HIF1α is dispensable for oocyte development and female fertility in mice
- Published
- Accepted
- Received
- Academic Editor
- Qing-Yuan Sun
- Subject Areas
- Biochemistry, Cell Biology, Developmental Biology, Molecular Biology
- Keywords
- HIF, Hypoxia, Oocyte, Embryo, Fertility
- Copyright
- © 2022 Chen et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. HIF1α is dispensable for oocyte development and female fertility in mice. PeerJ 10:e13370 https://doi.org/10.7717/peerj.13370
Abstract
Background
It has been thought that oocyte may develop in a low oxygen environment, as changes in follicle structure and formation of a fluid-filled antrum. The survival of hypoxic tissues is controlled by hypoxia-inducible factors (HIFs) that are activated in a low oxygen state. HIF1α is expressed in mature mouse oocytes and continues to be expressed after fertilization, from the 2-cell to blastocyst stage. However, the physiological roles of HIF pathway during oogenesis and embryogenesis have still not been elucidated in detail.
Methods
Mutant mice with oocyte-specific HIF1α deletion were generated by crossing Hif1αfl/fl mice with transgenic mice expressing Gdf9-promoter-mediated Cre recombinase. Breeding assay was carried out to detect female fertility. In vitro fertilization and embryo culture were used to assess early embryo development. Oocyte meiotic progression was also examined. Quantitative RT-PCR was used for analyzing of candidate genes expression.
Results
We successfully generated mutant mice with oocyte-specific deletion of HIF1α. Oocytes loss of HIF1α did not affect female fertility, ovulation and early embryo development. Moreover, oocytes can mature in vitro, and form well-organized spindle in the absence of HIF1α. In addition, pronounced differences in Hif2α and Hif3α mRNA expression were not observed in HIF1α-deleted oocytes. These results revealed that HIF pathway in oocytes is not essential for female fertility.
Introduction
Oxygen occupies a central role in the maintenance of life as we known it, perhaps most prominently in aerobic metabolism, where O2 serves as the terminal electron acceptor in oxidative phosphorylation (Kaelin Jr & Ratcliffe, 2008). Inadequate oxygen availability can lead to cellular dysfunction and, if sufficiently profound, cell death, in aerobic organisms (Kaelin Jr & Ratcliffe, 2008). As organisms become larger and more active, oxygen transport by simple diffusion becomes limiting. To maintain oxygen homeostasis, mammals have evolved specialized networks to maintain oxygen homeostasis at the tissue level. One of the critical aspects of this network is oxygen-dependent posttranslational hydroxylation of a transcription factor called hypoxia-inducible factor (HIF) (Giaccia, Simon & Johnson, 2004; Kaelin Jr & Ratcliffe, 2008). HIF is a heterodimer of bHLH-PAS (basic-helix-loop-helix, per-ARNT sim) proteins and consists of an unstable α-subunit and a constitutively and ubiquitously expressed β-subunit HIF1β (Bruick, 2003; Semenza, 2001). HIFα subunits exist as a series of isoforms encoded by distinct genetic loci. Three HIFα proteins have been found in higher metazoans, and HIF1α and HIF2α are able to interact with hypoxia response elements to activate transcription. Under well-oxygenated conditions, hydroxylation of one (or both) of two highly conserved prolyl residues located near the NTAD by prolyl hydroxylase domain-containing (PHD) proteins mediates interactions with the von Hippel-Lindau (VHL) E3 ubiquitin ligase complex that targets HIFα for proteasomal destruction (Ivan et al., 2001; Maxwell et al., 1999). Accordingly, hypoxia can inhibit HIFα hydroxylation, which leads to HIFα accumulation and nuclear translocation, and consequently activating genes involving in widespread biological processes (Kaelin Jr & Ratcliffe, 2008).
The ovarian follicle provides the oocyte with ideal environment for growth and development in preparation for ovulation and fertilization. In ovary, as the follicle enlarges, a basement membrane separates theca and granulosa cell layers, and thus antral follicles are not directly adjacent to vascular support. In addition, the formation of a fluid-filled antrum physically separates the cumulus-oocyte complex (COC) from granulosa layers (Redding, Bronlund & Hart, 2007; Rodgers & Irving-Rodgers, 2010). Consequently, the oocyte itself might develop in a potential hypoxia environment. There exists a paradox in that while the maturing oocyte resides in an avascular environment, it also relies heavily on oxidative phosphorylation which requires oxygen (Thompson et al., 2015). The activation of HIF target genes had been implicated in resolving this paradox. During follicle development and corpus formation, VEGF (vascular endothelial growth factor), the main inducer of angiogenesis, was upregulated by HIF. In addition, ovulation is also intrinsically linked to HIF activity through the ovulatory luteinizing hormone surge increasing HIF expression. Furthermore, HIF1α is presented in mature mouse oocytes and continues to be expressed from fertilization to blastocyst stage (Takahashi et al., 2016).
However, the physiological roles of HIF pathway during oogenesis and embryogenesis have still not been elucidated in detail. In this study, we delete Hif1α specifically in oocytes by Cre-loxp conditional knockout (Yu et al., 2013) and show that HIF1α deletion exerts little effect on oocyte maturation and embryo development in vivo and in vitro.
Materials and Methods
Mice
Animal care and use were carried out in accordance with the guiding principles of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (Approval No. IACUC-1806013). Mice housed in 12–12 h light-dark cycle, with constant temperature and with food and water provided ad libitum under SPF (specified pathogen free) conditions. Mice were randomly divided into cages, and each cage was capable of housing 4–5 mice. All cages were maintained in similar conditions, including cages density, bedding, and sanitation frequency. Mice were anesthetized with carbon dioxide for oocyte collection. No animals survived at the end of study.
Mice possessing loxP sites flanking exon 2 of the Hif1α gene were kindly provided by Dr. Jin Hou at Second Military Medical University (Liu et al., 2019). Gdf9-Cre transgenic mice were a gift from Dr. Heng-Yu Fan (Yu et al., 2013). To generate Hif1αfl/fl; Gdf9-Cre mice (referred to as Hif1α-cKO), female mice carrying the Hif1α floxed alleles were mated with Gdf9-Cre males (Yu et al., 2013). The Hif1 αfl/fl female mice were used as the control group (referred to as Control). Genotyping for LoxP and Cre were carried out using PCR amplification. Primers for Hif1α Loxp (Forward: 5′—AGTTACAGGTATTTATGAGCCA—3′, Reverse: 5′—CTAGTTGATCTTTCCGAGGAC—3′), and Gdf9-Cre (Forward: 5′—GGCATGCTTGAGGTCTGATTAC—3′, Reverse: 5′—CAGGTTTTGGTGCACAGTCA—3′) were used at 10 pmol using Vazyme PCR mix following manufacture’s protocol.
Fertility test
To perform fertility test, seven pairs of 8 weeks Control and Hif1α-cKO female mice were randomly selected and continually bred with WT male mice which have been confirmed fertility for 6 months. The mice were checked every week and the date and number of pups and litter size was recorded for each litter.
Oocyte collection and in vitro maturation
To collect GV oocytes, 6-8 weeks of females (n = 5) were stimulated with 5 IU PMSG (pregnant mare serum gonadotropin) (Ningbo, No. 2 hormone factory, Zhejiang, China). After 44–48 h, GV oocytes were carefully isolated from antral ovarian follicles by manual rupturing of antral ovarian follicles. For in vitro maturation, Oocytes were cultured in M2 media under mineral oil at 37 °C in a 5% of CO2 incubator.
To obtain MII oocytes, mice (n = 5) were induced to superovulate by IP injection of 5 IU PMSG followed 48 h later by injection of 5 IU hCG (Human Chorionic Gonadotropin). 16 h after hCG injection, mice were sacrificed and the oviducal ampullae were broken to release the cumulus-oocyte complexes. MII oocytes were freed of cumulus cells by exposure to 0.2% hyaluronidase.
In vitro fertilization and embryo culture
IVF assays were conducted as we described previously (Han et al., 2018). Briefly, normal sperm were isolated from the dissected epididymis of C57BL/6 male mice aged 10–20 weeks and left to capacitate for 1 h in HTF fertilization medium (Millipore, Merck) supplemented with 10 mg/ml BSA (bull serum albumin). Cumulus–oocyte complexes (COCs) were isolated from oviduct ampullae, and placed in other HTF fertilization medium (Millipore, Merck) supplemented with 10 mg/ml BSA. Then, dispersed spermatozoa were added to HTF drops containing COCs for fertilization in a 37 °C incubator. After co-incubation for 6∼9 h, presumptive zygotes were washed to remove cumulus cells and excess sperm, and then transferred into KSOM medium (Merck Millipore, Burlington, MA, USA) for further culture. Early embryo development potential was assessed at the indicated time points during culture.
Western blot
Samples each containing 100 MII oocytes from 5 mice were collected in SDS sample buffer and heated for 5 min at 100 °C. The proteins were separated by 10% SDS-PAGE and then transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, Darmstadt, Germany) by electrophoresis. After transfer, the membranes were blocked in PBST buffer containing 5% skimmed milk for 1 h, followed by incubation overnight at 4 °C with 1:2,000 anti-HIF1α antibody (ab237544; Abcam, UK) and 1:1,000 anti-α-Tubulin antibody (ab7291; Abcam, UK). After multiple washes, the membranes were incubated with horseradish peroxidase conjugated antibody for 1 h at room temperature. Finally, the bands were visualized using an ECL Plus Western Blotting Detection System (GE Healthcare, Little Chalfont, UK).
Immunofluorescence
MII oocytes from mice were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature, and permeabilized with 0.5% Triton X-100 for 20 min. Following incubation in 1% BSA blocking buffer for 1 h at room temperature, oocytes were incubated with FITC-conjugated anti-Tubulin antibody (F2168; Sigma-Aldrich) at 4 °C overnight. After rinsing with PBS, the oocytes were stained with propidium iodide (PI; 10 µM in PBS). Then the oocytes were mounted on glass slides and examined with a confocal laser scanning microscope (LSM 710; Carl Zeiss, Jena, Germany).
RNA extraction and Real-time quantitative PCR
Total RNA was extracted from 50 oocytes using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems, CA, USA). Then RNA from each group was reverse transcribed into cDNA using a QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. qRT-PCR was conducted using AceQ qPCR SYBR Green Master Mix (High ROX Premixed) (Vazyme) with Applied Biosystems StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, Massachusetts, USA). Data were normalized against Gapdh and quantification of the fold change was determined by the comparative CT method, as previously described (Li et al., 2020). The related primers are listed below: Hif1α F: 5′-GCACCGATTCGCCATGGAG-3′, R: 5′-TCTAGACCACCGGCATCCAG-3′; Hif1α F: 5′-TCCTTCGGACACATAAGCTCC-3′, R: 5′-GACAGAAAGATCATGTCACCGT-3′; Hif1α F: 5′-GAAGTTCACATACTGCGACGA-3′, R: 5′-GTCCAAAGCGTGGATGTATTCAT-3′; Gapdh: 5′-AGGTCGGTGTGAACGGATTTG-3′, R: 5′-TGTAGACCATGTAGTTGAGG TCA-3′.
Statistical analysis
All experiments were repeated at least three times. GraphPad Prism 7 software (GraphPad Software, San Diego, CA, USA) was used to analyze data and draw graphs. Student’s t test was used for statistic comparison. Data were reported as mean ± SD, and p < 0.05 were considered statistically significant. n.s., not significant.
Results
Generation of mutant mice with oocyte-specific deletion of Hif1α
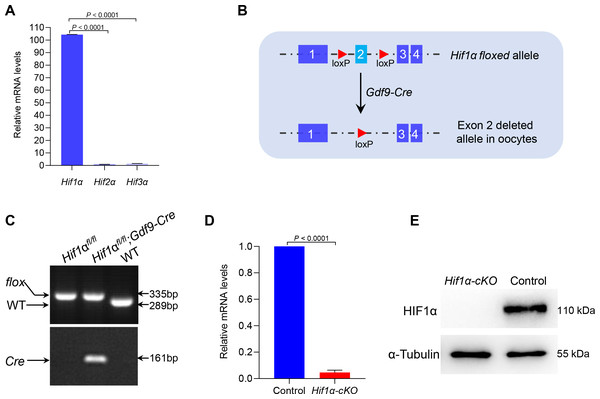
So far, three different alpha subunits have been known to exist in higher metazoans, termed HIF1α, HIF2α and HIF3α (Webb, Coleman & Pugh, 2009). By performing quantitative real-time PCR, we showed that Hif1α was predominantly expressed in oocytes (Fig. 1A). This observation suggests that HIF1α may have an important role in oocyte development. To investigate this, we generated mutant mice in which exon 2 of the Hif1α gene was targeted (Fig. 1B). This was achieved by crossing Hif1αfl/fl mice with transgenic mice expressing Gdf9 promoter-mediated Cre recombinase, which mediates recombination in mouse oocytes at the primordial stage, to knock out Hif1α specifically in oocytes (Yu et al., 2013). Hereafter, Hif1αfl/fl; Gdf9-Cre and Hif1αfl/fl mice are referred to as Hif1α-cKO and Control mice, respectively. Mice homozygous for this floxed allele and positive for Gdf9-Cre transgene were validated by genotyping PCR (Fig. 1C). qRT-PCR confirmed that oocytes from Hif1α-cKO mice had undetectable Hif1α mRNA as compared to those from Control mice (Fig. 1D). Moreover, immunoblotting result showed the absence of HIF1α protein in Hif1α-cKO oocytes (Fig. 1E), suggesting that Gdf9- mediated Cre excision of Hif1α is sufficient to delete HIF1α protein in mouse oocytes.
Figure 1: Targeted deletion of the Hif1α in mouse oocytes.
(A) qRT-PCR analysis of Hifα subunits mRNA levels in oocytes from WT mice. The relative mRNA levels of Hif3α in WT oocytes were set to 1.0. Data represent the mean ± SD (n = 3). (B) Schematic representation of Hif1α exon 2 deletion by Gdf9-Cre-mediated recombinase in oocytes. (C) PCR genotyping results of Hif1αfl/fl mice and Gdf9-Cre recombinase mice from DNA obtained from tail samples. A single 289 bp band and a single 335 bp band corresponded to the WT and homozygous floxed mice (Hif1αfl/fl) respectively (top); a single 161 bp band indicated the Gdf9-Cre transgene (bottom). (D) qRT-PCR analysis of Hif1α mRNA levels in oocytes from Control and Hif1α-cKO females. The relative mRNA level of Hif1α in Control oocytes was set to 1.0. Data represent the mean ± SD (n = 3). (E) Western blot showing the absence of HIF1α protein expression in Hif1α-cKO oocytes. MII oocytes were collected for analysis, and 100 oocytes were used for each sample. Level of α-Tubulin was used as an internal control. The experiments were repeated three times. Student’s t test (two-tailed) was used for statistical analysis.HIF1α was dispensable for female fertility
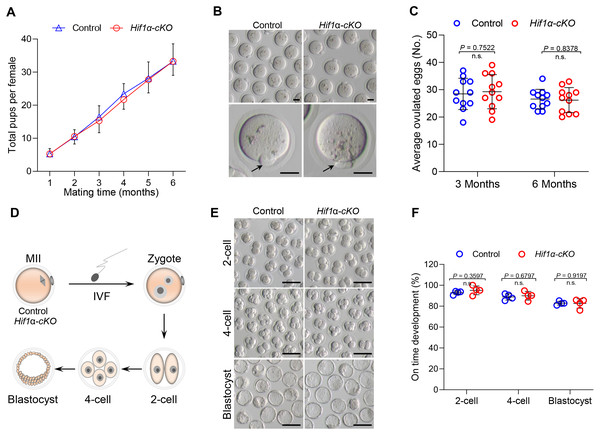
To detect the effect of HIF1α deletion on female fertility, breeding assay was carried out by mating Hif1α-cKO or Control mice with males of proven fertility for 6 months. As shown in Fig. 2A, female Hif1α-cKO mice were fertile and gave birth to pups comparable to that of Control mice. To assess oocyte quality, we superovulated Control and Hif1α-cKO females and collected oocytes at 16 h post-hCG injection. The oocytes collected from Hif1α-cKO ovaries were morphologically normal, displaying intact first polar body (Fig. 2B). In addition, there was no difference between the number of ovulated eggs from the Control and Hif1α-cKO ovaries (Fig. 2C). We then asked whether HIF1α deletion in oocytes would adversely affect the developmental competences of subsequent embryos. To do this, we carried out in vitro fertilization (IVF) of oocytes derived from Control and Hif1α-cKO mice, and cultured fertilized embryos in vitro to monitor early embryo development (Fig. 2D). Zygotic embryos from Hif1α-cKO oocytes exhibited similar on-time progression to 2-cell, 4-cell and blastocyst stages relative to Controls (Figs. 2E, 2F). These results show that loss of HIF1α in oocytes does not affect the fertility of female mice.
Figure 2: Loss of HIF1 α in oocytes had little effects on mouse fertility.
(A) Cumulative numbers of pups per female born from Control and Hif1α-cKO mice for 6 months (n = 7). (B) Representative bright-field images of ovulated MII oocytes from Control and Hif1α-cKO females. Arrows indicate the first polar body. Scale bars = 30 µm. (C) Average number of oocytes obtained after superovulation from Control and Hif1α-cKO females at indicated age (n = 11). (D) Schematic diagram of in vitro fertilization and embryo culture. (E) Representative bright-field images of Hif1α-cKO oocyte-derived E1.5, E2.5 and E4 embryos. Scale bars = 100 µm. Control, n = 89; Hif1α-cKO, n = 79. (F) The percentage of Hif1α-cKO oocyte-derived embryos that successfully progressed to the 2-cell, 4-cell and blastocyst stage at E1.5, E2 and E4 during in vitro culture. Data are expressed as mean ± SD. Student’s t test (two-tailed) was used for statistical analysis.HIF1α deletion exerts little effect on oocyte maturation and spindle organization in vitro
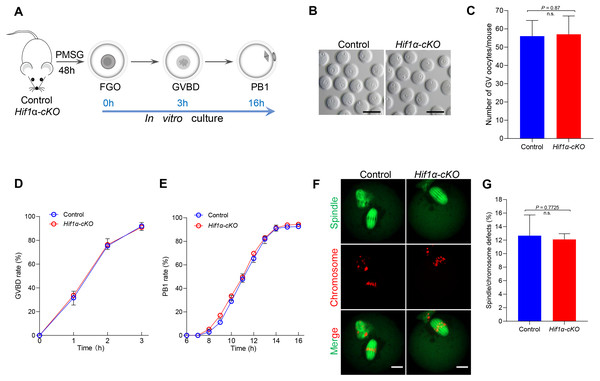
Given that HIF1α is not essential for oocyte maturation in vivo, we next studied how Hif1α-cKO oocytes matured in vitro. Fully-grown oocytes harvested from hormonally stimulated Control and Hif1α-cKO mice were cultured in maturation media to assess meiotic resumption within 3 h, and PB1 extrusion within 16 h (Fig. 3A). Comparable numbers of fully-grown germinal vesicle-stage (GV) oocytes were obtained from Control and Hif1 α-cKO mice ovaries (Figs. 3B, 3C), suggesting that the ovarian reserve is not compromised in the absence of HIF1α. Then, the collected GV oocytes were released for maturation in vitro. Meiotic resumption, as indicated by GV breakdown (GVBD), occurred in 80% of Hif1α-cKO oocytes during the first 2 hours’ culture, similar to the rate observed in Control oocytes (Fig. 3D). Next, we analyzed the exit from MI, marked by first polar body extrusion (PBE). We found that PBE began around 8–10 h for both Control and Hif1α-cKO oocytes and that both attained maximal PB1 rates of around 95% by 14 h (Fig. 3E). In addition, we examined spindle organization and chromosome alignment in Control and Hif1α-cKO oocytes that had extruded PB1. Immunofluorescence results showed that barrel-shape spindle and well-aligned chromosomes were readily observed in Hif1α-cKO oocytes, and that errors in meiotic apparatus were not markedly increased (Figs. 3F, 3G). Thus, loss of HIF1α does not compromise oocyte maturation and spindle organization in vitro.
Figure 3: HIF1α is not required for oocyte meiotic maturation in vitro.
(A) Schematic diagram of GV oocyte collection and in vitro maturation. (B) Representative bright-field images of GV oocytes collected from Control and Hif1α-cKO mice. Scale bar = 100 µm. (C) Mean number of GV stage oocytes obtained per mouse after priming with PMSG (n = 6 for each genotype). (D, E) GVBD and PB1 rates of oocytes from 8-week-old Control and Hif1α-cKO mice. Control, n = 67; Hif1α-cKO, n = 84. (F) Oocytes from Control and Hif1α-cKO mice were immunolabeled with α-Tubulin to label spindle (green) and co-stained propidium iodide to visualize chromosomes (red). Scale bar, 20 µm. (G) Quantification of Control (n = 78) and Hif1α-cKO oocytes (n = 69) with spindle defects or chromosome misalignment. Data are expressed as mean ± SD. Student’s t test (two-tailed) was used for statistical analysis. n.s., not significant.Loss of HIF1α did not disrupt the expression pattern of other HIF isoforms in oocytes
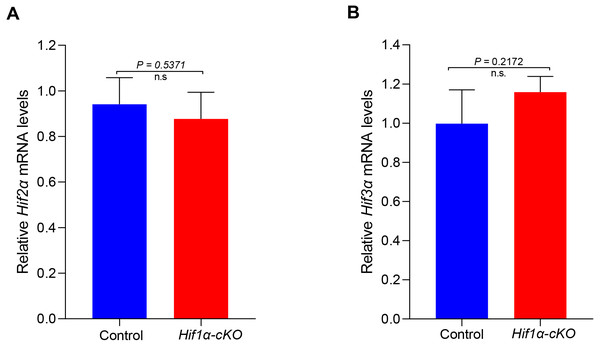
HIFα subunits exist as a series of isoforms encoded by distinct genetic loci. Among three HIFα isoforms, HIF1α and HIF2α appear closely related and can interact with hypoxia response elements (HREs) to activate transcription (Wiesener et al., 1998). We speculated that other subunits might compensate for the role of HIF1α in oocytes. However, the mRNA expression levels of Hif2α and Hif3α were not significantly changed in HIF1α-deleted oocytes (Fig. 4). This finding indicates that the null of HIF1α did not induce the compensatory expression of other isoforms, and demonstrates that HIF pathway is not required for oocyte development.
Figure 4: Expression analysis of other Hifα subunits in Control and Hif1α-cKO oocytes.
Relative mRNA levels of Hif2α and Hif3α are determined by real-time RT-PCR (n = 3). Data are expressed as mean ± SD. Student’s t test (two-tailed) was used for statistical analysis. n.s., not significant.Discussion
The adaptation of cells to the anaerobic environment is achieved by the transcriptional induction of genes that are involved in glycolysis, haematopoiesis, angiogenesis, invasion and regulation of vascular tone (Kaelin Jr & Ratcliffe, 2008). An evolutionarily conserved pathway mediated by oxygen-dependent posttranslational hydroxylation of a transcription factor called hypoxia-inducible factor (HIF) plays a pivotal role in this process (Giaccia, Siim & Johnson, 2003). However, because its deletion in mice causes embryonic lethality (Carmeliet et al., 1998), the in vivo roles of the HIF pathway in reproduction remain unclear. Our findings showed that Hif1α isoform was highly expressed in oocyte (Fig. 1A). We then constructed mice with oocyte-specific knockout of Hif1α at growing oocyte to investigate the potential role of Hif1α in oogenesis. Surprisingly, we found that HIF1α is dispensable for ovulation and female fertility in mice. Moreover, oocyte can mature in vitro, and form well-organized spindle in the absence of HIF1α (Fig. 3). In addition, it has been reported that other HIFα subunits appear closely related and are able to interact with hypoxia response elements (HREs) to induce transcriptional activity (Webb, Coleman & Pugh, 2009). However, we did not observe pronounced differences in Hif2α and Hif3α expression between Control and Hif1α-cKO oocytes (Fig. 4). Thus, our study shows that HIF pathway in oocyte is not required for female fertility.
It has been long thought that oocytes within antral follicles are limited access to oxygen. However, data accurately reflecting the O2 concentration adjacent to an oocyte are lacing (Thompson et al., 2015). A study has shown that oocytes within antral follicles exist in a relatively low oxygen tension, about 11–51 mm Hg (Redding, Bronlund & Hart, 2008). However, considering the potential for low O2 concentration within antral follicles, there is a paucity of evidence that HIF is involved in mammalian oogenesis and folliculogenesis. By developing a HIF reporter mouse, researchers found no HIF reporter activity in mural granulosa, cumulus layers, or oocytes from preovulatory follicles (Tam et al., 2010). This suggests HIF activity is dispensable for the development of growing antral follicle. Here, by using oocyte-specific HIF1α deletion mice, we further demonstrated this notion. There exists a paradox for the developing follicle. One possible explanation is that a low but not hypoxic environment exists in the follicle, most likely for protecting the oocyte from oxidative damage while offering enough O2 to meet its oxidative phosphorylation demands. Meanwhile, following the LH (luteinizing hormone) surge, HIF activity is dramatically increased and plays a key role in luteinization (Kim, Bagchi & Bagchi, 2009). In addition, echinomycin, an HIF1α inhibitor significantly decreased bovine oocyte maturation and subsequent blastocyst formation in vitro through modulating cumulus cell function (Turhan et al., 2021). Thus, more studies are required to determine the significance of HIF pathway in granulosa cells.
Conclusions
In conclusion, by generating mutant mice with oocyte-specific deletion of HIF1α, we provided genetic evidence that the HIF pathway in oocyte is not required for ovulation and female fertility. Our results further support the notion that oocyte resides in follicle with a low O2, but not a hypoxic environment.
Supplemental Information
Raw data of Hifα subunits expression in oocytes
qRT-PCR analysis of Hifα subunits mRNA levels in oocytes from WT mice. The relative mRNA levels of Hif3α in WT oocytes were set to 1.0. Data represent the mean ± SEM (n = 3)
Raw data for PCR genotyping
PCR genotyping results of Hif1αfl/fl mice and Gdf9-Cre recombinase mice from DNA obtained from tail samples. A single 289 bp band and a single 335 bp band corresponded to the WT and homozygous floxed mice (Hif1αfl/fl) respectively (Top); a single 161 bp band indicated the Gdf9-Cre transgene
Raw data of Hif1α expression in cKO oocytes.
qRT-PCR analysis of Hif1α mRNA levels in oocytes from Control and Hif1α-cKO females. The relative mRNA level of Hif1α in Control oocytes was set to 1.0.
Raw data for WB
Western blot showing the absence of HIF1α protein expression in Hif1α-cKO oocytes.
Raw data for fertility test
Cumulative numbers of pups per female born from Control and Hif1α-cKO mice for 6 months.
Raw data for superovulation
Average number of oocytes obtained after superovulation from Control and Hif1α-cKO females at indicated age
Raw data for early embryo on time development
The percentage of Hif1α-cKO oocyte-derived embryos that successfully progressed to the 2-cell, 4-cell and blastocyst stage at E1.5, E2 and E4 during in vitro culture.
Raw data for the number of GV oocyte
Mean number of GV stage oocytes obtained per mouse after priming with PMSG (n = 6 for each genotype).
Raw data for GVBD rate
GVBD rates of oocytes from 8-week-old Control and Hif1 α-cKO mice.
Raw data for PB1 rates
PB1 rates of oocytes from 8-week-old Control and Hif1 α-cKO mice.
Raw data for meiotic defects
Quantification of Control (n = 78) and Hif1α-cKO oocytes (n = 69) with spindle defects or chromosome misalignment. Data are expressed as mean ± SD.
Raw data for Hifα subunits expression
Relative mRNA levels of Hif2α and Hif3α are determined by real-time RT-PCR. Data are expressed as mean ± SD.