022). The ratio of nonsynonymous to synonymous substitutions reflects, to some extent, the selective pressure of gene evolution. Ka/Ks >1 represents positive selection for accelerated evolution, and Ka/Ks <1 represents the presence of purifying selection for gene duplication (Wang et al., 2010). The HVA22 paralogous homologous gene pairs in all three wild tomatoes had Ka/Ks <1, which further suggests that these homologous gene pairs underwent more intense environmental selection pressure and exhibited functional homogeneity during evolution. Genome-wide duplication events (WGD) have long been recognized as an important evolutionary force in species formation, adaptation to the environment, and shaping of species diversity (Wood et al., 2009; Soltis & Soltis, 2016). Dicotyledons experienced γ events in gene duplication; however, Solanaceae members experienced another WGD event (T event) about 65 million years ago (Knapp, 2012; Wu, Han & Jiao, 2020). HVA22 genes were added or lost in Solanaceae driven by genome-wide duplication events. As the Solanaceae diverged during evolution, the HVA22 family gene members showed a decreasing trend and then an increasing trend in Solanaceae (Fig. 5A). This might be related to the contraction and expansion of family genes in Solanaceae when they experienced the most recent T event. The HVA22 gene on SlChr 03 in tomatoes is the most conserved member of the Solanaceae family (Fig. 5B). For HVA22 family genes present only in tomatoes but not in other members of Solanaceae, this might be due to gene deletions caused by genome-wide duplication events (Fig. 5C). The result that HVA22 family genes were highly conserved in the four species of Lycopersicon also indicated the importance of HVA22 genes in the life activities of tomatoes.
The ubiquitin–proteasome system (UPS) regulates various biological functions in plants, such as hormonal responses (Chen et al., 2018; He et al., 2018), abiotic stress responses (Kim, Jang & Seo, 2016; Shu & Yang, 2017), plant growth and development (Cho et al., 2011; Koops et al., 2011), circadian rhythms (Gil et al., 2017), and plant immune responses (Lin et al., 2008; Luo et al., 2010). The predicted results of tomato HVA22 protein interactions indicated that the SlHVA22 protein was involved in plant signaling and the regulation of plant growth and development (Fig. 6). Among these, the tomato HVA22 protein was co-expressed with ubiquitin-binding enzymes, suggesting that the tomato HVA22 gene might be involved in the ubiquitination and regulation of tomato growth and development and abiotic stresses. In addition, the vesicle sorting protein (VPS) (Xiang, Etxeberria & Ende, 2013) plays an important role in the protein sorting pathway as an important protein for the formation of endosomal sorting complex protein (ESCRT) (Gao et al., 2017). Previous studies showed that the growth hormone transporter proteins PIN1, PIN2, and AUX1 were the cargo proteins of ESCRT. To some extent, this suggested that VPS played a role in hormone signaling (Spitzer et al., 2009). The results of the co-expression of HVA22 protein with VPS suggested that the HVA22 protein might be a cargo membrane protein of ESCRT and played an important role in intracellular vesicle transport. RNA-binding proteins interacted with RNA through the RNA-binding domain to regulate RNA metabolism and function. Conversely, RNA could bind to RAN-binding proteins and affect their lifespan and function (Hentze et al., 2018). The co-expression of RNA-binding proteins with the tomato HVA22 protein might indicate an important contribution of RNA-binding proteins in maintaining the function and longevity of the HVA22 gene. The results of co-expression of the tomato HVA22 gene with eukaryotic translation initiation factor 1A also further confirmed the conclusion that the HVA22 gene was present only in eukaryotes. Recently, that the AtGCN2 activation of eukaryotic translation initiation factor 2 phosphorylation was shown to be another key component in response to endoplasmic reticulum stress in A. thaliana, and it played an important role in the signaling process of the unfolded protein response (Afrin et al., 2020; Howell, 2021; Liu, Afrin & Pajerowska-Mukhtar, 2019).
The HVA22 gene was differentially expressed in different tissues of S. pimpinellifolium, while the expression of individual HVA22 genes showed an increasing trend during S. pimpinellifolium fruit development (Fig. 7). The root system, as the main tissue directly sensing drought and salt ions, has a high sensitivity to drought and salt ions. The high expression of the HVA22 gene in roots indicated that the HVA22 gene played an important role in plant root development and resistance to abiotic stress response. The expression of HVA22 family genes showed an increasing trend as the fruit grew during fruit development. The same result was shown in the HVA22 family genes in cultivated tomatoes (Wai et al., 2022). This might be related to the involvement of HVA22 family genes in tomato fruit growth and development. We treated S. pimpinellifolium seedlings with NaCl, PEG6000, and hormones, thereby performing qRT-PCR analysis of the HVA22 family genes in S. pimpinellifolium (Fig. 8). The results showed that SpiHVA22d, SpiHVA22g, SpiHVA22k, and SpiHVA22c genes responded positively to salt and drought stresses. This was also similar to the characterization of HVA22s described in barley, A. thaliana, C. clementina, and cultivated tomatoes (Wai et al., 2022). In addition, the responses of SpiHVA22k, SpiHVA22c, and SpiHVA22d genes to ABA and MeJA also suggested that SpiHVA22k, SpiHVA22c, and SpiHVA22d genes played important roles in the ABA and MeJA pathways. Interestingly, the SpiHVA22j gene in the S. pimpinellifolium HVA22 family of genes showed negative regulation under MeJA treatment. This might be related to the fact that the SpiHVA22j gene was not involved in the plant jasmonic acid pathway. Previous studies in barley also pointed to the role of the HVA22 gene as an early ABA-inducible gene (Shen et al., 2001). Our qRT-PCR results further concluded that the S. pimpinellifolium HVA22 gene might be involved in the plant ABA pathway, which in turn responded to plant regulation of abiotic stresses.
Conclusions
In the present study, we systematically identified HVA22 family genes in tomatoes. We used a bioinformatics approach to describe the physicochemical properties, gene structure, cis-acting elements, and protein interactions of different HVA22 genes. The expansion and contraction of HVA22 family genes during the evolution of Solanaceae species were also discussed. The expression profile data of different tissues of HVA22 family genes in tomatoes showed that the expression of HVA22 family genes was mainly concentrated in roots, flowers, and developing fruit. We validated five genes in the tomato HVA22 family using qRT-PCR. Four of these genes were involved in ABA- and MeJA-mediated regulatory pathways, and played important roles in tomato resistance to abiotic stresses (salt and drought). These results laid the foundation for further investigation of the function of HVA22 family genes in tomatoes.