Enhanced biomass and thermotolerance of Arabidopsis by SiERECTA isolated from Setaria italica L
- Published
- Accepted
- Received
- Academic Editor
- Brenda Oppert
- Subject Areas
- Agricultural Science, Bioinformatics, Genomics, Molecular Biology, Plant Science
- Keywords
- SiERECTA family, Expression characteristics, Thermotolerance, Biomass, Foxtail millet
- Copyright
- © 2022 Zheng et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. Enhanced biomass and thermotolerance of Arabidopsis by SiERECTA isolated from Setaria italica L. PeerJ 10:e14452 https://doi.org/10.7717/peerj.14452
Abstract
Foxtail millet is commonly used as a food and forage grass. ERECTA (ER) is a receptor-like kinase that can improve plant biomass and stress resistance. The sorghum SbER10_X1 gene was used as a probe to identify ER family genes on the Setaria italica genomes (SiERs), and determine the characteristics of the SiERs family. Herein, the structural features, expression patterns, and thermotolerance of SiERs function were identified by bioinformatics analysis, real-time PCR and transgenesis estimation. Results showed that SiERs had four members: two members were located on chromosome 1 with a total of six copies (SiER1_X1, SiER1_X2, SiER1_X3, SiER1_X4, SiER1_X5, and SiER1_X6), and two were on chromosome 4, namely, SiER4 (SiER4_X1 and SiER4_X2) and SiERL1. Among them, SiER1_X4 and SiER4_X1 were expressed highest in above-ground organs of foxtail millet, and actively responded to treatments with abscisic acid, brassinolide, gibberellin, and indole acetic acid. After overexpression of SiER1_X4 and SiER4_X1 in Arabidopsis, the plant height and biomass of the transgenic Arabidopsis significantly increased. Following high-temperature treatment, transgenic seedlings survived better compared to wild type. Transgenic lines showed higher SOD and POD activities, and expression level of AtHSF1 and AtBl1 genes significantly increased. These results indicated that SiER1_X4 and SiER4_X1 played important regulatory roles in plant growth and thermotolerance. The two genes provide potential targets for conventional breeding or biotechnological intervention to improve the biomass of forage grass and thermotolerance of field crops.
Introduction
Foxtail millet is an annual C4 crop that can be used as a food and forage grass (Singh, Muthamilarasan & Prasad, 2021). In arid and semi-arid regions, foxtail millet shows strong tolerance to various abiotic stresses such as drought, salinity, high temperature (Aidoo et al., 2016). However, the natural conditions experienced by field crops are increasingly complex. In addition, the human demand for food and energy is intensified by the global population growth and per capita income increase. To cope with this severe challenge, crop varieties need to be improved by traditional breeding, functional gene screening, genome editing, or other technologies. The foxtail millet genome contains many excellent genes related to drought resistance, high yield, and high light efficiency (Zhang et al., 2012). The rational utilization of foxtail millet functional genes is an important strategy to ensure future food security.
ERECTA (ER) belongs to the receptor-like kinases (RLKs), which are involved in the regulation of plant photosynthesis and transpiration efficiency, thereby increasing biomass and plant resistance (Masle, Gilmore & Farquhar, 2005; Van Zanten et al., 2009). Overexpression of SbER2-1 in maize conferred increased drought tolerance, expecially in regard to improved water-use efficiency (Li et al., 2019). When the Arabidopsis AtER gene was overexpressed in tomato and rice, the biomass of transgenic lines was increased and heat tolerance was enhanced (Shen et al., 2015). Further studies have shown that the fusion gene of chitin elicitor receptor kinase 1 and ER (CERK1n-ER) can induce the production of chitooligosaccharides and improve heat tolerance of Arabidopsis (Chen et al., 2020). Overexpression of poplar PdER gene in Arabidopsis resulted in reduced stomatal density, thereby influencing transpiration, water-use efficiency and drought tolerance (Li et al., 2021). Interference of MAPK cascade reaction through the interaction of ER with the BAK1 gene increased the resistance of Arabidopsis to the necrotrophic fungus, Plectosphaerella cucumerina BMM (PcBMM) (Jorda et al., 2016; Mei et al., 2021). These results demonstrated that the ER family has broad prospects for application in regulating plant development and stress tolerance.
In the current research, the characteristics of SiER family members (SiER s) in the foxtail millet were analyzed. SiER1_X4, and SiER4_X1 genes were isolated, and their biomass and thermotolerance of transgenic Arabidopsis were evaluated. The findings provide the functional genes for potential use in improvement of production and stress resistance in gramineous crops.
Materials and Methods
Phylogenetic analysis of the SiERs family in Setaria italica
Two SiER gene tags from foxtail millet (Seita.4G086700.1 and Seita.1G338900.1) were obtained with the sorghum SbER10_X1 gene (XM_002437978.2) as a reference sequence after BLAST in the Phytozome v12.1 database. Four families of SiER members were obtained by searching the NCBI database with the two SiER tags to predict the complete CDS and chromosome-position information. The exon distribution (GSDS 2.0), cis-regulatory elements of promoters (Plant CARE), subcellular localization characteristics (Plant-mPLoc) and motif structure (MEME) of SiERs family were predicted. Moreover, the conserved functional domains (PROSITE and SMART databases), amino acid size, molecular weight, and isoelectric point (ProtParam) of SiERs proteins were analyzed. Table S1 lists all databases and their URLs available at the journal’s website.
Based on the functional domains of SiERs, the amino acid sequences of the published ER family in monocot and dicot plants with similarity above 80% were downloaded from NCBI database (Annex S2, SiER1_X4 gene was listed in Annex S4), to produce a SiERs phylogenetic tree by MEGA5.0 software with a threshold of 1,000 replications for bootstrap, according to the neighbor-joining method (Tamura et al., 2013).
Genes isolation and subcellular localization of SiER1_X4 and SiER4_X1
Due to the abundant transcription of SiERs in the pedicel tissue of the Dungu variety at the heading stage, total RNA from pedicel was extracted with RNAprep Pure Kit (DP432; Beijing, China), and cDNA was synthesized with a PrimeScript First-Strand cDNA Synthesis Kit (6110A; Shiga, Japan). Taking the pedicel cDNA as material, specific primers (SiER1_X4-F2/SiER1_X4-R2 and SiER4_X1-F3/SiER4_X1-R3 in Annex S3) were designed to separate SiER1_X4 and SiER4_X1 fragment, respectively. The PCR reaction (50 µL) was as follows: 25 µL of 2 × PCR buffer, 10 µL of dNTP (2 mM), 1.5 µL of Primer-F (10 µM), 1.5 µL of Primer-R (10 µM), 1 µL of KOD FX (1.0 U/mL, KFX-101; Toyobo, China), 5 µL of cDNA as template, 6 µL of ddH2O. The PCR procedure was as follows: 94 °C for 2 min, 40 cycles (98 °C for 10 s, 65 °C for renaturation in both SiER1_X4 and SiER4_X1 gene, lasting for 30 s, 68 °C for 4 min for extension), and 68 °C for 10 min.
The code fragment of SiER1_X4 and SiER4_X1 (without the stop codon) was separated through SiER1_X4-gfpF1/SiER1_X4-gfpR1 and SiER4_X1-gfpF1 /SiER4_X1-gfpR1 primers (Annex S3). The same PCR procedure and reaction system as above were used, except for the 62 °C and 61 °C for renaturation in SiER1_X4-gfp and SiER4_X1-gfp gene, respectively. The fusion-protein was generated as below: PCR products of SiER1_X4 and SiER4_X1 were differentially integrated into the N terminal of green fluorescent protein vector (pJIT16318-GFP), which included CaMV35S promoter. pJIT16318-SiER1_X4 and pJIT16318-SiER4_X1 were transferred into wheat mesophyll protoplasts (isolation from 10-day-old wheat seedlings) via the PEG4000-mediated method (Cui et al., 2019). The transformed cells were incubated at 22 °C in darkness for 18–20 h, and then observed and photographed under a confocal laser scanning microscope (LSM700; CarlZeiss, Germany).
Thermotolerance identification of transgenic Arabidopsis
SiER1_X4 and SiER4_X1 segments (without the stop codon for fusion-protein development) were separated by primers of SiER1_X4-1302F1/SiER1_X4-1302R1 and SiER4_X1-1302F1/ SiER4_X1-1302R1, respectively (Annex S3). The same PCR procedure and reaction system were used as above, except for 64 °C and 62 °C for renaturation in SiER1_X4-1302 and SiER4_X1-1302 gene, respectively. The PCR products of SiER1_X4 and SiER4_X1 were inserted into pCAMBIA1302 vector (CaMV35S promoter) to obtain the fusion vectors of pCAMBIA1302-SbER1_X4 and pCAMBIA1302-SbER4_X1, respectively. Using a Agrobacterium tumefaciens-mediated transformation system (Bradley et al., 1997), the targeted fusion vectors were transformed into Arabidopsis (Columbia ecotype). The offspring seeds were screened with antibiotics to obtain homozygous transgenic SiER1_X4 and SiER4_X1 lines. The test steps were described by Chen et al. (2020).
The stable transgenic lines overexpressing the target genes were cultivated on MS medium for 3 days (without antibiotics), and then moved into a light incubator for 7 days. Seedlings of the similar size were transplanted into pots (6.8 × 6.8 cm) with nine plants in each pot and ten pots per transgenic line. After 10 days of continuous growth in a greenhouse (26 °C growth with an 8 h/16 h dark/light, photon flux density of 525 µmol m−2 s−1), five pots per transgenic line were treated in a light incubator at 42 °C for 48 h and 60 h, and the five remaining pots were cultivated at 26 °C for later biomass investigation (control).
After the high-temperature treatment for 60 h, leaves of transgenic and wild-type (WT) lines were collected, and some samples were used to determine SOD and POD activity, as described by Zheng et al. (2020), the remainder was quickly frozen in liquid nitrogen, and stored at −80 °C for qRT-PCR. The remaining treated lines were transferred to the greenhouse to control conditions (26 °C) for 11 days, to observe the recovery growth of Arabidopsis plants, the number of plants with green leaves was counted to assess the survival rate of transgenic and WT lines after high-temperature treatment. Four individual plants from each line were served as biological replicates.
Plant material and hormone-induction treatment
Five foxtail millet germplasm varieties (Dabaigu, Dungu, Jingu21, Yugu1, and Kuanjiu) were pre-germinated for 4 days. Seedlings with the similar germination were transplanted to pots (35 × 35 cm) with forty plants in each pot, and the flower pots were placed in a light incubator for growth (humidity 60%; temperature 23 °C/20 °C day/night; 16 h/8 h light/dark; light intensity 525 µmol m−2 s−1). After 6 days, mixture of the stems and leaves from a single plant for each variety was collected. After culturing the remaining plants for 15 days, seedlings were removed along with the roots, rinsed off the soil, and placed briefly on filter paper to dry, and then cultured in hormone solution or deionized water (control). The concentrations of the hormone solution were as follows: abscisic acid (ABA) 100 µM, brassinolides (BRs) 0.75 µM, gibberellin (GA3) 30 mM and indole acetic acid (IAA) 10 µM (Zheng & Hu, 2016). Samples (mixture of stems and leaves) were separately collected for qRT-PCR. The treatment periods were 0, 1, 2, 4, 6, 12, 24, 48, and 60 h.
In May 2021, the Dungu variety was planted in the experimental field, and embryo and coleoptile were collected at the germination stage. Roots, stems, flag leaves, flag leaf sheaths, pedicels, and inflorescence samples were collected at the flowering stage. Seeds were collected at the maturity stage. All samples were quickly frozen in liquid nitrogen after collection and stored at −80 °C for later detection of SiERs expression patterns in diverse organs. Three individual plants were selected as biological replicates for each sample collection.
qRT-PCR analysis
Nine cDNA sequences of the SiERs family were aligned to design specific primers for SiER1_X4 and SiER4_X1 qRT-PCR expression. The high-temperature related gene, AtHSFA1a, and superoxide suppressor gene, AtBl1, were used to determine the molecular-response mechanism of SiER1_X4 and SiER4_X1 in transgenic Arabidopsis plants after high-temperature stress (Yoshida et al., 2011; Ishikawa, Uchimiya & Kawai-Yamada, 2013). The primers of SiER1_X4 (SiER1_X4–qRTF2/SiER1_X4–qRTR2), SiER4_X1 (SiER4_X1–qRTF1/ SiER4_X1–qRTR1), AtHSFA1a (AtHSFA1a-qRTF2/AtHSFA1a-qRTR2), and AtBl1 (AtBI1-qRTF1/AtBI1-qRTR1), as well as the reference genes (SiActin -qRT F1/SiActin -qRT R1 and AtActin-qRTF5/ AtActin-qRTR5), are listed in Annex S3. The target-gene-expression level was detected by qRT-PCR analysis with the ABI Prism 7500 system (Applied Biosystems, Waltham, MA, USA). Three technical replicates and three biological replicates were conducted for all experiments, and the 2−ΔΔCt method was used for quantification (Liu et al., 2013).
Data processing and statistical analysis
qRT-PCR data was analyzed in accordance with the procedure of Zheng & Hu (2016). Error analysis was conducted with SPSS Statistics Software version 18.0 (SPSS18.0, IBM, USA) based on the biological replicates of three individual plants. The related indicators of agronomic traits were also statistically analyzed using SPSS18.0 software. The data of all graphs was represented as the mean ± standard error. The graphics were analyzed and produced with OriginPro 2018C SR1 and Excel 2010 software.
Results
Characteristics and phylogenetic relationship of the SiERs of foxtail millet
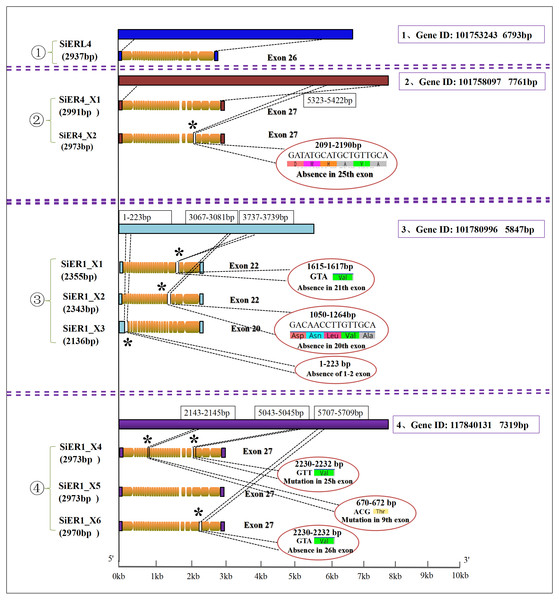
Four genes were found in the SiERs family of foxtail millet. Among them, SiERL4 (gene ID: LOC101753243) and SiER4 (gene ID: LOC10175555 8097) were distributed on chromosome 4, and SiER1 was located on chromosome 1 with two genes (gene ID: LOC101780996 and gene ID: LOC117840131) (Table 1). Further analysis (Fig. 1) showed that 1 copy and 26 exons were found in SiERL4 sequences (XM_004964364.4), and 2 copies and 27 exons in SiER4 sequences. In exon 25, 6 amino acids fewer were encoded in SiER4_X2 (XM_004964885.3) than in SiER4_X1 (XM_004964884.4). Three copies were found in the LOC101780996 gene of SiER1, exons 1 and 2 were lacking in SiER1_X3 (XM_014804622.2), 22 exons were found in the other two copies, 5 amino acids were lacking in exon 20 of SiER1_X2 (XM_014804623.2), and valine was lacking in exon 21 of SiER1_X1 (XM_014804625.2). Three copies were found in the LOC117840131 gene of SiER1, each of which contained 27 exons, compared with SiER1_X5 (XM_034720593.1), and mutations were found in exon 9 and 25 of SiER1_X4 (Annex S4), and one amino acid was lost in exon 26 of SiER1_X6 (XM_034720600.1). The amino acid structure prediction indicated that the proteins of the SiER4 family were larger, and the LOC101780996 of SiER1 was smaller. The nine copies of four genes in the SiERs family were all predicted to be transmembrane proteins, a typical feature of ER family proteins. In total 15 LRR tandem regions were detected in the SiERL4 protein, 13 LRR regions in SiER4, 9 LRR regions in LOC101780996 (SiER1), and 14 LRR regions in LOC117840131 (SiER1) (Annex S5).
| Name | Nucletide | Protein | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID(NCBI)DNA | Genelength(bp) | Locus(NCBI)mRNA | NumberofExons | Proteinaccession(NCBI) | pI | SubcellularLocation | Length ofProtein(AA) | MW(KDa) | Location | |
| SiER1_X1 | LOC101780996 | 5847 | XM_014804625.2 | 22 | XP_014660111.1 | 5.77 | Cell membrane | 786 | 86 | Chr. I |
| SiER1_X2 | XM_014804623.2 | 22 | XP_014660109.1 | 5.83 | Cell membrane | 782 | 85 | Chr. I | ||
| SiER1_X3 | XM_014804622.2 | 20 | XP_014660108.1 | 5.90 | Cell membrane | 713 | 78 | Chr. I | ||
| SiER1_X4 | LOC117840131 | 7319 | Annex 4 (OP492075) | 27 | Annex 4 listing | 5.45 | Cell membrane | 991 | 108 | Chr.I |
| SiER1_X5 | XM_034720593.1 | 27 | XP_034576484.1 | 5.50 | Cell membrane | 991 | 108 | Chr. I | ||
| SiER1_X6 | XM_034720600.1 | 27 | XP_034576491.1 | 5.50 | Cell membrane | 990 | 108 | Chr. I | ||
| SiER4_X1 | LOC101758097 | 7761 | XM_004964884.4 | 27 | XP_004964941.1 | 5.87 | Cell membrane. | 997 | 109 | Chr.IV |
| SiER4_X2 | XM_004964885.3 | 27 | XP_004964942.1 | 5.90 | Cell membrane. | 991 | 108 | Chr. IV | ||
| SiERL4 | LOC101753243 | 6793 | XM_004964364.4 | 26 | XP_004964421.1 | 5.55 | Cell membrane. | 979 | 106 | Chr. IV |
Notes:
pI is isoelectric point; MW is the molecular weight of amino acids. OP492075 is a GenBank accession number for SiER1_X4.
Figure 1: Nucleotide sequence characteristics of SiER family genes.
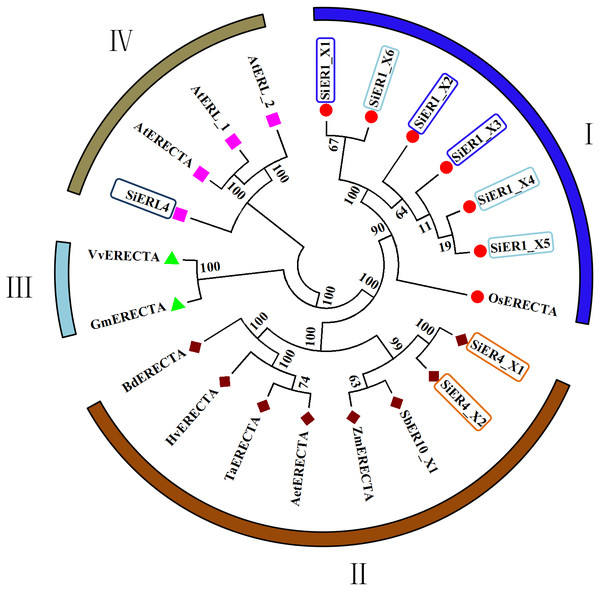
An asterisk (*) represents the mutation locus of amino acids.In the published ER family, cluster analysis showed four categories (Fig. 2): Category I and Category II contained the monocotyledonous plants, with the six copies of SiER1 family and rice ER protein constituting the first category, in which SiER1_X1 and SiER1_X6, and SiER1_X4 and SiER1_X5 were closely related. Category II was composed of two copies of SiER4 family, as well as ERs of sorghum, maize, goatgrass, wheat, barley, and brachypodium. SiER4 family was closely related to sorghum and maize. Category III was composed of ER family of dicotyledon as soybean and grape. Category IV was constituted by SiERL4 and Arabidopsis AtER and AtERL. These findings showed that in the evolution of ER families of different species, ERL was a seperate branching direction, the phylogenetic relationship of SbER1 family was close to modern aquatic plants, whereas that of SbER4 family was closer to field xerophytic plants.
Figure 2: Phylogenetic tree of ER family proteins in monocots and dicots.
Each category is represented by the same symbol with the same color. Numbers beside the branches represent bootstrap values based on 1,000 replications. Plant species and NCBI accession numbers of proteins in phylogenetic tree are listed in Annex S2.SiERs gene structure and its cis-regulatory elements
The cis-regulatory elements of SiERs family promoters were primarily involved in regulating three types of plant functional responses as follows (Table 2): (a) cell development process, including seed development, endosperm formation, meristem and mesophyll cell differentiation, cell-development cycle changes; (b) hormone-response mechanisms, including regulation pathways mediated by salicylic acid, methyl jasmonate, abscisic acid, gibberellin and auxin; (c) biological metabolic reactions, including light response, drought and low temperature induction, adversity defense, anaerobic induction, circadian rhythm regulation. These finding suggests that the SiERs could participate in the regulation of plant growth and development, and may increase plant resistance to external stress.
| Code | Functional elements of SiER promoters | Functional characteristic | Note | |||
|---|---|---|---|---|---|---|
| SiER1 (LOC101780996) | SiER1 (LOC117840131) | SiER4 (LOC101758097) | SiERL4 (LOC101753243) | |||
| 1 | RY-element | RY-element | RY-element | Seed-specific regulation | Cell development process | |
| 2 | GCN4_motif | Endosperm expression | ||||
| 3 | CAT-box | CAT-box | CAT-box | Meristem expression | ||
| 4 | HD-Zip 1 | The palisade mesophyll cells differentiation | ||||
| 5 | MSA-like | MSA-like | MSA-like | Involved in cell cycle regulation | ||
| 6 | TCA-element | TCA-element | TCA-element, | Salicylic acid responsiveness | Hormone-response mechanisms | |
| 7 | GARE-motif, TATC-box | GARE-motif, TATC-box | P-box | Gibberellin responsive | ||
| 8 | TGACG-motif, CGTCA-motif | TGACG-motif, CGTCA-motif | CGTCA-motif, TGACG-motif, | CGTCA-motif, TGACG-motif | Methyl jasmonate responsiveness | |
| 9 | ABRE | ABRE | ABRE | ABRE | Abscisic acid responsiveness | |
| 10 | TGA-element | TGA-element | Auxin responsive | |||
| 11 | TC-rich repeats | Defense and stress responsiveness | Biological metabolic reactions | |||
| 12 | Box 4, Sp1, GTGGC-motif, G-Box, TCCC-motif, GATA-motif, TCT-motif, ATCT-motif, GT1-motif | G-Box, Gap-box, GTGGC-motif, GT1-motif, TCCC-motif, | Box4, GT1-motif, G-Box, GTGGC-motif, Sp1, ATCT-motif, GATA-motif, TCCC-motif, TCT-motif, | TCCC-motif, Sp1, Box4, TCT-motif, L-box, G-Box, 3-AF1 binding site | Light responsive | |
| 13 | MBS | MBS | MBS | MBS | Drought inducibility | |
| 14 | LTR | LTR | LTR | LTR | Low temperature responsiveness | |
| 15 | ARE | ARE | ARE, GC-motif | ARE | The anaerobic induction | |
| 16 | GC-motif | GC-motif | GC-motif | Anoxic specific inducibility | ||
| 17 | Circadian | Circadian | Element involved in circadian control | |||
Notes:
Functional characteristics of cis-acting elements of SbER promoters were predicted in the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
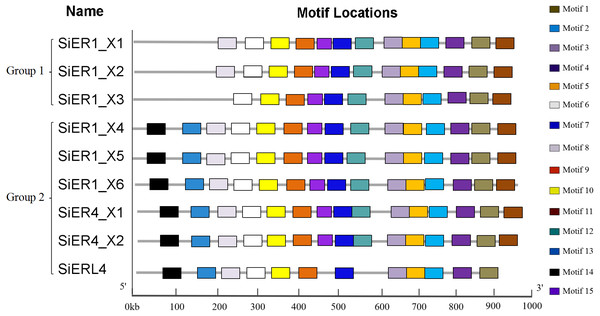
The SiERs is a typical receptor-like kinase (Annex S5), including the N-terminal signal-peptide region, the leucine tandem region (LRRs), the transmembrane region, and the C-terminal serine/threonine kinase domain. ER families of different species greatly differed in amino acid residues in the N-terminal signal-peptide region and transmembrane region (Annex S6). The 15 motif-conserved structures in the SiERs family can be divided into two categories (Fig. 3): The first category included SiER1_X1, SiER1_X2, and SiER1_X3, whereas the remaining six copies were classified into the second category. In the first category, motif 14 and 13, encoding the N-terminal signal-peptide region and the 1-3 LRR tandem domains, respectively, were lacking. Motif 8, encoding No. 4 and 5 of the LRR region, was additionally lacking in SiER1_X3. In the second category, except for SiERL4 that lacked motif 15 and 12 (encoding 13-14 LRR structures and transmembrane region, respectively), the other SiER proteins were all equipped with 15 completely conserved motif structures. This finding showed that no significant difference existed in the motif distribution of SiER family members, except for some amino acid change during the SiERs evolution, indicating that the function of SiERs could be conserved in the foxtail millet.
Figure 3: Motif analysis of SiERs amino acid sequence.
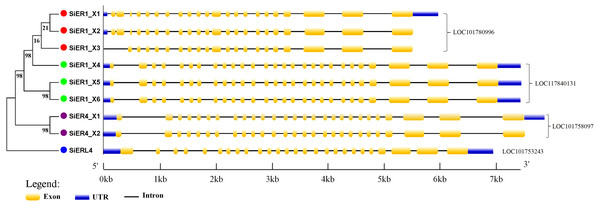
The gene-structure characteristics of different SiERs copies revealed the following (Fig. 4): SiERs exons differed in length: exons 25, 26, and 27 near the 3′-UTR region were larger, which encoded the threonine/serine kinase region of ER proteins. Exons near the 5′-UTR region had different cascade numbers, which mainly encoded the leucine tandem region of ER proteins. From these characteristics, it was speculated that SiERs proteins had similar regulatory functions, which received upstream signal and transmitted them into the cell, to induce downstream genes effects by phosphorylation. In the LOC101780996 genes, SiER1_X3 lacked the first two exons, and the distribution of other exons was similar. The first intron of SiER4 family (LOC101758097) was larger, resulting in the largest sequence of SiER4 family. SiERL4 (LOC101753243) had 26 exons and was divided into a separate branch. It was reported that the ER family often constituted 27 exons, and ERL belonged to the ERECTA-LIKE1 family (Masle, Gilmore & Farquhar, 2005; Pillitteri & Torii, 2012). In this study, both of SiER1_X4 and SiER4_X1 had 27 exons, showed typical gene-structure of the SiERs family, and were selected to to determine their functional characteristics.
Figure 4: Intron–exon structure of SiERs family in monocots and dicots.
Each gene is represented by the same symbol with the same color. Numbers beside the branches represent the bootstrap values based on 1,000 replications.Expression patterns of SiERs in different foxtail millet varieties and diverse organs
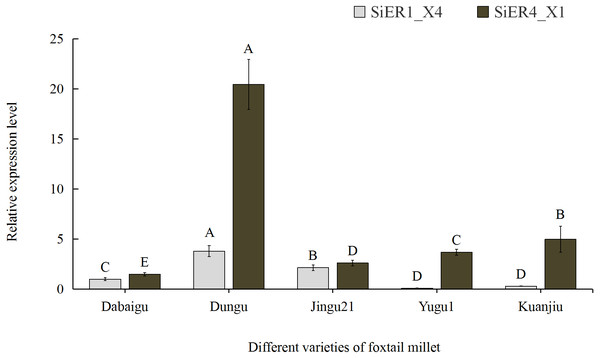
Among the five common foxtail millet varieties in China, SiER1_X4 and SiER4_X1 showed the highest expression levels in Dungu, whereas SiER1_X4 showed the lowest expression level in Yugu 1, as well as the lowest expression level of SiER4_X1 in Dabaigu (Fig. 5). Compared with SiER1_X4, SiER4_X1 showed a higher expression level in the five foxtail millet varieties. This finding showed that SiERs had different transcription levels in different foxtail millet varieties and SiER4_X1 may have a stronger regulatory function on the development of foxtail millet. Dungu was selected as an important material for subsequent gene-expression analysis.
Figure 5: Expression profiles of SiER1_X4 and SiER4_X1 gene in five varieties of foxtail millet (n = 9).
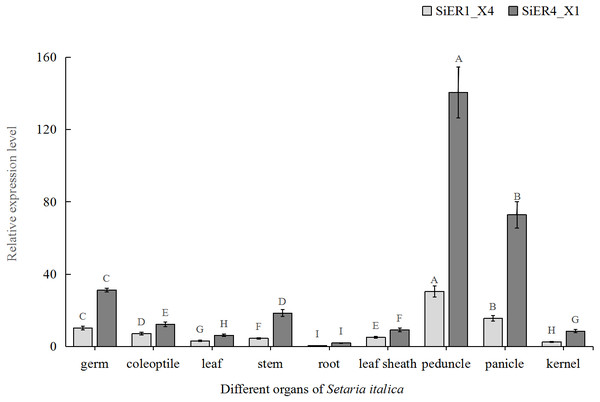
The capital letters represent the greatly significant difference of the same gene expression among different foxtail millet varieties (P < 0.01). The primers of SiER1_X4 gene (SiER1_X4–qRTF2/SiER1_X4–qRTR2), SiER4_X1 gene (SiER4_X1–qRTF1/SiER4_X1–qRTR1) and reference gene (SiActin-qRTF1/SiActin-qRTR1) are listed in Annex S3.In the different organs of Dungu, SiER1_X4 and SiER4_X1 genes were highly expressed in above-ground organs but rarely expressed in underground roots (Fig. 6). Taking root organ as a reference, the expression level of the two genes in the pedicel were both the highest, reaching 70 and 61 times of that in the roots, respectively. The expression level in panicle ranked the second (only 36 and 31 times, respectively). The expression levels in leaves and kernels were similar, both of which were at a low level. Thus, the functional roles of SiERs probably differed in regulating the development of different organs of foxtail millet, and the transcription levels of SiER4_X1 gene in different organs were significantly higher than those of SiER1_X4.
Figure 6: Expression profiles of SiER1_X4 and SiER4_X1 genes during foxtail millet growth stages (n = 9).
Foxtail millet variety Dungu cDNA was used to detect expression patterns of the two genes. The capital letters represent the greatly significant difference of the same gene expression among different foxtail millet growth stages (P < 0.01). The primers of SiER1_X4 gene (SiER1_X4–qRTF2/SiER1_X4–qRTR2), SiER4_X1 gene (SiER4_X1–qRTF1/SiER4_X1–qRTR1) and reference gene (SiActin-qRTF1/SiActin-qRTR1) are listed in Annex S3.Expression patterns of SiER1_X4 and SiER4_X1 under hormone induction and subcellular localization analysis
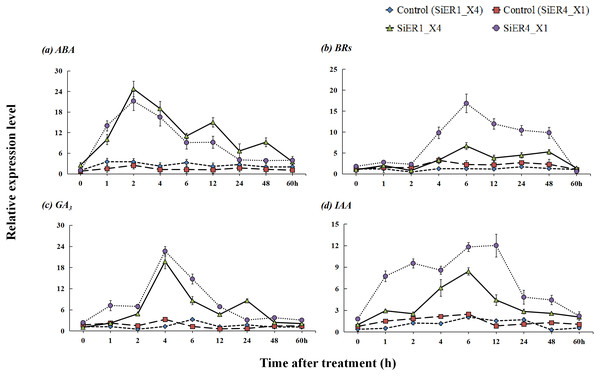
Upon treatments with the hormones abscisic acid, brassinolides, gibberellin, and indole acetic acid, SiER1_X4 and SiER4_X1 established stable expression levels in the respective control samples, whereas a significantly increased expression level was observed in the treated samples (P < 0.01). With prolonged hormone-treatment time, the expression levels of the two genes showed a response pattern of initial increase and then decrease (Fig. 7). After treatment with ABA, the expression levels of the two genes rapidly increased. At 2 h, the expression reached the highest level, those of SiER1_X4 and SiER4_X1 were 7.1 and 8.6 times of the respective controls, respectively. After treatment with BRs for 2 h, the expression levels of SiER1_X4 and SiER4_X1 gene gradually increased, the expression was the highest at 6 h. Upon treatment with GA3, the expression levels of the two genes rapidly increased after 2 h, and the expression was the highest at 4 h, which were 15.9 and 7.0 times of the control, respectively, after which the expression level rapidly decreased. After auxin (IAA) treatment, the expression of SiER1_X4 slowly increased, whereas the expression of SiER4_X1 rapidly increased. At 6 h and 12 h respectively, the expression of the two genes reached their highest levels, respectively. Thus, compared with IAA treatment, the transcription level of the SiER4_X1 gene was higher under the other three treatments. These findings showed that SiERs actively respond to hormone induction and might participate in the regulation of millet development and stress-resistance related physiological processes.
Figure 7: Expression patterns of SiER1_X4 and SiER4_X1 genes after hormone induction.
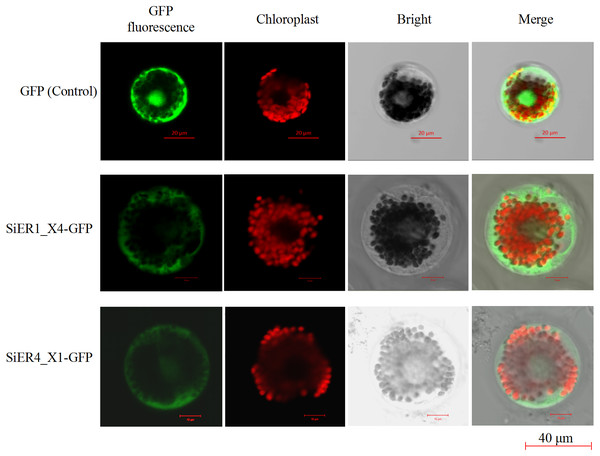
Foxtail millet variety Dungu cDNA was used to detect expression patterns of the two genes (n = 9). (A) Abscisic acid (ABA) treatment (100 µM); (B) brassinolide (BR) treatment (0.75 µM); (C) gibberellin (GA3) treatment (30 mM); and (D) auxin (IAA) treatment (10 µM).The ORF fragments of SiER1_X4 and SiER4_X1 were 2973 bp and 2991 bp, respectively (Annex S7). The subcellular localization analysis showed that the fluorescence signals of the two fusion proteins were located on the cell membrane and chloroplast of wheat mesophyll protoplasts, whereas the control pJIT16318-GFP was distributed on the cell membrane, cytoplasm and nucleus (Fig. 8). This result indicated that SiER1_X4 and SiER4_X1 primarily acted on cell membranes and chloroplasts, which was consistent with the above-mentioned prediction of SiERs as transmembrane proteins.
Figure 8: Subcellular localization of SiER1_X4 and SiER4_X1 fusion proteins in wheat mesophyll protoplasts.
SbER1_X4-GFP, SbER4_X1-GFP, and pJIT16318-GFP (control) were transiently expressed in wheat mesophyll protoplasts, respectively. Images were captured using a confocal microscope (scale bar = 40 µm).Overexpression of SiERs in Arabidopsis thaliana increased the biomass
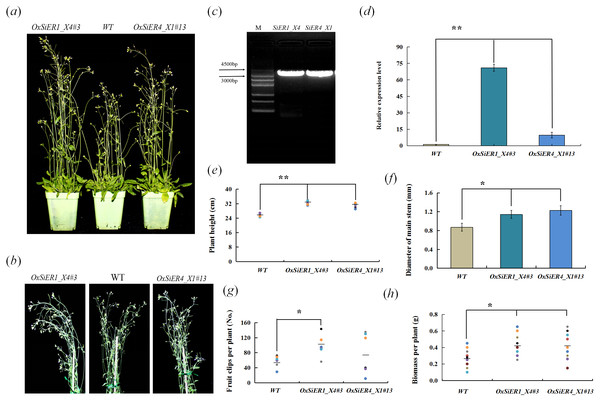
SiER1_X4 and SiER4_X1 were transformed into Arabidopsis, and the T4 generation plants were investigated (Fig. 9). In the transgenic lines OxSiER1_X4#3 and OxSiER4_X1#13, the expression levels of SiER1_X4 and SiER4_X1 were 66 and nine times those of control lines (WT), respectively. Compared with WT, the plant height of the two transgenic lines significantly increased (P < 0.01), and the main stem diameter and the biomass per plant were significantly higher than those of WT lines (P < 0.05), indicating that overexpression of SiER1_X4 and SiER4_X1 gene could enhance the biomass of Arabidopsis. It had significant implications for improving the biomass of forage crops, such as sorghum and foxtail millet. Among them, the total number of siliques per plant of the SiER1_X4 transgenic lines was significantly more than those of WT lines (P < 0.05), whereas silique number was only slightly increased for SiER4_X1 lines. Meanwhile, plant height, total number of siliques per plant, and biomass per plant of SiER1_X4 transgenic Arabidopsis were higher than those of SiER4_X1 transgenic lines.
Figure 9: Biomass-related traits of transgenic Arabidopsis.
WT is the wild Arabidopsis lines, OxSiER4_X1#13 and OxSiER1_X4#3 are Arabidopsis lines transfected from SiER4_X1 and SiER1_X4 genes, respectively. (A) Arabidopsis plants grown for 30 days; (B) plant stalk of Arabidopsis grown for 30 days; (C) the fragment isolated from SiER1_X4 and SiER4_X1 genes (Annex S7); (D) detection of overexpression level of transgenic Arabidopsis (n = 9); (E) plant height of transgenic Arabidopsis (n = 6); (F) main stem diameter of transgenic Arabidopsis (n = 7); (G) total number of siliques per plant of transgenic Arabidopsis (n = 6); and (H) biomass per plant of transgenic Arabidopsis (n = 9). Asterisks represent a significant difference (* P < 0.05; ** P < 0.01).Thermotolerance of Arabidopsis thaliana overexpressing SiERs genes
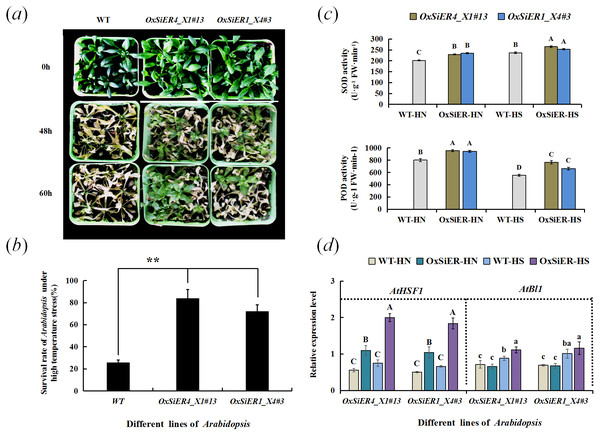
After treating Arabidopsis overexpressing SiER1_X4 and SiER4_X1 genes at elevated temperature (42 °C), the plant leaves withered and several plants showed local necrosis. After recovering for 11 days at 26 °C, only a few plants of WT lines showed vital signs, and the others all died; whereas the survival rate of transgenic Arabidopsis was extremely and significantly higher than that of WT (P < 0.01), expecially the SiER4_X1 transgenic plants, which showed a stronger ability to restore growth (Figs. 10A and 10B). Further determination of the antioxidant-enzyme activity of Arabidopsis showed that the SOD activity of SiER1_X4 and SiER4_X1 lines before and after high-temperature treatment was significantly higher than that of WT plants, as well as the POD activity of both transgenic lines (P < 0.01) (Fig. 10C). Under high-temperature stress, the SOD and POD activities of SiER4_X1 lines were slightly higher than those of SiER1_X4 lines.
Figure 10: Detection of thermotolerance of transgenic Arabidopsis.
WT is the wild type of Arabidopsis lines, OxSiER4_X1#13 and OxSiER1_X4#3 are Arabidopsis lines transfected from SiER4_X1 and SiER1_X4 genes, respectively. HN and HS represent well-culture and high-temperature stress plants, respectively. (A) Restored culturing for 11 d after high-temperature stress of transgenic Arabidopsis; (B) survival rate of transgenic Arabidopsis after high-temperature stress (n = 5); (C) SOD and POD activity of transgenic Arabidopsis (n = 4); and (D) expression identification of AtHSF1 and AtBl1 gene in transgenic Arabidopsis (n = 9). Capital and lowercase letters represent a significant difference at 0.01 and 0.05 level, respectively.Analysis of expression of the high-temperature regulation gene, AtHSF1, and the superoxide suppressor gene, AtBl1, showed that the expression level of AtHSF1 in the transgenic lines was extremely and significantly higher than those of WT lines (P < 0.01) (Fig. 10D). Particularly, after high-temperature induction, the AtHSF1 expression level of transgenic lines significantly increased. Before high-temperature treatment, the expression level of AtBl1 did not significantly differ between the transgenic lines and WT. After high-temperature treatment, the expression level of AtBl1 increased and reached a significant difference in the SiER4_X1 lines (P < 0.05). Before and after high-temperature treatment, the AtHSF1 expression level of WT lines did not change significantly, whereas the expression level of AtBl1 significantly increased (P < 0.05). These findings suggested that overexpression of SiER1_X4 and SiER4_X1 genes may improve the high-temperature tolerance of Arabidopsis, which may be due to the influence of heat-related gene expression in the regulatory pathway and induction of the variable activity of related antioxidant enzymes. Moreover, SiER4_X1 showed a better regulatory function than SiER1_X4.
Discussion
The characteristics of gene families have become an important means to analyze their function. The accuracy and reliability of analysis on the evolutionary features depend on genome-sequencing information. This study found that the foxtail millet genome contained four SiER family members, two genes were distributed on the first chromosome, with a total of six copies, and two genes were distributed on the fourth chromosome, with three copies. In rice, wheat, sorghum, cotton, tobacco crops, the ER family also had two members, and each member had different spliceosomes, resulting in an uneven distribution of the number of introns and exons in the genome (Liu et al., 2019). In foxtail millet, the spliceosomes in different copies of SiERs had obvious different forms, indicating that the relationship of the SiERs family was more complicated in the evolutionary process. In eukaryotes, the gain or loss of introns is one of the evolutionary mechanisms of creation of a gene family (Roy & Penny, 2007), and furthermore, the difference in the number of introns affected the target-gene expression level. The introns of AtER genes were absent in Arabidopsis, leading to the reduced target protein by 500–900 times (Karve et al., 2011). With decreased LRR in the extracellular region of soybean GmER (decreased exons), shading treatment increased the hypocotyl length, leaf area, and petiole length of Arabidopsis (Du et al., 2018). We speculate that different spliceosomes of SiERs result in differences in regulatory functions.
Before the emergence of monocotyledonous and dicotyledonous plants, the ER family evolved into two large subfamilies, namely, ER and ERL. Later, with the occurrence of gene-replication events, multiple copies of ER and ERL families gradually formed (Liu et al., 2019). In the present study, the ER family can be clearly divided into four categories: aquatic monocot, terrestrial monocot, dicot, and Arabidopsis ER and ERL families. Among them, six copies on the first chromosome were closely related to aquatic monocots (rice), and two copies on the fourth chromosome were closely related to terrestrial monocots. Further analysis of amino acid sequences of SiERs in other species showed that different ER families greatly differed in amino acid residues in the N-terminal signal-peptide recognition and transmembrane regions. ER family proteins are transmembrane proteins that can sense external stimuli, activate the expression of intracellular signal factors, and regulate the physiological response of cells (Shpak et al., 2004). The most important function of ER was phosphorylation. The amino acid position difference in the transmembrane region influenced the phosphorylation event, and the N-terminal extension region was one of the components of overall kinase folding that was critical to the kinase activity (Kosentka et al., 2017).
The ER family was reported to involve in light-induced under growth (Van Zanten et al., 2010), improve drought resistance of maize (Li et al., 2019), and participate in the regulation of non-host resistance of rice blast disease, and coordinately regulate the resistance of Arabidopsis to the quantitative traits of Verticillium wilt together with ABA and methyl jasmonate (Häffner et al., 2014). Moreover, it inhibited cell division and promote cell elongation (Qu, Zhao & Tian, 2017). As determined in the current research, SiER promoters contained core elements related to abscisic acid, low temperature, drought, methyl jasmonate, anaerobic induction, and light response, suggesting that SiERs may played an important roles in plant resistance and photosynthesis. However, no study has reported regarding the mechanism of low-temperature and anaerobic-induced responses. Van Zanten et al. (2009) also reported that ER affected the photoelectron-transfer capacity and carboxylation rate of ribulose diphosphate carboxylase (Rubisco), thereby increasing the photosynthetic capacity of Arabidopsis. SiERs had two common cis-acting elements, G-Box and TCCC-motif, which were involved in the light-response process. Moreover, SiER1_X4 and SiER4_X1 were both located on chloroplasts, implying that SiERs are involved in the photosynthetic function. These results indicated great application potential for improving foxtail millet photosynthesis and plant biomass.
Overexpression of SiER s could promote Arabidopsis biomass accumulation, which was primarily due to the increase of stem thickness and plant height of transgenic plants, whereas the amount of pod numbers was uncertain. This finding was similar to previous results (Xing et al., 2011; Masle, Gilmore & Farquhar, 2005). Under high-temperature treatment, Arabidopsis overexpressing SiERs had strong survival ability, and the SOD activity of transgenic lines significantly increased. Increased SOD activity could eliminate the damage to cells inflicted by reactive oxygen species produced by plants under high-temperature stress (De-Pinto, Locato & De-Gara, 2012). SiERs may be involved in the regulation of SOD synthesis or activity at high temperature and alleviate the damage to cells inflicted by O2− and H2O2 during adversity. Moreover, the expression levels of the high-temperature regulation gene AtHSF1 and superoxide suppressor gene AtBl1 confirmed the above statement, and the specific mechanism requires further study. In Arabidopsis, transforming CERK1n-ERc complex factors showed that under high-temperature stress, the H2O2 and related electrolyte content in transgenic Arabidopsis were less, and the ability to withstand high temperatures was significantly increased (Chen et al., 2020). This finding was similar to our current results, thereby providing an important basis for the next step to reveal the molecular mechanism of high-temperature tolerance of crops.
Conclusions
This study analyzed the characteristics of SiER family members (SiER s) in foxtail millet. The foxtail-millet genome contained four SiERs member. Among them, SiER1_X4 and SiER4_X1 actively responded to the induced reaction of ABA, BRs, GA3, and IAA, with a higher expression level in above-ground organs of foxtail millet. Compared to wild type, the transgenic Arabidopsis lines overexpressing the two genes enhanced the plant height and biomass accumulation, and showed the higher SOD and POD activities under high temperature, reflecting an increased thermotolerance in Arabidopsis plants. These results provided potential targets for conventional breeding or biotechnological methods to improve forage crop production under harsh environments.
Supplemental Information
Alignment of ERECTA family in N-terminal and transmembrane domains
Raw data of nucleotides, amino acids, promoters sequence of SiER family
SiER1_X4 and SiER4_X1 expression patterns data. Overexpression of SiERs increased the biomass data of sorghum. Overexpression of SiERs enhanced the thermotolerance data of Arabidopsis.