Genome-wide identification and expression pattern analysis of lipoxygenase gene family in turnip (Brassica rapa L. subsp. rapa)
- Published
- Accepted
- Received
- Academic Editor
- Ivo Feussner
- Subject Areas
- Agricultural Science, Bioinformatics, Genomics, Molecular Biology, Plant Science
- Keywords
- Turnip, Lipoxygenase, Phylogenetic analysis, Gene expression, Abiotic stress
- Copyright
- © 2022 Yan et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. Genome-wide identification and expression pattern analysis of lipoxygenase gene family in turnip (Brassica rapa L. subsp. rapa) PeerJ 10:e13746 https://doi.org/10.7717/peerj.13746
Abstract
Turnip (Brassica rapa L. subsp. rapa) is an important crop with edible and medicinal values, and various stresses, especially salt stress and drought stress, seriously threaten the yield of turnips. LOXs play important roles in regulating plant growth and development, signal transduction, and biotic and abiotic stress responses through secondary metabolites produced by the oxylipin metabolic pathway, and although the turnip genome has been published, however, the role of LOX family genes in various abiotic stress responses has not been systematically studied in turnips. In this study, a total of 15 LOX genes (BrrLOX) were identified in turnip, distributed on six chromosomes. Phylogenetic tree analysis classified these LOX genes into two classes: three 9-LOX proteins and 12 13-LOX type II proteins. Gene duplication analysis showed that tandem and segmental duplication were the main pathways for the expansion of the BrrLOX gene family. The Ka and Ks values of the duplicated genes indicate that the BrrLOX gene underwent strong purifying selection. Further analysis of the cis-acting elements of the promoters suggested that the expression of the BrrLOX gene may be influenced by stress and phytohormones. Transcriptome data analysis showed that 13 BrrLOX genes were expressed at one or more stages of turnip tuber development, suggesting that LOX genes may be involved in the formation of turnip fleshy roots. The qRT-PCR analysis showed that four stresses (salt stress, drought stress, cold stress, and heat stress) and three hormone treatments (methyl jasmonate, salicylic acid, and abscisic acid) affected the expression levels of BrrLOX genes and that different BrrLOX genes responded differently to these stresses. In addition, weighted gene co-expression network analysis (WGCNA) of BrrLOX revealed seven co-expression modules, and the genes in these co-expression modules are collectively involved in plant growth and development and stress response processes. Thus, our results provide valuable information for the functional identification and regulatory mechanisms of BrrLOX in turnip growth and development and stress response.
Introduction
Lipoxygenases (LOXs; EC 1.13.11.12) are non-heme iron-containing dioxygenases capable of generating fatty acid hydroperoxides by specifically catalyzing the oxygenation of polyunsaturated fatty acids (PUFAs) with a (1Z, 4Z)-pentadiene system (Feussner & Wasternack, 2002). These enzymes play important roles in plant growth, development, and stress response (Wasternack & Hause, 2013). Typically, LOXs are divided into two main categories depending on the oxygenation site on the PUFA chain: 9-LOX and 13-LOX, (oxygenation at the 9th or 13th carbon). Here, 9-LOX catalyzes the production of 9-HPOT (9-hydroperoxy-(10E,12Z)-octadecatrienoic acid), involved in stress (Ul Hassan, Zainal & Ismail, 2015), while 13-LOX catalyzes the production of 13-HP?T (13-hydroperoxy-(9Z,12E)-octadecatrienoic acid), which mediates the synthesis of jasmonic acid derivatives and participates in various life processes in various plants (Viswanath et al., 2020). The LOXs contain two structural domains, a conserved PLAT/LH2 (polycystin-1, lipoxygenase, alpha-toxin/lipoxygenase homology) structural domain at the N-terminal end and a highly conserved LOX structural domain at the C-terminal end. The PLAT/LH2 structural domain plays an important role in membrane binding. The LOX structural domain contains a highly conserved [His-(X)4-His-(X)4-His-(X)17-His-(X)8-His] region, which is rich in histidine (His) residues and plays an important role in the binding of LOX to iron and in exerting enzymatic activity (Andreou & Feussner, 2009; Steczko et al., 1992).
Many LOX genes have been cloned and functionally characterized, for example, in Arabidopsis thaliana, 6 AtLOX genes have been identified. AtLOX1 and AtLOX5 have been observed to be involved in lateral root development (Vellosillo et al., 2007). AtL0X2, AtL0X3, AtLOX4 and AtLOX6 are involved in JA synthesis, among them AtLOX2 can be induced by the damage response (Bell, Creelman & Mullet, 1995), and AtLOX6 is involved in crustacean defence and drought stress (Grebner et al., 2013), and AtLOX3 and AtLOX4 can regulate flower development (Caldelari et al., 2011). The ectopic expression of the DkLOX3 gene of persimmon in tomato promoted ripening (Hou et al., 2015) and senescence, while the CsLOX1 gene of cucumber in Arabidopsis regulated stamen development (Yong et al., 2020). The CaLOX1 gene of Capsicum annuum ectopic expression in Arabidopsis enhances the tolerance of transgenic plants to high salt stress and severe drought stress (Lim et al., 2015). The ectopic expression of the oriental melon CmLOX10 gene in Arabidopsis enhanced drought tolerance (Xing et al., 2020). These studies suggest that LOX genes play an important role in plant growth and development and in diverse stresses. Although LOX family genes have been identified in several plants such as radish (Wang et al., 2019), pepper (Sarde et al., 2018), poplar (Chen et al., 2015), watermelon (Liu et al., 2020), and pear (Li et al., 2014), however, no systematic analysis of the LOX family in turnips has been reported so far. With the completion of whole genome sequencing of turnips, it has become possible to identify and characterise the LOX family of genes in turnips.
Turnip (Brassica rapa L. subsp. rapa) is a biennial herb of the cruciferous family. They are important root vegetables in China and are grown mainly in Xinjiang and Qinghai (Paul et al., 2019). The Xinjiang region, however, is located in a relatively arid geographical environment with saline soils, and turnip seedlings are often affected by drought, salinity and other stresses during growth, resulting in reduced turnip yields (Jia et al., 2020). Therefore, in-depth studies on genes related to turnip stress signaling are important to improve stress resistance, reduce stress impact, and ultimately increase yield.
The study identified 15 members of the turnip LOX gene family based on available genomic information and analyzed the structure and evolution of BrrLOXs through phylogenetic analysis, gene structure, conserved structural domains, and gene duplication events. In addition, the expression patterns of the BrrLOX genes in the roots at different developmental stages were analyzed based on publicly available RNA-seq data. Further, the response of the BrrLOX genes to abiotic stresses was determined by qRT-PCR analysis. Genome-wide characterisation, evolutionary and expression pattern analysis of turnip LOX genes could provide the basis for future gene function studies and new ideas for improving turnip stress resistance.
Materials and Methods
Identification of the turnip LOX genes
The genome, CDS, and protein sequence files of turnip were downloaded from the Brassicaceae database (http://brassicadb.cn/#/) (Chen et al., 2021). The Hidden Markov Model file for lipoxygenase (PF00305) was downloaded from the Pfam database (https://pfam.xfam.org/) (Mistry et al., 2020), and the turnip genome was searched using HMMER 3.0 software and candidate LOX proteins with an e value < 1.0E−05 were screened (Wheeler & Eddy, 2013) and submitted to the conserved domains database (CDD, https://www.ncbi.nlm.nih.gov/cdd/?term=) to confirm the presence of lipoxygenase and the presence of PLAT/LH2 structural domain (Lu et al., 2020). After excluding sequences without the lipoxygenase and PLAT/LH2 structural domains, the non-redundant sequences were named according to their chromosomal position order. ExPASy (https://web.expasy.org/protparam/) was used to analyze the basic physicochemical properties of LOX proteins. The chloroplast transit peptides and the subcellular localization were predicted using TargetP 2.0 Server (https://services.healthtech.dtu.dk/service.php?TargetP-2.0) and WoLF PSORT (https://psort.hgc.jp/), respectively. Global sequence alignment was performed using the EMBOSS Needle tool (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) to determine the similarity and identity between the LOX members. The conserved motifs of LOX (the number of motifs is set to 20) were analyzed using the MEME suite (https://meme-suite.org/meme/tools/meme) (Bailey et al., 2015), and the gene structure was mapped using TBtools software (Chen et al., 2020).
Phylogenetic analysis
The published LOX protein sequences of Arabidopsis (Umate, 2011), radish (Wang et al., 2019), watermelon (Liu et al., 2020), cotton (Shaban et al., 2018), sorghum (Shrestha, Pant & Huang, 2021), buckwheat (Ji et al., 2020), banana (Liu et al., 2021), and poplar (Chen et al., 2015) were retrieved from (https://www.arabidopsis.org/), BRAD (http://brassicadb.cn/#/), CuGenDB (http://cucurbitgenomics. org/organism/1), CottonGen (https://www.cottongen.org/), NCBI (https://www.ncbi.nlm.nih.gov/), MBKBASE (http://www.mbkbase.org/Pinku1/), Banana Genome Hub (https://banana-genome-hub.southgreen.fr), and Phytozome (http://www.Phytozome.net/), respectively. Multiple sequence alignment was performed using MUSCLE software (Edgar, 2021) and phylogenetic trees were constructed by the neighbor-joining method using MEGA X software (Partial deletion; 95% site coverage cutoff, and 1,000 bootstrap replicates) (Kumar et al., 2018) and visualized using iTOL (https://itol.embl.de/) (Letunic & Bork, 2021).
Chromosome localization and gene duplication analysis
Whole-genome duplication and tandem repeat relationships between BrrLOX genes and genetic covariance between species were detected using MCScanX software (https://github.com/wyp1125/MCScanX) (Wang et al., 2012). Chromosome location was obtained from the gene feature format (GFF) gene annotation files and Visualized gene co-linear relationships using Circos software (http://circos.ca) (Krzywinski et al., 2009). Then, the synonymous substitution rate (Ka) and the non-synonymous substitution rate (Ks) were calculated for gene pairs using KaKs_Calculator 2.0 software (Wang et al., 2010).
Analysis of cis-acting elements in the promoters of turnip LOXs
A 2,000 bp sequence upstream of the start codon of each turnip LOX gene was extracted from the turnip genome database, and the cis-acting elements of the promoter were predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Expression analysis of RNA-seq data from turnip fleshy roots
The raw transcriptome data of the three developmental periods (each with two independent biological replicates) of turnip fleshy roots were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA273340/) (Li et al., 2015), the transcriptome data were remapped to the turnip reference genome using HISAT2 (Pertea et al., 2016), and the TPM values (Transcripts Per Kilobase of exon model per Million mapped reads) of turnip LOX were calculated using the featureCounts tool (Liao, Smyth & Shi, 2014), and finally the TPM values were normalized by a logarithmic scale with base 2 and visualized in a heat map using the TBtools software (Chen et al., 2020).
Plant materials
We performed a transcriptome analysis of ‘Qiamagu’ turnip seedlings showing the transcriptome changes caused by four treatments (mock control; 100 µM MeJA spray; 100 mM NaCl watering; 100 µM MeJA spray + 100 mM NaCl watering) on leaves of four-leaf stage seedlings (Table S1), with three biological replicates of each treatment. The plant material used for the stress and hormone treatments was the Xinjiang cultivar ‘Qiamagu’. Turnips were grown at 25 °C in a light incubator with 16 h light/8 h dark photoperiod and 60% relative humidity. Seedlings of uniform size and two weeks old were selected for stress and hormone treatments. To examine the effect of methyl jasmonate (MeJA), salicylic acid (SA), and abscisic acid (ABA) treatments on turnip LOX, leaves were sampled at 0, 4, 8, 12, and 24 h after spraying 100 µM of MeJA, SA, and ABA, respectively. To investigate the effect of abiotic stress on turnip LOX, turnip plants at the four-leaf stage were watered with 100 mM NaCl and 20% PEG6000 to simulate salt stress and drought stress, respectively, and exposed to 4 °C and 40 °C to simulate low and high temperatures, respectively. The leaves were sampled at 0, 4, 8, 12, and 24 h after treatment, immediately frozen in liquid nitrogen, and stored at −80 °C for further use.
Real-time quantitative PCR
Total RNA was extracted from the leaf samples using RNAprep pure Plant Kit (Tiangen, Beijing, China) and reverse-transcribed to generate the first-strand cDNA using RT reagent Kit with gDNA Eraser (Takara, Beijing, China). The cDNA was diluted and used for qRT-PCR. Primers for BrrLOX family genes used in qRT-PCR were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi) with turnip β-Actin as the internal reference gene (Ye et al., 2012). All primers used in this study are listed in Table S2. Relative expression levels of the BrrLOX genes were calculated following the 2−ΔΔCt method. Three biological replicates were set up for each time point per treatment, with each repetition consisting of nine plants.
Weighted gene co-expression network analysis (WGCNA)
Functional annotation of turnip CDS sequences using EggNOG-mapper (version 2.1, http://eggnog5.embl.de/#/app/emapper) (Huerta-Cepas et al., 2017) and WGCNA(version 1.69) analysis of successfully annotated genes in transcripts (Zhang & Horvath, 2005) (Mock-control; 100 µM MeJA spray; 100 mM NaCl watering; 100 µM MeJA spray + 100 mM NaCl watering). In the analysis, the parameters were set as follows. an optimal soft threshold (power) value of 7; minModuleSize of 30, and mergeCutHeight of 0.25. Finally, all co-expression networks associated with BrrLOXs were extracted, and the edges with weights below 0.2 were filtered out. We visualized the network connections using Cytoscape software (version 3.8.0, https://cytoscape.org/) (Shannon et al., 2003). Further, the functional annotation of BrrLOXs and co-expressed genes was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) using ggplot2 to identify the top 10 GO annotations and the top 20 KEGG functional annotations (Kanehisa et al., 2016).
Results
Identification of turnip LOX gene family members
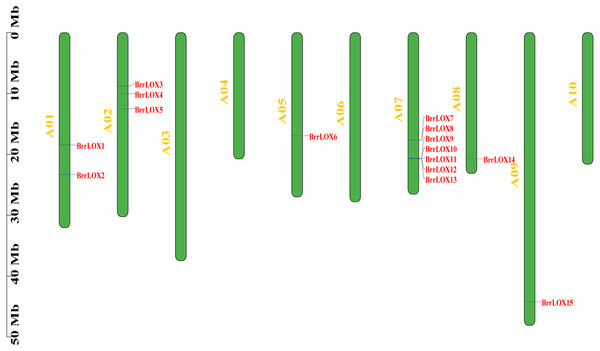
A total of 15 BrrLOX candidate genes were identified in the turnip genome (Table 1, Table S3) and named sequentially from BrrLOX1 to BrrLOX15 based on their chromosomal location. The distribution of BrrLOX genes on the chromosomes is shown in Fig. 1. The BrrLOX genes were distributed on 6 out of the 10 turnip chromosomes, of which A07 contained seven LOX genes, A02 contained three, A01 contained two, and A05, A08, and A09 contained only one each.
| Gene name | Gene ID | Chromosome location | CDS/bp | Size/aa |
Molecular weight/kD |
PI |
Chloroplast transit peptides |
Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| BrrLOX1 | A01p27670.1_BraTUA | chr01:18452597-18458178 ( +) | 2,574 | 857 | 97.55 | 5.18 | – | Cytoplasm |
| BrrLOX2 | A01p33650.1_BraTUA | chr01:23308367-23312630 ( +) | 2,499 | 832 | 94.97 | 6.12 | – | Cytoplasm |
| BrrLOX3 | A02p16520.1_BraTUA | chr02:8763241-8766727 ( +) | 2,703 | 900 | 102.92 | 5.74 | Yes | Chloroplast |
| BrrLOX4 | A02p18430.1_BraTUA | chr02:10053798-10060145 ( +) | 2,742 | 913 | 104.28 | 7.37 | Yes | Chloroplast |
| BrrLOX5 | A02p22030.1_BraTUA | chr02:12517721-12521356 ( +) | 2,760 | 919 | 103.87 | 6.57 | – | Cytoplasm |
| BrrLOX6 | A05p23650.1_BraTUA | chr05:16936753-16942334 ( −) | 2,499 | 832 | 94.98 | 6.06 | Yes | Cytoplasm |
| BrrLOX7 | A07p26570.1_BraTUA | chr07:17655268-17659036 ( −) | 2,715 | 904 | 103.10 | 5.53 | Yes | Chloroplast |
| BrrLOX8 | A07p26590.1_BraTUA | chr07:17673223-17673580 ( −) | 2,715 | 904 | 103.10 | 5.53 | Yes | Chloroplast |
| BrrLOX9 | A07p32380.1_BraTUA | chr07:20618987-20624363 ( +) | 2,676 | 891 | 101.59 | 5.59 | Yes | Cytoplasm |
| BrrLOX10 | A07p32390.1_BraTUA | chr07:20631218-20636163 ( +) | 2,598 | 865 | 98.50 | 5.26 | Yes | Chloroplast |
| BrrLOX11 | A07p32400.1_BraTUA | chr07:20641143-20646177 ( +) | 2,679 | 892 | 101.41 | 5.29 | Yes | Chloroplast |
| BrrLOX12 | A07p32410.1_BraTUA | chr07:20652307-20656259 ( +) | 2,700 | 899 | 102.4 | 5.61 | Yes | Chloroplast |
| BrrLOX13 | A07p32610.1_BraTUA | chr07:20757766-20762464 ( −) | 2,757 | 918 | 104.76 | 7.9 | Yes | Chloroplast |
| BrrLOX14 | A08p30720.1_BraTUA | chr08:20717446-20721543 ( −) | 2,760 | 919 | 103.69 | 6.33 | Yes | Cytoplasm |
| BrrLOX15 | A09p62970.1_BraTUA | chr09:44213441-44218856 ( −) | 2,760 | 919 | 103.85 | 7.16 | Yes | Chloroplast |
Figure 1: Chromosome localization of BrrLOX genes.
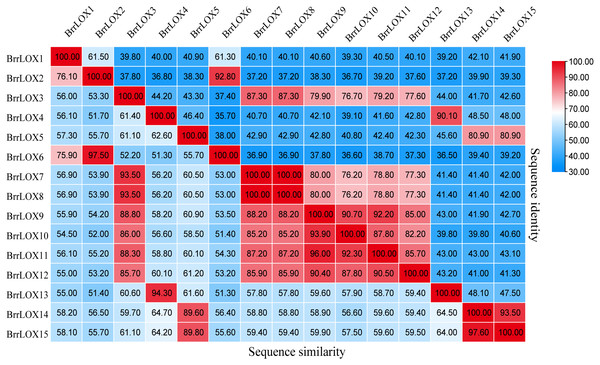
The CDS lengths of the 15 predicted BrrLOX family genes ranged from 2,499 to 2760 bp, respectively, and encoded BrrLOXs proteins from 832 to 919 amino acids in length. The predicted molecular weight of BrrLOX proteins ranged from 94.97 to 104.76 kDa, with a theoretical isoelectric point range of 5.18 to 7.9. The chloroplast transit peptide is present in 11 members of BrrLOXs. Subcellular localisation predicted by WoLF PSORT shows that nine of the BrrLOXs proteins are localised in the chloroplast and the remaining six are localised in the cytoplasm. The protein sequence alignment revealed a sequence similarity of 51.30%–100% and a sequence identity of 35.70%–100% among the BrrLOX members (Fig. 2). Specifically, BrrLOX7 and BrrLOX8 showed the highest similarity and identity (100%). The lowest similarity (51.3%) was between BrrLOX4 and BrrLOX6, BrrLOX6 and BrrLOX13; BrrLOX4 and BrrLOX6 showed the lowest identity (35.70%).
Figure 2: Turnip LOX protein Sequence similarity and identity (%).
The high, medium and low similarity and identity of LOX genes are shown in red, white and blue, respectively.Phylogenetic relationship of LOX family members in different species
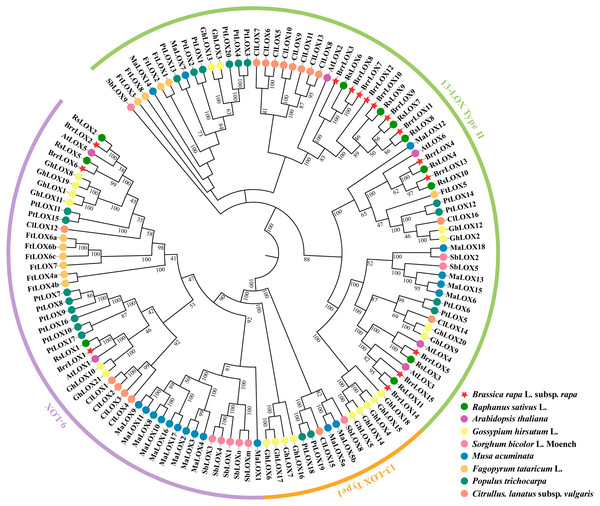
We constructed phylogenetic trees based on 128 LOX protein sequences from 9 different species, including turnip (Brassica rapa L. subsp. Rapa), radish (Raphanus sativus L.), Arabidopsis (Arabidopsis thaliana), cotton (Gossypium hirsutum L.), sorghum (Sorghum bicolor L.Moench), banana (Musa acuminata), buckwheat (Fagopyrum tataricum L.), poplar (Populus trichocarpa) and watermelon (Citrullus. lanatus subsp. vulgaris), using the neighbor-joining algorithm in MEGA X software (Fig. 3 and Table S4) (Kumar et al., 2018). The LOX protein sequences were classified into 9-LOX, 13-LOX type I, and 13-LOX type II. Of the 15 BrrLOXs identified, BrrLOX1, BrrLOX2, and BrrLOX6 belong to the 9-LOX subfamily, while the other 12 to 13-LOX type II. The BrrLOX gene family analyzed lacked the 13-LOX type I, a result also found in Arabidopsis thaliana and other Brassica species (Brassica rapa, Brassica oleracea and Raphanus sativus) (Wang et al., 2019).
Figure 3: Phylogenetic relationships of LOX proteins of turnip with other plant species.
The phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap repetitions. The 9-LOX branch is shown in purple, the 13-LOX type I branch is shown in orange, and the 13-LOX type II branch is shown in green.Conserved motifs and gene structure of turnip LOXs
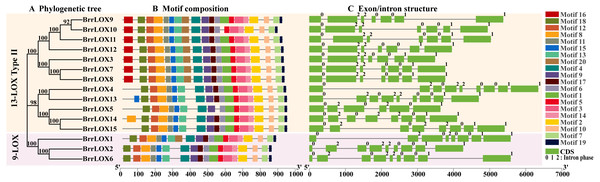
The protein motifs of the turnip BrrLOX proteins were analyzed using MEME suite. The analysis revealed 16 conserved motifs (motif2–15, motif18, motif19) widely present in all BrrLOX proteins (Supplementary Figure_S1). The BrrLOXs showed some degree of specificity in motif distribution, with all LOXs except BrrLOX5 containing motif17 (Fig. 4B). Meanwhile, all LOXs except BrrLOX10 have motif1, and motif1 contains a histidine-rich (His) 38 amino acid residue motif consisting of His-(X)4-His-(X)4-His-(X)17-His-(X)8-His. In addition, BrrLOX13 has an additional motif15 at the N-terminus, and BrrLOX14 has an additional motif8 at the N-terminus. These differences in motif arrangement may be responsible for the functional differentiation of the turnip LOX proteins. Our analysis of the gene structure showed that the BrrLOX genes consist of 5–8 introns, with BrrLOX5 having the fewest introns (five) and BrrLOX2, BrrLOX4, BrrLOX6, and BrrLOX13 having the most (eight) (Fig. 4C).
Figure 4: Phylogenetic analysis (A), motif composition (B), gene structure (C), of BrrLOX genes.
Numbers (0, 1, and 2) indicate intron phases.Gene duplication and collinearity analysis of the BrrLOX genes
In the evolution of plants, gene duplication regulates the acquisition of new genes and the creation of new genetic traits (Zhang, 2003). The present study explored the gene duplication events of the LOX family of turnips using the MCScanX software (Wang et al., 2012). In the BrrLOX gene family, seven tandem replication gene pairs (BrrLOX7/BrrLOX8, BrrLOX9/BrrLOX10/BrrLOX11/BrrLOX12) and four whole gene replication (WGD) or fragment replication (SD) gene pairs (BrrLOX2/BrrLOX6, BrrLOX4/BrrLOX13, BrrLOX5/BrrLOX14, BrrLOX14/BrrLOX15) were identified. Furthermore, Ka and Ks were calculated to estimate the evolutionary selection pressure on the duplicate gene pairs of turnip LOX genes.
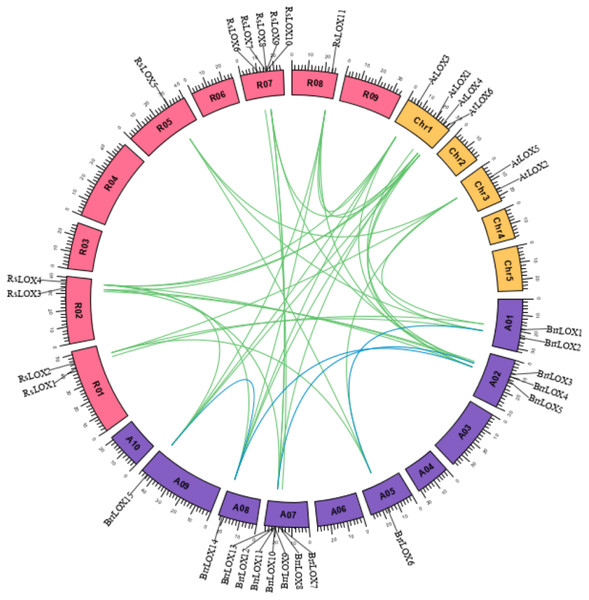
The Ka/Ks values for all 11 LOX duplicate gene pairs were less than one (Table 2), indicating that LOX family genes are mainly influenced by purifying selection during evolution. Analysis of the collinearity of LOX genes in Arabidopsis, radish, and turnip by interspecies collinearity (Fig. 5) identified that 8 members of the 15 BrrLOX genes were collinear with 5 AtLOX genes, giving rise to a total of 11 interspecies collinear gene pairs, mainly on chromosome 2 of turnip. Ten members were collinear with nine RsLOX genes (Fig. 5; Table S5), yielding a total of 17 interspecies collinear gene pairs, mainly on chromosome 2. These collinear gene pairs provide a basis for functional studies on the BrrLOX genes.
| Gene name | Gene name | Ka | Ks | Ka/Ks | Duplication type |
|---|---|---|---|---|---|
| BrrLOX7 | BrrLOX8 | 0 | 0 | NA | Tandem duplication |
| BrrLOX9 | BrrLOX10 | 0.031829569 | 0.1236034 | 0.257513701 | Tandem duplication |
| BrrLOX9 | BrrLOX11 | 0.039085368 | 0.140370417 | 0.278444483 | Tandem duplication |
| BrrLOX9 | BrrLOX12 | 0.063227762 | 0.13126891 | 0.481665936 | Tandem duplication |
| BrrLOX10 | BrrLOX11 | 0.048024707 | 0.128237559 | 0.374497979 | Tandem duplication |
| BrrLOX10 | BrrLOX12 | 0.066902608 | 0.117764339 | 0.568105835 | Tandem duplication |
| BrrLOX11 | BrrLOX12 | 0.057649265 | 0.131984388 | 0.436788515 | Tandem duplication |
| BrrLOX2 | BrrLOX6 | 0.034172652 | 0.327625452 | 0.10430402 | WGD or segmental duplication |
| BrrLOX4 | BrrLOX13 | 0.047264236 | 0.380220011 | 0.124307597 | WGD or segmental duplication |
| BrrLOX5 | BrrLOX14 | 0.107959478 | 0.941074251 | 0.114719405 | WGD or segmental duplication |
| BrrLOX14 | BrrLOX15 | 0.033123836 | 0.41651106 | 0.079526906 | WGD or segmental duplication |
Notes:
- Ka
-
non-synonymous substitution rate
- Ks
-
synonymous substitution rate
- Ka/Ks
-
the average number of non-synonymous sites
Figure 5: Collinear relationships of Arabidopsis, radish and turnip LOX genes.
The pink rectangle represents the chromosomes of radish, the yellow rectangle represents the chromosomes of Arabidopsis, and the purple rectangle represents the chromosomes of turnips.The blue lines indicate whole gene duplication (WGD) or fragment duplication (SD) gene pairs of BrrLOX, and the green lines indicate LOX genes that are homologous between the two species.Analysis of cis-elements in BrrLOX gene promoters
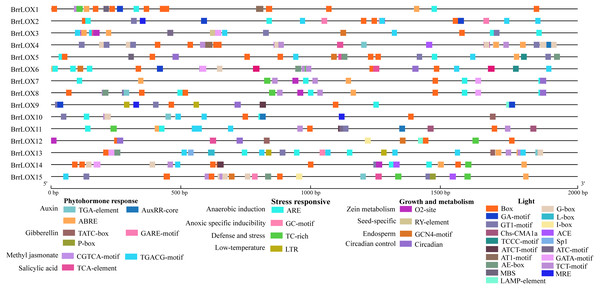
We retrieved a 2,000 bp sequence upstream of the start codon of the BrrLOX genes and performed a cis-regulatory element prediction analysis. The analysis identified 38 cis-acting elements classified into four main categories: phytohormone responsive elements, growth and metabolic responsive elements, stress responsive elements, and light responsive elements (Fig. 6). The main phytohormone response elements included TGA-element associated with auxin, GARE-motif, P-box, and TATC-box with gibberellin, ABRE with abscisic acid, CGTCA-motif and TGACG-motif with methyl jasmonate, and TCA-element with salicylic acid, suggesting multiple phytohormones regulate the BrrLOX genes. The primary growth and metabolism response elements included circadian, GCN4-motif, Ry-element, and O2-site involved in circadian control, endosperm expression, seed-specific regulation, and zein metabolism regulation, suggesting the roles of BrrLOX genes in plant growth and metabolism. The major stress response elements included TC-rich repeats, LTR, ARE, and GC-motif related to defense and stress, low-temperature response, anaerobic induction, and anoxic-specific inducibility. Light response elements, mainly Box 4, G-box, and GT1-motif, exist in all BrrLOX promoters regions.
Figure 6: Cis-acting elements identified in BrrLOX gene family promoters.
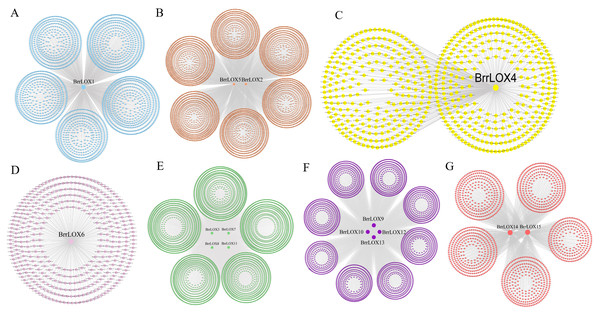
Weighted gene co-expression network analysis (WGCNA) of BrrLOXs
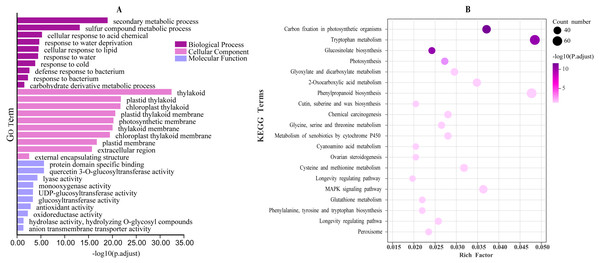
There were 42,877 functional genes in the turnip CDS sequence that were successfully annotated by EggNOG-mapper (version 2.1) and a co-expression network was constructed using WGCNA based on these functional genes and four transcriptomic data. Co-expression networks associated with BrrLOX genes were visualized using Cytoscape software (version 3.8.0), and a total of seven co-expression modules, named modules A to G, were obtained (Fig. 7). The largest of these modules is module B, with 5,426 genes co-expressed with BrrLOX2 and BrrLOX5. The smallest is module D with 383 genes co-expressed with BrrLOX6. Furthermore, GO enrichment analysis showed that genes co-expressed with BrrLOXs were mainly enriched in the secondary metabolic process, cellular response to lipid, response to a bacterium, sulfur compound metabolic process, and carbohydrate derivative metabolic process in the biological process category (Fig. 8A). Gene annotations were mainly enriched in the cellular components in the thylakoid, extracellular region, plastid thylakoid, chloroplast thylakoid, and plastid membrane. From molecular functions, genes were primarily enriched in tryptophan metabolism, phenylpropanoid biosynthesis, carbon fixation in photosynthetic organisms, MAPK signaling pathway-plant, and 2-oxocarboxylic acid metabolism. According to KEGG functional enrichment analysis, these genes co-expressed with BrrLOXs were mainly enriched in tryptophan metabolism, phenylalanine biosynthesis, carbon fixation in photosynthetic organisms, MAPK signaling pathway-plant, and 2-oxocarboxylic acid metabolism (Fig. 8B).
Figure 7: Analysis of BrrLOX gene co-expression network.
The middle nodes indicate BrrLOX genes and all other colored nodes indicate genes co-expressed with BrrLOX.Figure 8: GO and KEGG analyses of genes in the BrrLOX gene co-expression network.
(A) GO functional annotation; (B) KEGG pathway enrichment.Transcriptome analysis of the LOX family genes of turnip
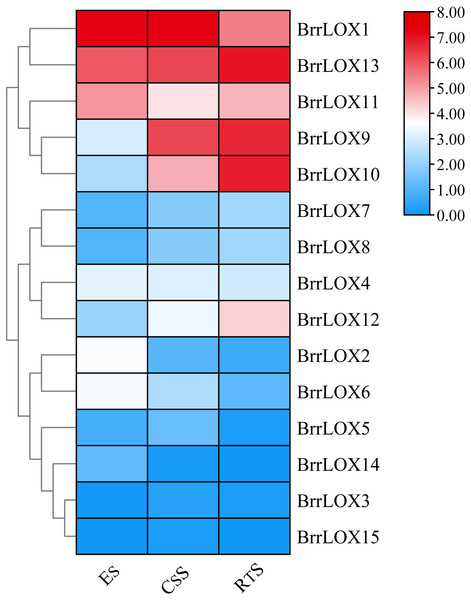
To investigate the expression of turnip LOX family genes during fleshy roots development, we reanalyzed the expression levels of BrrLOX genes using publicly available RNA sequence data from three fleshy roots development stages, including ES (early stage before cortex splitting), CSS (cortex-splitting stage) and RTS (secondary root-thickening stage) (Fig. 9; Table S6), in which BrrLOX1 and BrrLOX13 were highly expressed at all three stages; BrrLOX11 expression was higher in ES and RTS than in CSS, while BrrLOX9 and BrrLOX10 expression levels were higher in RTS than in ES and CSS. BrrLOX12 was highly expressed in RTS. However, BrrLOX3 and BrrLOX15 were expressed at 3 periods with little or no expression.
Figure 9: Expression of the BrrLOX gene family in the fleshy roots of turnips at different developmental stages.
The heat map was created based on publicly available turnip flesh root transcriptome data. In the heat map, the color bars represent the average of the TPM values of log-transformed BrrLOX genes in two biological replicate samples. Red represents high expression level, blue represents low expression level. ES, early stage before cortical division; CS, cortical division stage; RTS, secondary root thickening stage.Expression analysis of BrrLOX family genes in turnip under abiotic stress and exogenous hormone treatment
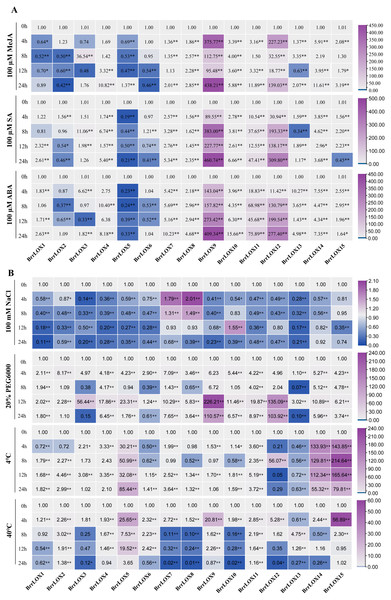
We further performed qRT-PCR analysis to detect the changes in the transcript levels of LOX genes in turnip leaves under different abiotic stresses (salt stress, drought stress, cold stress, and heat stress) (Fig. 10A and Table S7) and exogenous hormone treatment (MeJA, SA, and ABA) (Fig. 10B and Table S7). Under NaCl treatment, the expression of most genes (BrrLOX1-6, BrrLOX9, BrrLOX11-15) was downregulated, while that of BrrLOX7 and BrrLOX8 was upregulated at 4 h and 8 h after exposure; BrrLOX10 was upregulated at 12 h. Under drought stress, 10 LOX genes were upregulated, among which BrrLOX9 was most significantly expressed at 12 h. Under low-temperature treatment, BrrLOX3-5, BrrLOX11, BrrLOX14, and BrrLOX15 genes were upregulated at all time points, with BrrLOX15 showing a significant high expression at 8 h; BrrLOX13 was downregulated at all time points. Under high-temperature treatment, BrrLOX2, BrrLOX5, and BrrLOX11 were upregulated at all time points, with the most significant expression for BrrLOX15 at 4 h.
Figure 10: Expression analysis of BrrLOX family genes under (A) exogenous hormonal (MeJA, SA, and ABA) and (B) abiotic stress (NaCI, PEG6000, 4 °C and 40 °C) treatments.
The relative expression levels of BrrLOX genes in turnip leaves were analysed at 4, 8, 12 and 24 h of treatment and compared with their values at 0 h. The gene relative expression was calculated using the 2−ΔΔCT method with β-Actin as an internal reference, and values represent mean of three biological replicates. Purple indicates up-regulated expression levels, blue indicates down-regulated expression levels (* p < 0.05, ** p < 0.01, one-way ANOVA).Furthermore, the BrrLOX genes showed different expression patterns under various hormonal treatments (Fig. 10B). BrrLOX1 gene expression was downregulated under the MeJA treatment and upregulated under the ABA treatment. BrrLOX4, BrrLOX7-12, and BrrLOX14 were upregulated with three exogenous hormones, with the highest effect for BrrLOX9 at 24 h.
Discussion
Oxidized lipid compounds (e.g., jasmonic acid, green leaf aldehydes, etc.) synthesized via the LOX pathway play important roles in plant growth and development and stress responses, such as seed germination (Li et al., 2018), fruit development (Contreras et al., 2017), drought (Sofo et al., 2004), injury (Xu et al., 2003), and pests and diseases (Christensen et al., 2013). However, fewer studies have been reported on the analysis of the functions of members of the turnip LOX gene family, therefore, this study identified the LOX family members in turnip based on the turnip reference genome and analyzed their structure and evolution. Further, transcriptome data was analyzed to understand their role during development, and qRT-PCR was used to study their roles in response to abiotic stresses (cold, heat, simulated drought, and salinity) and exogenous hormone treatments (MeJA, ABA, and SA).
The present study identified 15 BrrLOX family genes in turnip by BLASTP search and CDD structural domain test, which was more than in Arabidopsis (six) (Umate, 2011) and radish (11) (Wang et al., 2019), but less than in cucumber (23) (Liu, Liu & Jiang, 2011) and cotton (21) (Shaban et al., 2018). These observations revealed that the number of LOXs is not proportional to genome size and suggested that the LOX genes are not conserved across dicotyledons and have changed during evolution. Phylogenetic tree analysis divided the turnip LOX family into two subfamilies, 9-LOX (BrrLOX1, BrrLOX2, BrrLOX6) and 13-LOX Type II (BrrLOX3, BrrLOX4-5, BrrLOX7-15), lacking 13-LOX Type I. This result is consistent with the classification of the LOX gene family of other Brassica species (Wang et al., 2019). Typically, 13-LOX could be divided into two subfamilies, 13-LOX Type I and 13-LOX Type II; based on the presence or absence of chloroplast transit peptide, where 13-LOX Type I lacked chloroplast transit peptide and had >75% sequence similarity, while LOX of 13-LOX type II had chloroplast transit peptide and had >35% sequence similarity (Bell & Mullet, 1993). In the present study, LOX belonging to 13-LOX type II, with the exception of BrrLOX5, contained chloroplast transit peptides. Sequence similarity analysis revealed that, with the exception of tandem replication genes, the sequence similarity of the few belonging to 13-LOX type II was still greater than 80%, which was significantly higher than 35%. This finding is not entirely consistent with the above classification, but such results have been found in banana (Liu et al., 2021), so we think it may be related to the structural diversity of LOX genes.
Gene duplication is a major mechanism that increases the number of gene family members and functional diversity (Magadum et al., 2013). Therefore, we analyzed the gene duplication events in the turnip genome to understand the formation and functional diversity of the LOX gene family. Previous studies identified two tandem repeat gene pairs and four fragment repeat gene pairs in the LOX family genes of banana and two tandem repeat gene clusters in watermelon (Liu et al., 2021; Liu et al., 2020). The present study found that four pairs were formed by whole gene duplication (WGD) or fragment duplication (SD), accounting for 46.67% (7/15) of the total number of LOX genes, and seven pairs by tandem duplication, accounting for 40.00% (6/15) of the total number of LOX genes. Our results suggest that tandem replication and fragment replication are the predominant replication events of the BrrLOX genes. In addition, Ka/Ks values for the homologous gene pairs were less than one, indicating that the turnip LOX genes have undergone purifying selection, promoting the stability of gene function. Eight of the 15 BrrLOX genes showed collinearity with Arabidopsis LOX genes and 10 with radish. The presence and identification of these collinear gene pairs will facilitate studies on turnip genes compared with the known functional genes of Arabidopsis and radish.
Weighted gene co-expression network analysis (WGCNA) can cluster genes with similar expression patterns into the same module, and analysis of genes in these modules can provide a better understanding of gene interaction networks and biological functions. Therefore, in our study, a total of seven co-expression modules associated with the BrrLOX gene were screened using WGCNA analysis. Most of the neighboring genes in the BrrLOX gene co-expression network were associated with sub-metabolic processes, bacterial defense, drought stress, cold stress, and photosynthesis. In addition, the BrrLOX promoters contain a large number of cis-acting elements associated with growth and development and stress response. These results suggest that BrrLOX genes are extensively involved in the growth and development of turnips and in response to various biotic-abiotic stress pathways.
In plants, LOXs play important roles in regulating the development of various organs, including roots. In Arabidopsis, oxidized lipids produced by the 9-LOX pathway regulate root development by affecting transcriptional changes in cell wall modification genes (Vellosillo et al., 2012). In potato, the antisense expression suppressor mutant POTLX-1 inhibited LOX activity and root tuber development by inhibiting the expression of LOX1-like genes (Kolomiets et al., 2001). In this study, transcriptome data analysis of three fleshy roots developmental stages revealed that LOX family genes were differentially expressed in turnip fleshy roots, with three LOX genes highly expressed in the ES phase, five in the CSS phase, and six in the RTS phase, suggesting their roles in the development of turnip fleshy roots. Specifically, BrrLOX11 was more expressed in RTS and ES than in CSS, BrrLOX9 and BrrLOX10 in RTS than in CSS and ES phases, implying that different BrrLOX members perform different functions at different developmental phases.
Furthermore, the LOX pathway produces secondary metabolites involved in plant responses to various abiotic stresses. In tomato, overexpression of the TomLOXD gene increased resistance to high-temperature stress by catalyzing the α-linolenic acid pathway (Hu et al., 2013). The melon CmLOX10 gene is heterologously overexpressed in Arabidopsis thaliana and improves drought tolerance in transgenic plants by increasing the synthesis of endogenous jasmonic acid (Xing et al., 2020), the pepper CaLOX1 gene increased tolerance under high salt stress and severe drought stress by regulating lipid peroxidation and various other pathways (Lim et al., 2015). The present study analyzed the expression profiles of BrrLOX genes under different stress treatments by qRT-PCR and found altered expression of all genes. Here, 12 LOX genes were downregulated under NaCl stress, and only 3 (BrrLOX7, BrrLOX8, BrrLOX10) were upregulated, consistent with the observations in radish under salt stress (Wang et al., 2019). 10 LOX genes were expressed higher than the control under drought stress, 6 under low temperature, and 3 under high temperature, indicating the role of BrrLOX genes in multiple stress responses and this result was also found in tomato (Upadhyay, Handa & Mattoo, 2019). The present study also revealed that 4 tandemly replicated genes, BrrLOX9, BrrLOX10, BrrLOX11 and BrrLOX12, had significant differences in gene expression levels although they were structurally similar. We speculate that the turnip LOX gene family has undergone gene subfunctionalization during the evolutionary process, leading to differences in the roles of homologous genes under different stresses.
Several studies have found that exogenous JA/MeJA treatment strongly induces LOX gene expression. For example, in watermelon, the expression of CILOX genes was significantly up-regulated after exogenous jasmonic acid treatment, with the expression of CILOX7 being 60-fold higher than that of the control (Liu et al., 2020). In banana, the expression of eight MaLOX genes was up-regulated after exogenous MeJA treatment (Liu et al., 2021). Our findings are consistent with these results; here, MeJA upregulated the expression of nine BrrLOX genes, with BrrLOX9 being the most significantly upregulated compared to the control (> 90-fold). In addition, among the nine up-regulated BrrLOX genes, only the promoter regions of four LOX genes (BrrLOX10, BrrLOX11, BrrLOX14, BrrLOX15) contained MeJA cis-acting regulatory elements, and there was no direct correlation between the number of these cis-acting elements and gene expression patterns. Therefore, the potential role of MeJA cis-acting elements in gene regulation needs to be verified in future studies.
ABA is another important plant stress hormone. A recent study showed that the melon CMLOX13 gene expression in Arabidopsis enhanced tolerance to severe drought by regulating ABA accumulation and stomatal closure (Xing et al., 2019). In the present study, nine BrrLOX (BrrLOX1, BrrLOX4, BrrLOX7, BrrLOX9-12, BrrLOX14, BrrLOX15) genes were up-regulated in expression under ABA and drought stress treatments, among which, BrrLOX9 had the highest relative expression levels under both stresses, therefore, in the next study, we will select BrrLOX9 as a candidate gene to explore the role of BrrLOX gene in drought stress. In addition, we identified defense and stress response elements, low temperature response elements and light response elements in the promoter region of the LOX genes of the turnip. These results suggest that LOX genes may be involved in a variety of biological processes during the growth and development of turnips.
Conclusions
The present study identified 15 LOX genes in the turnip genome with intact structural domains of lipoxygenases, which were unevenly distributed on six chromosomes of the turnip genome. Phylogenetic analysis classified turnip LOX genes into two subgroups, 9-LOX and 13-LOX type II. Gene structure analysis indicates that BrrLOX genes are highly conserved in the same subfamily or evolutionary branch, which may indicate that they have similar biological functions. Gene replication analysis indicates that tandem and fragment replication play a dominant role in the expansion of the BrrLOX family and the generation of new BrrLOX genes. Further qRT-PCR showed that different LOX genes responded to abiotic stresses, such as salt, drought, low and high temperature, suggesting their roles in abiotic stress tolerance. The WGCNA constructs suggest that BrrLOX may be involved in plant growth and development, and biotic and abiotic stress responses. Thus, our results will help to investigate further the key functions of LOX genes in evolution, root development, and abiotic stress tolerance.