Comparative genomic analysis of the COBRA genes in six Rosaceae species and expression analysis in Chinese white pear (Pyrus bretschneideri)
- Published
- Accepted
- Received
- Academic Editor
- Genlou Sun
- Subject Areas
- Bioinformatics, Genomics, Molecular Biology, Plant Science
- Keywords
- Phylogenetic analysis, COBRA, Rosaceae species, Pyrus bretschneideri, Secondary cell wall (SCW)
- Copyright
- © 2022 Zhao et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. Comparative genomic analysis of the COBRA genes in six Rosaceae species and expression analysis in Chinese white pear (Pyrus bretschneideri) PeerJ 10:e13723 https://doi.org/10.7717/peerj.13723
Abstract
COBRA-Like (COBL) genes encode a glycosylphosphatidylinositol (GPI) anchoring protein unique to plants. In current study, 87 COBRA genes were identified in 6 Rosaceae species, including Pyrus bretschneideri (16 genes), Malus domestica (22 genes), Fragaria vesca (13 genes), Prunus mume (11 genes), Rubus occidentalis (13 genes) and Prunus avium (12 genes). We revealed the evolution of the COBRA gene in six Rosaceae species by phylogeny, gene structure, conservative sequence, hydrophobicity analysis, gene replication events and sliding window analysis. In addition, based on the analysis of expression patterns in pear fruit combined with bioinformatics, we identified PbCOBL12 and PbCOBL13 as potential genes regulating secondary cell wall (SCW) formation during pear stone cell development. This study aimed to understand the evolutionary relationship of the COBRA gene in Rosaceae species, clarify the potential function of COBRA in pear fruit development, and provide essential theoretical basis and gene resources for improving pear fruit quality through genetical modification mechanism.
Introduction
Plant secondary wall is mainly composed with cellulose, hemicellulose and lignin. In the development of fruit growth of Dangshan su pear, the continuous accumulation of secondary walls will form stone cells near the core (Su et al., 2019b). The diameter and number of stone cell clusters greatly influence the fruit quality of Dangshan su pear (Zhang et al., 2017). The formation of the secondary wall is a complex dynamic process (Yan et al., 2014). Identifying the primary genes involved in the lignin and cellulose production pathway is crucial for comprehending the secondary wall biosynthesis. In the Dangshan su pear, the critical genes of the lignin biosynthesis pathway are completely characterized, but the cellulose biosynthesis genes are poorly recognized (Su et al., 2019b; Cheng et al., 2018; Cheng et al., 2017; Cheng et al., 2020; Cheng et al., 2019a; Cheng et al., 2019b; Li et al., 2019; Li et al., 2020b). Since then, 36 members of the CesA gene family have been isolated from Dangshan su pear, of which four genes may be involved in secondary wall formation (Li et al., 2020a). Except for cellulose synthase, members of the COBRA gene family, which encode glycosylphosphatidylinositol (GPI)-anchored proteins, have been identified as new players in the regulation of the orientation of cell expansion in the plant cell wall, these proteins were involved in cell elongation, secondary wall thickening, plant root development and seed coat morphological changes (Roudier et al., 2005; Schindelman et al., 2001).
The COBRA gene, which encodes 454 amino acids, plays an important role in plant cell elongation cellulose synthesis and cellulose deposition. The AtCOBL4 gene was involved in the synthesis of cellulose in the cell wall, although the phenotype of Atcobl4 was normal, the stem was easy to break (Brown et al., 2005). In the COBRA family reported in Arabidopsis, AtCOBL2 was involved in depositing cellulose in seed coat cells (Ben-Tov et al., 2015). In Arabidopsis thaliana, when AtCOBL5 was functionally absent, growth and development were affected, and a large amount of stress-responsive substances were produced, while defense-related gene expression appeared up-regulated (Ko et al., 2006). AtCOBL9 gene deletion resulted in shorter and less numerous root hairs (Jones, Raymond & Smirnoff, 2006). COBL (COBRA like) family members have similar functions in rice. The rice BC1 (Brittle culm 1) gene was up to 60.7% homologous to the Arabidopsis COB gene. bc1 plants had reduced cell wall thickness, reduced cellulose content, and reduced stalk stiffness (Dai et al., 2009; Li et al., 2003). The bc1 gene affected cellulose assembly by binding microfibrils and ultimately regulated cellulose crystallite size (Liu et al., 2013). A previous study found that the OsBC1L4 gene was mainly localized in the cell wall and cell membrane, and its mutants had abnormal cell swelling and reduced cellulose content (Dai et al., 2011). The OsBC1L5 gene was involved in secondary wall synthesis in thick-walled tissues of stem nodes. At the same time, Osbc1l5 loss of function resulted in severely impaired male gametophyte transport (Dai et al., 2009). OsBC1L6 regulated β-glucan synthesis during endosperm cell wall formation by interacting with cellulose moieties on the plasma membrane during seed maturing (Midorikawa et al., 2019). The COBRA family has been reported in maize, and maize BK2 can ultimately affect stalk strength by influencing cellulose deposition in the secondary wall (Ching et al., 2006). Maize Roothairless3 (Rth3) was homologous to rice BC1L1, and RTH3 was highly expressed in root epidermal cells, root hair cells and lateral root primordia. The mutants of this gene do not have normal root hair development along with reduced field yield (Hochholdinger et al., 2008). Heterologous overexpression of cotton GhCOBL9A plants upregulated CESA gene expression and cellulose deposition while promoting longitudinal elongation of hypocotyl and root cells during early development (Niu et al., 2018).
Rosaceae plants are widely distributed in China and have significant economic value, Such as pear (Pyrus bretschneideri), strawberry (Fragaria vesca), black raspberry (Rubus occidentalis), sweet cherry (Prunus avium), apple (Malus domestica), Japanese apricot (Prunus mume) belong to Rosaceae family. Previous studies have found that the COBRA gene family was identified in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays) and cotton (Gossypium raimondii) with 12, 11, nine, and 19 members, respectively (Borner, Lilley & Stevens, 2003; Dai et al., 2009; Dai et al., 2011; Sindhu et al., 2007; Niu et al., 2015). Most of the COBRA genes were involved in plant growth and development by regulating cellulose which eventually affected the formation of secondary walls. To further understand the COBRA gene family in Rosaceae, we identified all 87 members in six Rosaceae species and evaluated their phylogenetic relationships, hydrophobicity, gene structures, cis-regulatory elements, and tissue expression patterns. Our study will facilitate further functional studies of specific genes in the COBRA family.
Materials and Methods
Identification of COBRA genes in six Rosaceae species
In this work, the Pyrus bretschneideri genome was downloaded from GIGADB datasets (http://gigadb.org/dataset/100083), and five Rosaceae genomes (Fragaria vesca, Rubus occidentalis, Prunus avium, Malus domestica, Prunus mume) were obtained from the following website (https://www.rosaceae.org/). Arabidopsis COBRA gene family members amino acid sequences were used as the query sequence for BlastP (Protein BLAST: search protein databases using a protein query (nih.gov)) search (E = 0.001) from the local protein database. The SMART online software program (http://smart.embl-heidelberg.de/) was used to screen the genes (Letunic, Doerks & Bork, 2012). The protein sequences lacking a whole COBRA domain and redundant sequences were discarded. We used the ExPASY online website to predict the molecular weights, isoelectric points, and GRAVY values of COBRA protein (http://web.expasy.org/protparam/) (Artimo et al., 2012). Signal peptides were analyzed and predicted by SignalP 4.1 server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) software.
Evolutionary analysis of COBRA gene family
Sequence alignment of all COBRA proteins was done using the ClustalW tool in MEGA 7.0 software. The phylogenetic tree was constructed with MEGA 7.0 software using the NJ (Neighbor-Joining) (bootstrap = 1,000). The sequences of A. thaliana, O. sativa, Z. mays, and Populus were obtained from the article (Roudier et al., 2002; Li et al., 2003; Sindhu et al., 2007; Zhang et al., 2010).
COBRA gene structures and conserved motif prediction
The Gene Structure Display Server (http://gsds.gao-lab.org/) was used to investigate the gene structures (introns/exons) of the COBRA gene family members (Guo et al., 2007). The conserved sequences of the COBRA gene family were analyzed by MEME online (https://meme-suite.org/meme/) software (Bailey et al., 2015). The parameter settings are as follows: The number of identified motifs were 20 with Parameters for the conserved motif prediction were motif width greater than 6 and less than 200.
Promoter analysis and cis-acting element analysis of PbCOBL genes
In this study, we found the 1,500 bp–2,000 bp upstream of the start codon in the pear genome database, which was the promoter sequence of the gene. The online software PlantCare (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to examine cis-acting elements.
Chromosomal location, gene duplication, and Ka/Ks ratio analysis
Mapinspect software was used to map the position of COBRA genes on chromosomes (Zhu et al., 2015; Su et al., 2019a). The determination of gene replication events of COBRA genes in 6 species mainly followed the following principles: First, the similarity of the two genes was greater than 80%. The distance between two genes on the same chromosome was more than 200 kb, which was tandem-duplicated genes. Two genes located on different chromosomes were defined as segmentally duplicated genes. Finally, DnaSP v5.0 software calculated non-synonymous (Ka) and synonymous substitution (Ks) values and performed a sliding window analysis. The parameter was set to window size 150 bp and step size 9 bp (Zhao et al., 2021).
RNA extraction and qRT-PCR analysis
In this study, Dangshan su pear was used as the material, which growed in Dangshan County, Anhui Province. Samples of current-year flowers, buds, stems, mature leaves and fruits were obtained. The fruits were picked 15 days after pollination (DAP), 23 DAP 39 DAP, 47 DAP, 55 DAP, 63 DAP, 79 DAP, and 102 DAP. 39 DAP fruits were selected for different tissue expression analyses. We used the RNA extraction kit of Tiangen (Beijing, China) to extract RNA from different materials. Reverse transcription was performed using a PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa, China). The qRT-PCR primers were designed using Beacon Designer 7 software (Table S1). The pear Tubulin gene (No. AB239680.1) was used as an internal reference (Wu et al., 2012). Each 20 µL qRT-PCR system included 10 µL of SYBR Premix Ex Taq TM II, 2 µL cDNA, 0.8 µL of Forward primer and Reverse primer, 6.4 µL of water. This study was conducted according to the procedures in the instruction manual, and three biological repetitions were run for each sample. The relative expression level of genes was calculated by the 2−ΔΔCt method (Livak & Schmittgen, 2001).
Results
Identification, characterization analysis of COBRA genes
Using the amino acid sequences of Arabidopsis COBRA gene family members as probes, we identified 87 COBRA proteins in six Rosaceae species. Including 16 COBRA proteins (PbCOBL1-PbCOBL16) in Pyrus bretschneideri, 22 in Malus domestica (MdCOBL1-MdCOBL22), 13 in Fragaria vesca (FvCOBL1-FvCOBL13), 11 in Prunus mume (PmCOBL1-PmCOBL11), 13 in Rubus occidentalis (RoCOBL1-RoCOBL13) and 12 in Prunus avium (PaCOBL1-PaCOBL12). The detailed information (gene name, gene identifiers, amino acid number, signal peptide, molecular weight, theoretical isoelectric point, Grand average of hydropathicity and subdivided subgroup) of each COBRA was presented in Table 1 and Table S2. These results showed that the largest molecular weight among the six Rosaceae species was MdCOBL12, which was 131.89 kDa. The smallest was PaCOBL4, which was 12.59 kDa. Except for MdCOBL17 and PaCOBL3, the grand average of hydropathicity of other genes were negative, indicating that most members of COBRA genes were hydrophilic proteins. In these Rosaceae species, the lowest pI value was 5.04 (MdCOBL16), whereas the highest pI value was 9.59 (MdCOBL1). Among the six Rosaceae species, about 74% of COBRA proteins contained signal peptides, and 26% of proteins did not contain a signal peptide.
| Gene name | Gene ID | AA | KD | pI | GRAVY | Signal peptide | Subdivided subgroup |
|---|---|---|---|---|---|---|---|
| PbCOBL1 | Pbr039918.1 | 435 | 49.18 | 8.46 | −0.206 | Yes | COBRA |
| PbCOBL2 | Pbr033684.1 | 674 | 75.58 | 8.93 | −0.318 | No | COBL7 |
| PbCOBL3 | Pbr028526.1 | 335 | 38.19 | 6.64 | −0.296 | No | COBRA |
| PbCOBL4 | Pbr026558.1 | 432 | 48.97 | 8.06 | −0.339 | Yes | COBRA |
| PbCOBL5 | Pbr020181.1 | 633 | 70.78 | 9.00 | −0.240 | No | COBRA |
| PbCOBL6 | Pbr020180.1 | 456 | 50.99 | 8.92 | −0.105 | Yes | COBRA |
| PbCOBL7 | Pbr016608.1 | 457 | 51.64 | 9.10 | −0.157 | Yes | COBRA |
| PbCOBL8 | Pbr011992.1 | 657 | 71.63 | 5.70 | −0.124 | Yes | COBL7 |
| PbCOBL9 | Pbr010999.1 | 311 | 35.17 | 6.30 | −0.367 | No | COBRA |
| PbCOBL10 | Pbr009004.1 | 241 | 27.20 | 8.78 | −0.425 | No | COBRA |
| PbCOBL11 | Pbr008592.1 | 610 | 66.55 | 5.07 | −0.070 | No | COBRA |
| PbCOBL12 | Pbr007186.1 | 456 | 50.89 | 8.92 | −0.084 | Yes | COBRA |
| PbCOBL13 | Pbr007185.1 | 445 | 49.81 | 8.73 | −0.287 | Yes | COBRA |
| PbCOBL14 | Pbr007184.1 | 423 | 46.82 | 9.00 | −0.203 | Yes | COBRA |
| PbCOBL15 | Pbr004198.1 | 674 | 75.60 | 8.99 | −0.337 | Yes | COBL7 |
| PbCOBL16 | Pbr001136.1 | 692 | 75.24 | 5.96 | −0.071 | Yes | COBL7 |
Phylogenetic and hydrophobic analysis of COBRA genes
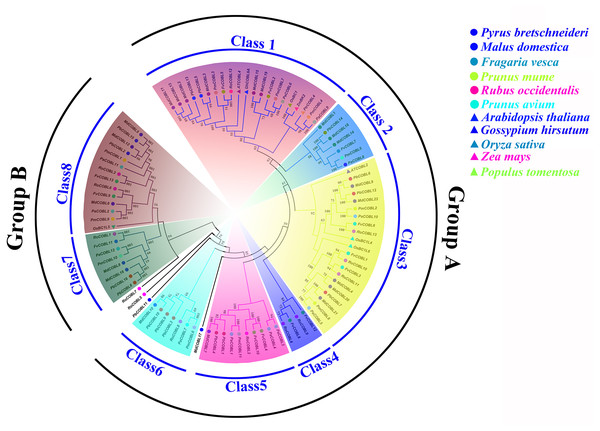
Phylogenetic analysis showed that the 87 COBRA genes were classified into two subclasses (Group A and Group B), similar to the Arabidopsis (Fig. 1, Table S5). Group A was structurally similar to AtCOBRA, and Group B had a higher similarity to AtCOBL7. We divided Group A into Class1–Class6 and Group B into Class7–Class8. MdCOBL17, RoCOBL7, RoCOBL5, and PbCOBL11 were independent branches in the evolutionary tree, and no genes related to them have been identified. Most PbCOBL genes were more tightly grouped with MdCOBLs. In Class1, AtCOBL4 and GhCOBL9A were clustered with MdCOBL11, MdCOBL13, MdCOBL2, MdCOBL3, PbCOBL13, PbCOBL5, PmCOBL3, FvCOBL5, RoCOBL12. OsBC1, ZmBK2, PtrCOBL4 with PmCOBL4, PaCOBL9 in one branch. AtCOBL2, OsBC1L4, and OsBC1L6 appeared in Class3, and OsBC1L5 was alone in Class8.
Figure 1: Phylogenetic relationships and subfamily designations in COBRA proteins from Pyrus bretschneideri, Fragaria vesca, Prunus mume, Rubus occidentalis, Prunus avium, Malus domestica, Arabidopsis thaliana, Oryza sativa, Zea mays, Gossypium hirsutum, Populus tomemtosa.
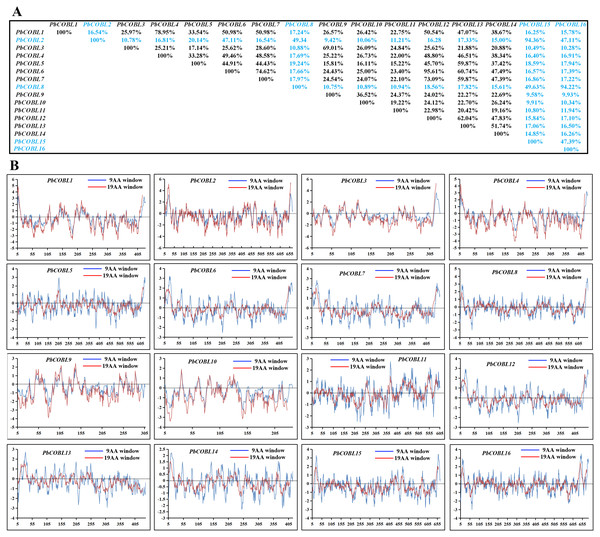
We compared the similarity of COBRA proteins of six Rosaceae species (Fig. 2, Figs. S1–S5). In Pyrus bretschneideri, comparisons among the Group B subgroup genes showed identity in the range of 47.11% to 94.36%. Within the Group A subgroup, the proteins are 15.22% to 95.61% identical. Protein similarity between the two subgroups ranged from 9.58% to 47.11% (Fig. 2). In Prunus mume, the similarities among the three members of the Group B subfamily were 53.3% for PmCOBL1-PmCOBL9, 50.66% for PmCOBL1-PmCOBL10, and 49.32% for PmCOBL9-PmCOBL10, respectively. The similarity between Group A and Group B subfamilies ranged from 12.28% to 19.25%, and the similarity between Group A subfamily members ranged from 15.27% to 76.75% (Fig. S1). In Rubus occidentalis, the similarity with other members of RoCOBL7 was low because RoCOBL7 was a separate branch in the evolutionary tree. The similarities between the three members of the Group B subclade, RoCOBL1, RoCOBL3, and RoCOBL6, were 49.78%, 56.63%, and 50.6%, respectively. The protein similarity between the two members of the two subclades ranged from 5.86% to 16.59%. The similarity of Group A subclade members ranged from 7.63%–85.40% (Fig. S2). In Fragaria vesca, Group B, Group A and two subclades individual member similarities ranged from 47.96% to 56.29%, 15.4%–93.21%, and 7.49%–19.45%, respectively (Fig. S3). In Prunus avium, the similarity of three members PaCOBL2, PaCOBL11, and PaCOBL12 was 23.45%, 41.84%, and 28.14%, respectively. The results of the comparison between subgroups were 8.05%–18.39%. Protein similarity among Group A members ranged from 6.64%–72.44% (Fig. S4). In Malus domestica, Group B, Group A and two subclades, individual member similarities ranged from 25.68%–79.05%, 5.98%–94.04%, 5.98–94.04% (Fig. S5).
Figure 2: Characteristics of COBRA proteins in Pyrus bretschneideri.
(A) COBRA member similarity comparison. (B) Comparison of hydrophobicity of COBRA members.We studied their hydrophobicity to determine if the 87 proteins were likely to have a GPI anchor similar to COBRA. We performed the hydrophobic analysis of 87 COBRA genes from six Rosaceae species. As shown in Fig. 2 and Figs. S1–S5, most amino acids showed a similar trend, with the middle part hydrophilic and the ends hydrophobic. The GPI modification sites and potential w-cleavage sites of 87 proteins were predicted using big-PI. Among the 87 proteins, the program found 34 significant potentials for GPI modification using the default parameters and proposed a possible GPI addition for the remaining three proteins (FvCOBL1, PbCOBL1, PbCOBL4, PmCOBL5, PmCOBL11, RoCOBL2, PaCOBL1). All proteins have potential w-cleavage sites.
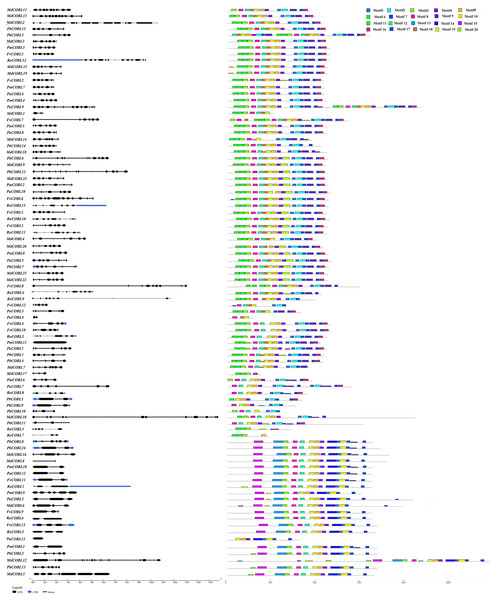
Figure 3: Predicted Pyrus bretschneideri, Fragaria vesca, Prunus mume, Rubus occidentalis, Malus domestica and Prunus avium COBRA protein conserved motifs and exon-intron structures.
(A) Gene structures of the COBRA genes. (B) Distribution of MEME motifs in COBRA genes. (C) The color and corresponding number of each motif box.Structural and conserved motif analysis of COBRA proteins
To gain a comprehensive understanding of the diversity of COBRA genes in the six Rosaceae families, we performed gene structure and conserved sequence analysis of 87 genes (Fig. 3). Group A had 66 members with the number of exons ranging from one to 12, of which 30 members had six exons, five members contained 12 exons, and one member contained 13 exons. Group B had 21 members, of which eight members contained only two exons, three members contained three exons, two members contained one exon, and one member contained four exons. In addition, PbCOBL8, PmCOBL9, PbCOBL2 contained six exons. MdCOBL16, PbCOBL15, MdCOBL5 contained seven exons and MdCOBL12 contained 13 exons. We performed a conserved structure analysis of 87 genes using MEME online software. We found that motif 8, motif 6, and motif 10 were relatively conservative, motif 15 and motif 13, and motif 14, were specific to Group B members (except for PaCOBL11), and motif 2 was specific to Group A.
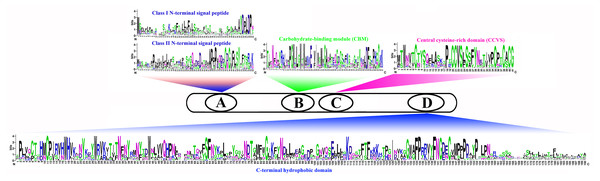
We compared the sequences of six species (Fig. 4, Figs. S6–S11). Four conserved structural domains were identified in 87 genes, N-terminal signal peptide, Carbohydrate-binding module (CBM), central cysteine-rich domain(CCVS), and C-terminal hydrophobic domain, respectively. Sequence comparison revealed that GroupB members generally had more amino acids than Group A, and the N-terminal signal peptide was different in the two subgroups. Analysis of the conserved regions of the six species COBRA members revealed that the central cysteine-rich domain (CCVS) was the relatively conserved region in both subclades, and almost all members contained the CCVS structural domain. The Central cysteine-rich domain contained a consensus N-glycosylation site, this site was mainly associated with post-translational modifications of GPI-anchored proteins and more generally with extracellular proteins. There was also an N-glycosylation site in the C-terminal hydrophobic domain. N-terminal signal peptide and C-terminal hydrophobic domain played an important role in function. However, the sequence comparison results showed that the similarity between N-terminal signal peptide and C-terminal hydrophobic domain in each subgroup was not high, suggesting that there was no solid selective pressure on these areas as long as their hydrophobic nature was considered.
Figure 4: Conserved COBRA domain composition. All 87 COBRA genes had a characteristic ID domain.
Alignment analysis of the 87 COBRA gene sequences using the ClustalW tool in MEGA 7.0 software. These domain diagrams were plotted using the online WebLogo tool.Chromosomal location and duplication events of COBRA family genes in six Rosaceae
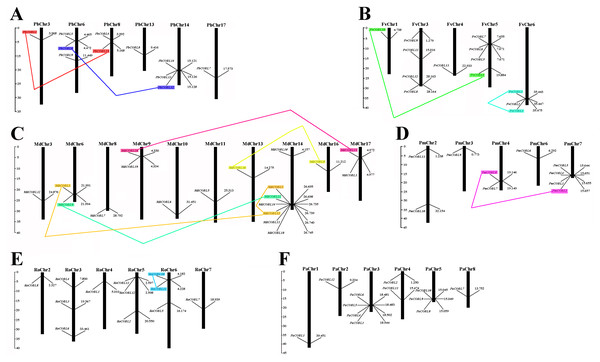
Based on the genome-wide data of pear, strawberry, black raspberry, sweet cherry, japanese apricot, and apple, all COBRA genes exact chromosomal physical localization was determined, as shown in Fig. 5. In pear, 11 PbCOBL genes were distributed on six chromosomes (Chr3, Chr6, Chr8, Chr13, Chr14, Chr17), and five genes were not localized on any chromosome. In strawberry, 13 genes were located on five chromosomes (except Chr2 and Chr7). In Japanese apricot, four genes were distributed on chromosomes 7; two genes on chromosomes 2 and 4; and one gene on chromosomes 3 and 6, respectively. In black raspberry, there were no genes on chromosome 1; three genes on chromosomes 3, 5, and 6, and one gene on chromosomes 2, 4 and 7. In apple, 18 genes were distributed on chromosomes 3, 6, 8, 9, 10, 11, 13, 14, 16 and 17 (except Chr1, 2, 4, 5, 7, 12 and 15). In sweet cherry, 12 genes were distributed on six chromosomes (except Chr6 and 7), of which there were four genes on chromosome 3, only one gene on chromosome 1, 2 and 8, two genes on chromosome 4 and three genes on chromosome 5.
Figure 5: Chromosomal locations of six Rosaceae species.
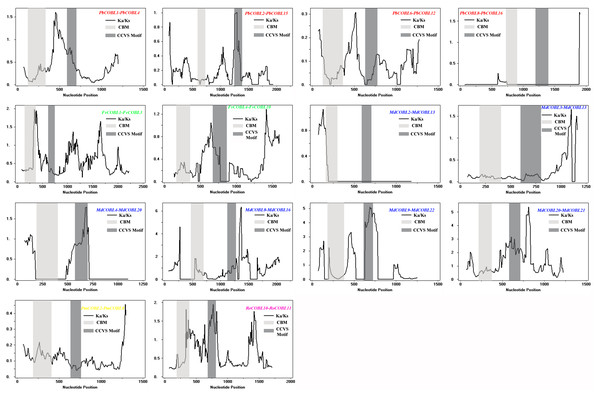
Chromosomal locations of COBRA genes in Pyrus bretschneideri (A), Fragaria vesca (B), Malus domestica (C), Prunus mume (D), Rubus occidentalis (E), and Prunus avium (F). Duplicated gene pairs are connected with colored lines.To understand the role of drivers of gene duplication in the evolutionary process, we calculated Ka, Ks, and Ka/Ks ratios of duplicated gene pairs in six Rosaceae species (Fig. 5, Table S3). Ka/Ks = 1 is the cut-off value that indicates neutral selection, Ka/Ks < 1 represents negative selection and Ka/Ks > 1 represents a positive selection. All 14 duplicated gene pairs were identified in pear, strawberry, apple, Japanese aprico, and black raspberry. There were four duplicated gene pairs in pear, two pairs in strawberry, six pairs in apple, one pair in plum and 1 pair in black raspberry. Among the 14 replicated gene pairs, only MdCOBL3-MdCOBL13 and RoCOBL10-RoCOBL11 Ka/Ks > 1 were 1.272 and 1.141, respectively, and the remaining replicated gene pairs had Ka/Ks < 1. Among the 14 replication gene pairs, 12 gene pairs experienced segmental duplication, and only two pairs of genes (RoCOBL10-RoCOBL11, FvCOBL1-FvCOBL3) experienced tandem duplication. To gain a comprehensive understanding of the selection pressure on COBRA genes, we performed a sliding window analysis (Fig. 6), which showed that in 14 pairs of gene replication events, most of the Ka/Ks loci were less than 1, and only very few loci had Ka/Ks values greater than 1.
Figure 6: Sliding window plots of duplicated COBRA genes.
Analysis of cis-acting elements of COBRA gene family promoter in Pyrus bretschneideri
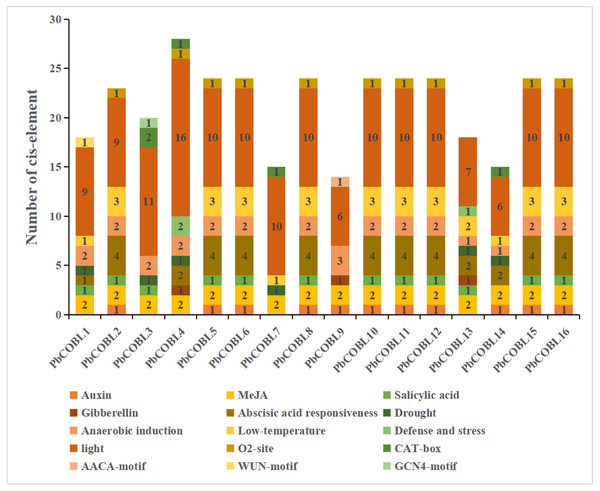
In order to study the specific expression of the COBRA gene, We used the plant care database to study cis-acting elements of promoters of 16 COBRA genes in Pyrus bretschneideri (Fig. 7, Table S4). The promoter of COBRA genes contained many cis-acting elements related to hormones, including responses to Auxin-responsive element (TGA-box, AuxRE), MeJA-responsiveness (CGTCA-motif, TGACG-motif), Salicylic acid responsiveness (TCA-element), Gibberellin-responsiveness, Abscisic acid responsiveness. CGTCA-motif and GACG-motif cis-acting elements were identified in all 16 genes. A total of 11 cis-acting elements associated with auxin were found in 11 members. Among them, AuxRE was only present in PbCOBL14, TGA-box was not contained in PbCOBL1,3,4,7,13,14, and all other genes were contained. TCA-element, which are cis-acting elements involved in salicylic acid responsiveness, were identified in PbCOBL1, 2, 3, 5, 6, 8, 10, 11, 12, 13, 15 and 16. Gibberellin-responsiveness (P-box, GARE-motif, TATC-box) was identified only in PbCOBL4,9 and 13. Abscisic acid responsiveness (ABRE) were more prominentin numbers, 43 ABRE cis-acting elements were identified in 13 genes, and only PbCOBL3,7,9 was not identified with ABRE.
Figure 7: Promoter cis-elements of the 16 PbCOBLs.
It is related to plant growth and development, including participating in light response elements (G-box, GT1 motif, GATA motif, Sp1, Box 4, TCT-motif, AE-box, Lamp element, CHS-cma1a, GA motif, I-box, 3-af1 binding site, ACE, TCCC motif) were identified 154 times in 16 genes. The cis-acting regulatory element O2-site involved in the regulation of maize alcohol-soluble protein metabolism was identified in 10 members (PbCOBL2,4,5,6,8,10,11,12,15 and 16). Meristem expression (CAT-box) was identified in PbCOBL3,4,7 and14. Endosperm specific negative expression (AACA motif). Wound-responsive element (WUN-motif) and Endosperm expression (GCN4-motif) were only identified in PbCOBL9. cis-acting elements associated with biotic and abiotic stress responses were also identified in COBRA genes. Six PbCOBL genes (PbCOBL1, 3, 4, 7, 13, and 14) had MBS, and two members (PbCOBL3,14) contained TC-rich repeats. Anaerobic induction (ARE) was identified in 15 genes (Except PbCOBL7). A total of 32 identifications in 13 members of the low-temperature responsiveness (LTR). COBRA family promoters were mostly engaged in hormone response and light response processes. Plant growth and development are facilitated by the reaction to various hormones and light responses.
Expression characteristics of Chinese white pear COBRA genes
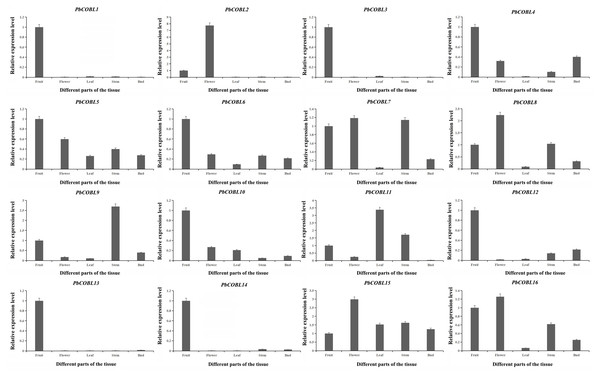
To gain a deeper understanding of the COBRA gene family, we performed expression pattern analysis in the stems, leaves, fruits, flowers and buds of Dangshan su pear (Fig. 8). The results showed that PbCOBL1,3,10,12,13,14 were highly expressed only in fruits and almost not in other tissues. PbCOBL2 was highly expressed in flowers. PbCOBL5,15 were expressed at relatively high levels in several tissues. PbCOBL9 was highly expressed in stems. PbCOBL4,6,7,8,16 were expressed at low levels in leaves and at high levels in other tissues.
Figure 8: Expression patterns of COBRA genes of Chinese white pear in different organs.
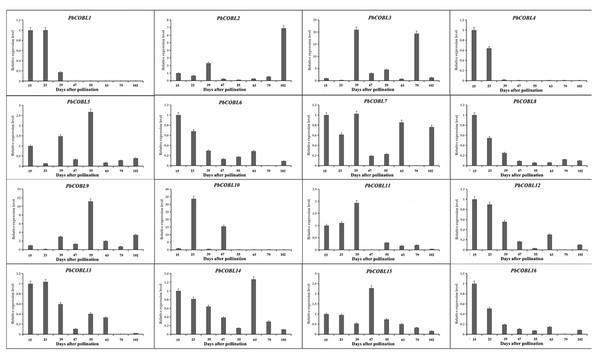
We examined the expression pattern of the COBRA gene family in eight periods of fruit development of Dangshan su pear (Fig. 9). As shown in the figure, PbCOBL1 and PbCOBL4 were substantially expressed at 15 and 23 DAP and non significant expression were found on remaining time interval. PbCOBL6,8,11,12,16,13 showed similar expression patterns, with higher expression in early fruit development (15 DAP, 23 DAP, 39 DAP) and a decreasing trend in late fruit development. PbCOBL10 expression showed two peaks at 23 DAP and 47 DAP during eight periods of fruit development, and PbCOBL2 showed a similar expression pattern with PbCOBL10, with two peaks at 39 DAP and 102 DAP. PbCOBL7 was barely expressed in 79 DAP. PbCOBL5 and PbCOBL9 showed a peak at 55 DAP, with lower expression in other periods. PbCOBL14,15 were expressed at each period of fruit development, with PbCOBL14 highly expressed at early fruit development (15,23,39 DAP) and 63 DAP, PbCOBL15 peaking at 47 DAP, and low expression at other periods. PbCOBL3 was mainly highly expressed in 39 DAP and 79 DAP.
Figure 9: Expression patterns of COBRA genes of Chinese white pear in fruit at different developmental stages.
Discussion
COBRA, a glycosyl-phosphatidyl inositol-anchored protein (GPI), generally had an N-terminal signal peptide, Carbohydrate-binding module (CBM), central cysteine-rich domain (CCVS), and a hydrophobic C-terminal hydrophobic domain. Among them, CBM was a functional structural domain with strong binding to crystalline cellulose. The CCVS region was mainly involved in forming disulfide bonds or metal ion binding with cysteine-rich features (Roudier et al., 2002; Liu et al., 2013; Niu et al., 2018). Previous studies found that the COBR family was involved in cell wall biosynthesis-related to plant roots, stems, leaves and fruit ripened (Dai et al., 2009; Brown et al., 2005; Persson et al., 2007).
In this study, a total of 87 COBRA genes were identified in six Rosaceae species. The pI values of 87 members ranged from 5.04 (MdCOBL16) to 9.59 (MdCOBL1). The proteins encoded by the COBRA gene include acidic protein and basic protein. Except for MdCOBLl17 and PaCOBL3, the other 85 genes were negative, indicating that most COBRA proteins were hydrophilic proteins (Table S2). Among Rosaceae species, pear had the second-highest number of COBRA members after apples, a phenomenon that might be related to whole-genome duplication (WGD). Studies have shown that WGD and chromosome rearrangements were accompanied by chromosome doubling, altered gene sequences, and also extensive gene loss (Tang et al., 2008). The common ancestor of Rosaceae has nine chromosomes. Apple and pear experienced WGD twice in 130 million years ago (Mya) and 30–45 Mya, while the other four species only experienced WGD of 130 million years ago (Mya) (Shulaev et al., 2011; Wu et al., 2013). The long process of biological evolution was accompanied by chromosome doubling, breaking, and rearrangement, resulting in 17 chromosomes for pears and apples, eight for japanese apricot and sweet cherry, and seven for strawberry and black raspberry. During the evolution of the COBRA gene family, the number of COBRA genes in pears might be less than that of apples due to the loss of members. For phylogenetic analysis, the 87 COBRA members were divided into two subclades which was further subdivided Subclade A (Class1–Class6) and subclade B (Class7–Class8). Among them, MdCOBL17, RoCOBL7, RoCOBL5, and PbCOBL11 were independent branches in the evolutionary tree, and no genes related to them were identified. Interestingly, the COBRA genes of pear and apple were present in Class1–Class8, at least one gene, and this phenomenon might be due to the involvement of pear and apple in the second WGD.
Gene function was strongly related to gene structure and conserved sequence, with similar conserved structural domains in the same family and higher similarity in the same subclade, implying that members of the same subclade may have similar functions to each other. For example, the FvCOBL5 and PmCOBL3 genes were structurally similar (similar number and length of exons). In terms of gene structure, Group A members had a high number of exons, and analysis of conserved regions revealed that the same subgroup had approximately the same conserved protein regions, with differences between different subgroups. A previous study found that COBL7 differs from COBRA in the N-terminal signal peptide region, and a specific 170 amino acid sequence of the COBL7 subfamily was found in several species, which overlaps with the COBRA subfamily N-terminal signal peptide after 170 amino acids (Roudier et al., 2002). This difference might result in differences in the function of the two subgroups. A similar phenomenon was found in 87 COBRA genes of six Rosaceae (Figs. S6–S11). Gene duplication events were often followed by the differentiation of gene functions, such as the creation of new functions and the loss of old ones. Thus, gene duplication events were the driving force of biological evolution, allowing organisms to become more adapted to the diversity of their environment (Chao et al., 2017; Tang et al., 2016). Previous studies found that gene replication events have been found in multiple gene families, such as the PKS gene family in cotton and the MADS-box gene family in pear (Su et al., 2017; Meng et al., 2019). In current study, 14 pairs of replication gene pairs were identified, including four pairs of pear, six pairs of apple, one pair of strawberry, one pair of Japanese aprico, and one pair of black raspberry. Among them, FvCOBL1-FvCOBL3 and RoCOBL10-RoCOBL11 underwent tandem duplication, and the remaining 12 gene pairs underwent segmental duplication. Apples and pears had significantly more replicate gene pairs than the other four Rosaceae species, probably because pears and apples experienced two WGDs while the other four Rosaceae species experienced one WGD (Wu et al., 2013; Zhang et al., 2012). We calculated the Ka, Ks and Ka/Ks values for 14 replicated gene pairs. We found that the Ka/Ks values of MdCOBL3-MdCOBL13 and RoCOBL10-RoCOBL11 were greater than 1, indicating that these gene pairs underwent rapid evolutionary diversification after a duplication event during evolution. The Ka/Ks of other genes is less than 1, implying that these gene pairs have been experiencing a markedly purifying selection during evolution (Table S3).
The overall analysis of expression profiles in different tissues will contribute to studying the tissue-specific and dynamic expression of COBRA genes in pear. The high expression of PbCOBL15, PbCOBL5 in different tissues suggested that PbCOBL15, PbCOBL5 played an important role in the growth and development of Dangshan su pear (Fig. 9). A previous study found that COBRA gene family was involved in the synthesis of secondary wall cellulose (Dai et al., 2011; Liu et al., 2013; Brown et al., 2005). In our study, we found that PbCOBL1,3,12,13,14 were only highly expressed in fruits and hardly expressed in other tissues. It was also found that the cis-acting elements of their promoter contained many hormone response elements and light response elements. Interestingly, only the PbCOBL13 promoter contained the gibberellin responsiveness element, but not in the other three genes, which might be the reason for the functional difference (Fig. 7).
Previous studies found that the development of stone cells in Dangshan su pear fruit increased initially and then decreased, starting from 7 DAP and reaching the peak at 55 DAP (Zhang et al., 2017; Su et al., 2019b). The expression patterns of these 16 PbCOBLs at eight developmental stages of fruit showed that the expression pattern of any one gene was not consistent with the trends obtained for the Dangshan su pear fruit stone cell. However, there were two special genes, PbCOBL12 and PbCOBL13, which were highly expressed in the early stage of fruit development (15 DAP, 23 DAP, and 39 DAP). The 15 DAP-39 DAP is a process of massive accumulation of lithocytes accompanied by high expression of PbCOBL12 and PbCOBL13 (Fig. 8). According to the phylogenetic results, PbCOBL13 clustered with AtCOBL4 as a branch, and PbCOBL12 was in Class3 with AtCOBL2 (Fig. 1). Previous studies illustrated that AtCOBL4 and AtCOBL2 ultimately affect secondary wall formation by regulating the expression of cellulose synthase (Ben-Tov et al., 2015; Brown et al., 2005). Protein three-dimensional structures prediction showed that PbCOBL12 was similar to AtCOBL2 and PbCOBL13 was similar to AtCOBL4 (Fig. S12). We speculate that PbCOBL12, and PbCOBL13 are mainly expressed in fruit and have similar functions to AtCOBL2 and AtCOBL4 in regulating SCW formation in pear fruit cells by regulating the expression of the genes encoding key cellulose synthesis enzymes.
Conclusions
In this study, 87 COBRA genes were identified in six Rosaceae species. We analyzed the evolutionary relationship between COBRA in six species using evolutionary analysis, hydrophobicity analysis, gene structure, and conservative sequence analysis, cis-acting element analysis, gene duplication and slide window analysis, spatiospatiotemporal expression pattern analysis, and screened PbCOBL12 and PbCOBL13 as key genes regulating secondary wall during Dangshan su pear fruit development.
Supplemental Information
Characteristics of cobra family proteins in Prunus mume. A: COBRA member similarity comparison. B: Comparison of hydrophobicity of COBRA members
Characteristics of cobra family proteins in Rubus occidentalis. A: COBRA member similarity comparison. B: Comparison of hydrophobicity of COBRA members
Characteristics of cobra family proteins in Fragaria vesca. A: COBRA member similarity comparison. B: Comparison of hydrophobicity of COBRA members
Characteristics of cobra family proteins in Prunus avium. A: COBRA member similarity comparison. B: Comparison of hydrophobicity of COBRA members
Characteristics of cobra family proteins in Malus domestica A: COBRA member similarity comparison. B: Comparison of hydrophobicity of COBRA members
Predicted three-dimensional structures of Pb COBRL1 3, PbCOBL12, AtCOBL 2, AtCOBL4
Basic information of COBRA genes in five Rosaceae species
The COBRA genes of Fragaria vesca, Prunus mume, Rubus occidentalis, Malus domestica and Prunus avium identified in this study are listed.