Taxonomic evaluation of Xylodon (Hymenochaetales, Basidiomycota) in Korea and sequence verification of the corresponding species in GenBank
- Published
- Accepted
- Received
- Academic Editor
- Madhava Meegaskumbura
- Subject Areas
- Bioinformatics, Ecology, Mycology, Taxonomy
- Keywords
- Hyphodontia, ITS, nrLSU, Phylogeny, Schizoporaceae, Schizopora, White-rot, Wood decay fungus
- Copyright
- © 2021 Cho et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Taxonomic evaluation of Xylodon (Hymenochaetales, Basidiomycota) in Korea and sequence verification of the corresponding species in GenBank. PeerJ 9:e12625 https://doi.org/10.7717/peerj.12625
Abstract
Genus Xylodon consists of white-rot fungi that grow on both angiosperms and gymnosperms. With resupinate and adnate basidiomes, Xylodon species have been classified into other resupinate genera for a long time. Upon the integration of molecular assessments, the taxonomy of the genus has been revised multiple times over the years. However, the emendations were poorly reflected in studies and public sequence databases. In the present study, the genus Xylodon in Korea was evaluated using molecular and morphological analyses of 172 specimens collected in the period of 2011 to 2018. The host types and geographical distributions were also determined for species delimitation. Furthermore, public sequences that correspond to the Xylodon species in Korea were assessed to validate their identities. Nine Xylodon species were identified in Korea, with three species new to the country. Morphological differentiation and identification of some species were challenging, but all nine species were clearly divided into well-resolved clades in the phylogenetic analyses. Detailed species descriptions, phylogeny, and a key to Xylodon species in Korea are provided in the present study. A total of 646 public ITS and nrLSU sequences corresponding to the nine Xylodon species were found, each with 404 (73.1%) and 57 (61.3%) misidentified or labeled with synonymous names. In many cases, sequences released before the report of new names have not been revised or updated. Revisions of these sequences are arranged in the present study. These amendments may be used to avoid the misidentification of future sequence-based identifications and concurrently prevent the accumulation of misidentified sequences in GenBank.
Introduction
The genus Xylodon (Pers.) Fr. is the oldest genus in Schizoporaceae Jülich of the Hymenochaetales. It is characterized by resupinate basidiomes with various hymenophore types, including grandinioid, odontioid, and poroid hymenophores, as well as monomitic to trimitic hyphal systems, different types of cystidia including capitate, clavate, moniliform, and subulate cystidia, basidia with four sterigmata and basal clamp connections, and narrow to wide subglobose or ellipsoid basidiospores that are inamyloid and colorless (Hjortstam & Ryvarden, 2009; Riebesehl & Langer, 2017). Many of these morphological characteristics are shared with other genera in Schizoporaceae. For a long time, odontioid resupinate species have been placed in Hyphodontia. However, phylogenetic studies have shown that many misaligned species with odontioid and poroid hymenophores belong to the genus Xylodon (Larsson et al., 2006; Hjortstam & Ryvarden, 2009; Yurchenko & Wu, 2016). Currently, Xylodon is the largest genus within the family Schizoporaceae and encompasses species within the genera Lagarobasidium (Viner et al., 2018), Odontiopsis, and Palifer (Riebesehl et al., 2019) as synonyms. The genus Schizopora has also been integrated into Xylodon because of its synonymous morphological characteristics and indifferentiable sequences (Riebesehl & Langer, 2017).
Xylodon species inhabit a wide range of niches worldwide, including temperate, tropical, or subtropical regions of America (Langer, 1994), Asia (Langer, 1994; Chen, Wu & Chen, 2018), and Europe (Eriksson & Ryvarden, 1976; Riebesehl et al., 2019). They grow on various angiosperm and gymnosperm species, as well as ferns such as Cyathea (Riebesehl et al., 2019). They play an important role in the ecosystem as white-rot fungi, degrading some of the most recalcitrant macromolecules with extracellular ligninolytic enzymes (Nguyen et al., 2021). These enzymes may be practically used in biotechnology. For example, X. paradoxus has been shown to have high oxidative enzyme activities (Volobuev, 2020), and X. flaviporus has potential bioremediation abilities to degrade organic pollutants such as polycyclic aromatic hydrocarbons (Lee et al., 2014). In addition to their enzymatic abilities, the compounds of X. flaviporus also have medicinal effects, inhibiting RANKL-stimulated osteoclastogenesis (Kwon et al., 2019).
Taxonomic studies on many Xylodon species in Korea were primarily based on morphology and less on genetic assessment (Lim & Jung, 2001; Lee & Jung, 2005). In addition, the transition of old names to the genus Xylodon has not been reflected in some species in Korea to date. The later integration of phylogenetic analyses has greatly increased the accuracy of the classification and identification of Xylodon species. However, public databases contain numerous reference sequences that differ in species identities, making it difficult to accurately identify query sequences with certainty (Nilsson et al., 2006; Jung et al., 2014; Jargalmaa et al., 2017). In the present study, we reflected the recent changes made in the classification and taxonomy of Xylodon in Korea and evaluated the public database to validate the sequence identities and rectify misidentified sequences. This study serves as a standard to accurately differentiate and identify Xylodon species in Korea, regardless of the methods (either molecular or morphological approach).
Materials & Methods
Specimen collection
A total of 172 specimens (Table 1) were collected in the period of 2011 to 2018 in South Korea. They were obtained from institutions for fungal collection in Korea, namely Korea National Arboretum (KA), National Institute of Biological Resources of Korea (NIBR), and Seoul National University Fungus Collection (SFC) as dried specimens. The host type of each species was analyzed based on the specimen collection information (Table 1).
| Identity | Strains | ||
|---|---|---|---|
| Angiosperms | Gymnosperms | No description | |
| Xylodon asperus | – | SFC20170209-01, SFC20170209-11, SFC20170209-13, SFC20180410-30, SFC20180426-01, SFC20110519-18 | SFC20121130-02 |
| X. flaviporus | SFC20120820-08, SFC20140313-22, SFC20140529-03, SFC20140529-14, SFC20140530-01, SFC20140530-04, SFC20140921-05, SFC20140926-17, SFC20150320-07, SFC20150404-03, SFC20150404-07, SFC20150514-06, SFC20150526-04, SFC20150625-24, SFC20150626-06, SFC20150707-62, SFC20150715-09, SFC20150716-10, SFC20150909-13, SFC20160114-06, SFC20160602-19, SFC20160614-40, SFC20160621-05, SFC20160629-02, SFC20160811-04, SFC20160812-38, SFC20160816-01, SFC20160906-02, SFC20160920-08, SFC20160922-02, SFC20160922-08, SFC20170209-03, SFC20170430-09, SFC20170524-07, SFC20170807-04, SFC20170808-21, SFC20170831-06, SFC20170908-67, SFC20180410-17, SFC20180704-47, SFC20180705-20 | SFC20120926-26, SFC20150407-05, SFC20150902-27, SFC20150908-38, SFC20160909-12, SFC20180710-24 | SFC20110921-10, SFC20110921-35, SFC20111001-50, SFC20111001-84, SFC20120409-11, SFC20120410-10, SFC20120508-01, SFC20120601-03, SFC20120601-11, SFC20120919-65, SFC20130315-25, SFC20130404-05, SFC20130521-43, SFC20130521-49, SFC20130719-41, SFC20130917-15, SFC20140412-07, SFC20140530-03, SFC20150129-03, SFC20150501-03, SFC20150701-12, SFC20160114-27, SFC20160114-32, SFC20160126-18, SFC20160127-07, SFC20160527-52, SFC20160726-31, SFC20170705-08, SFC20170920-29, SFC20180410-29, SFC20180524-06, SFC20180705-85, SFC20180802-03 |
| X. kunmingensis | SFC20160114-24, SFC20170317-07 | – | – |
| X. nespori | – | SFC20120601-18, SFC20150523-08 | – |
| X. niemelaei | KUC20160721B-26 | ||
| X. ovisporus | SFC20110823-19, SFC20120410-26, SFC20120726-01, SFC20121009-17, SFC20121009-34, SFC20130403-08, SFC20130521-61, SFC20130730-29, SFC20140410-02, SFC20140411-02, SFC20140911-31, SFC20140926-20, SFC20150516-05, SFC20150518-12, SFC20150527-11, SFC20150625-33, SFC20150707-80, SFC20150716-02, SFC20160114-21, SFC20160225-08, SFC20160225-14, SFC20160526-13, SFC20160527-02, SFC20160712-07, SFC20160811-11, SFC20160817-23, SFC20160908-39, SFC20170208-11, SFC20170221-03, SFC20170221-09, SFC20170228-01, SFC20170317-10, SFC20170430-11, SFC20170713-32, SFC20180207-01, SFC20180523-10, SFC20180720-01, SFC20180807-06, SFC20180810-02 | SFC20150407-06, SFC20160512-31, SFC20160811-36, SFC20160920-29, SFC20170718-08, SFC20171018-07 | SFC20110823-19, SFC20120410-26, SFC20120726-01, SFC20121009-17, SFC20130403-08, SFC20130521-61, SFC20130730-29, SFC20140410-02, SFC20140411-02, SFC20140926-20, SFC20150516-05, SFC20160114-21, SFC20170430-11, SFC20170713-32, SFC20180207-01, SFC20180523-10, SFC20180720-01, SFC20180807-06, SFC20180810-02 |
| X. serpentiformis | KUC20121019-31 | – | – |
| X. spathulatus | – | SFC20180710-20, SFC20180818-36 | – |
| X. subflaviporus | SFC20120821-53, SFC20150514-14, SFC20150522-08, SFC20160628-20, SFC20160708-32, SFC20161012-15, SFC20180818-15 | SFC20170316-24, SFC20170316-25, SFC20170426-14 | SFC20150701-67, SFC20150707-63, SFC20180808-08 |
Morphological observations
All specimens were preliminarily grouped with respect to their macromorphological characteristics, including hymenophore types, aculei length, and pore size. Subsequently, micromorphological features, including cystidia types and basidiospore size, were observed using one to six well-preserved specimens of each group. Pieces of dried specimens were mounted in 5% KOH. Observations were performed under a Nikon 80i compound light microscope (Nikon, Tokyo, Japan) at 400× to 1,000× magnification. For each specimen, 20 basidia and 30 basidiospores were measured when possible. Basidiospore dimensions were expressed as the range of minimum to maximum length and width. “Q” refers to the average length to width ratio of basidiospores.
DNA extraction, PCR, and sequencing
Small hymenophore pieces obtained from 1 to 10 representative specimens of each group were peeled off from the wood using sterile forceps, and each was placed in 200 μL of 2× CTAB buffer. Besides the representative specimens, other ambiguous specimens, including juvenile specimens or those too bad in state, were also analyzed. Genomic DNA extraction was conducted using the AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) according to the manufacturer’s protocol with a modification where sterile micropestles were used to grind the tissue samples. PCR was performed using a PCR premix (Bioneer, Daejeon, Korea). The internal transcribed spacer (ITS) region was amplified using primers ITS1F and ITS4B (Gardes & Bruns, 1993) under the following conditions: 95 °C for 5 min, 35 cycles of 95 °C for 40 s, 55 °C for 40 s, and 72 °C for 1 min, followed by 72 °C for 5 min. The nuclear large subunit ribosomal RNA (nrLSU) region was amplified with primers LR0R and LR7 (Hopple & Vilgalys, 1994) under the same PCR conditions as those for ITS. The PCR products were electrophoresed on 1% agarose gel to verify the results. They were then purified using the ExpinTM PCR Purification Kit (GeneAll Biotechnology, Seoul, Korea) following the manufacturer’s instructions. Sequencing was performed at Macrogen (Seoul, Korea) with the PCR primers using an ABI Prism 3700 Genetic Analyzer (Life Technologies, Gaithersburg, MD, USA). All ITS and nrLSU sequences were proofread and edited using MEGA X (Kumar et al., 2018).
Sequence analyses
All ITS and nrLSU sequences belonging to Schizoporaceae were downloaded from the National Center for Biotechnology Information (NCBI) to include sequences described by their former names, such as Hyphodontia and Schizopora. Hyphodontia has been transferred to Hyphodontiaceae (Wang et al., 2021), but the sequences in NCBI were still classified under Schizoporaceae at the time of analysis. Subsequently, sequences from the Basic Local Alignment Search Tool (BLAST) results that grouped phylogenetically with the validated sequences from the present study were analyzed to rectify the misidentifications in the public database. The geographical location for each GenBank sequence was also analyzed for species identification and differentiation. All sequences were obtained on the 2021–07–21.
For each genetic region (ITS and nrLSU), multiple alignments for the phylogenetic analyses were performed using MAFFT version 7 (Katoh & Standley, 2013) with the default settings. Manual trimming was performed at the ends of the alignments. Combined RAxML (Stamatakis, 2006) of ITS and nrLSU regions was constructed with the CIPRES web portal (Miller, Pfeiffer & Schwartz, 2011) using the GTR+G model with 1,000 bootstrap replicates for the maximum likelihood phylogenetic analyses. The sequences used in the present study are listed in Table 2. Representative sequences for each species were deposited in GenBank with accession numbers MZ520578–MZ520585 for ITS sequences and MZ520587–MZ520597 for nrLSU sequences (Table 2).
| Species | Strain | GenBank accession | Reference | |
|---|---|---|---|---|
| ITS | nrLSU | |||
| Xylodon asperus | 2004b | DQ873606 | DQ873607 | Larsson et al., 2006 |
| SFC20170209-01 | MZ520578 | MZ520587 | This study | |
| SFC20170209-11 | MZ520579 | MZ520588 | This study | |
| X. astrocystidiatus | TNM F24764 | NR_154054* | NG_068732* | Yurchenko & Wu, 2014 |
| X. australis | CANB569567 | MT158703 | MT158739 | Fernández-López et al., 2020 |
| X. borealis | Spirin 9416 | MH317760 | MH638259 | Viner et al., 2018 |
| X. cystidiatus | FR-0249200 | MH880195 | MH884896 | Riebesehl et al., 2019 |
| X. filicinus | MSK F 12869 | NR_163313* | NG_067836* | Riebesehl et al., 2019 |
| X. flaviporus | SFC20150211-16 | MZ520581 | MZ520590 | This study |
| SFC20170316-25 | MZ520582 | MZ520591 | This study | |
| SFC20180710-24 | MK992840 | MZ520592 | Lupala et al., 2019 | |
| X. follis | FR-0249814 | MH880204 | MH884902 | Riebesehl et al., 2019 |
| X. hyphodontinus | KAS-GEL9222 | MH880205 | MH884903 | Riebesehl et al., 2019 |
| X. kunmingensis | MSK-F 7381 | MH880196 | MH884897 | Riebesehl et al., 2019 |
| SFC20170317-07 | MZ520580 | MZ520589 | This study | |
| TUB FO 42565 | NR_163312* | MH884898* | Riebesehl et al., 2019 | |
| X. magallanesii | MA:Fungi:90391 | MT158720 | MT158756 | Fernández-López et al., 2020 |
| X. nespori | GEL3158 | DQ340310 | DQ340346 | Unpublished |
| GEL3290 | DQ340309 | DQ340343 | Unpublished | |
| SFC20150523-08 | MZ520583 | MZ520593 | This study | |
| X. niemelaei | LWZ20171015-12 | MT319625 | MT319361 | Wang et al., 2021 |
| KUC20160721B-26 | MF774798 | MZ520595 | This study | |
| GC 1512-1 | KX857808 | KX857813 | Chen, Wu & Chen, 2017 | |
| X. nothofagi | ICMP 13839 | AF145582 | MH260064 | Unpublished |
| X. ovisporus | FR-0249797 | MH880201 | MH884901 | Riebesehl et al., 2019 |
| KUC20130808-17 | KJ668462 | KJ668314 | Unpublished | |
| MA:Fungi:79440 | MH260071 | MH260066 | Unpublished | |
| SFC20170718-08 | MZ520584 | MZ520594 | This study | |
| X. paradoxus | MA-Fungi_70444 | MH260070 | MH260065 | Unpublished |
| X. pseudolanatus | CFMR FP-150922 | NR_163314* | NG_067837* | Riebesehl et al., 2019 |
| X. quercinus | MA:Fungi:27435 | MT158718 | MT158754 | Fernández-López et al., 2020 |
| X. raduloides | KAS-JR26 | MH880225 | MH884910 | Riebesehl et al., 2019 |
| MAF 75310 | KY962825 | KY962864 | Fernández-López et al., 2018 | |
| X. serpentiformis | KUC20121019-31 | KJ668517 | KJ668369 | Unpublished |
| TUB-FO 42688 | MH880229 | MH884913 | Riebesehl et al., 2019 | |
| X. spathulatus | SFC20180818-36 | MK992854 | MZ520596 | Lupala et al., 2019 |
| Wu 1407-105 | KX857804* | KX857811* | Chen, Wu & Chen, 2017 | |
| MSK-F 12931 | MH880231 | MH884914 | Riebesehl et al., 2019 | |
| X. subflaviporus | SFC20180818-15 | MZ520585 | MZ520597 | This study |
| Wu 0809-76 | KX857803* | KX857815* | Chen, Wu & Chen, 2017 | |
| X. taiwanianus | CBS 125875 | MH864080 | MH875537 | Vu et al., 2019 |
| Hyphodontia pallidula | GEL2097 | DQ340317 | DQ340372 | Unpublished |
Note:
Asterisks indicate type sequences, and bolded sequences are those newly generated in the present study.
Results
Morphological and molecular identification of Xylodon species in Korea
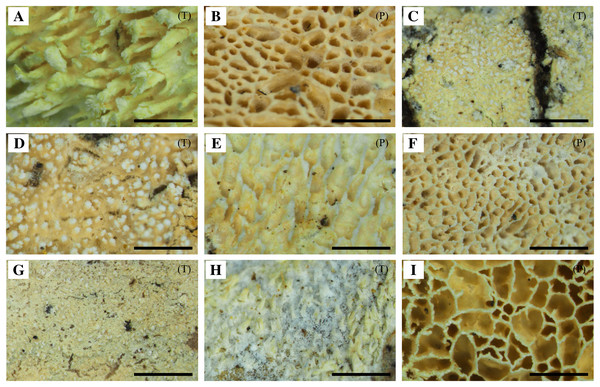
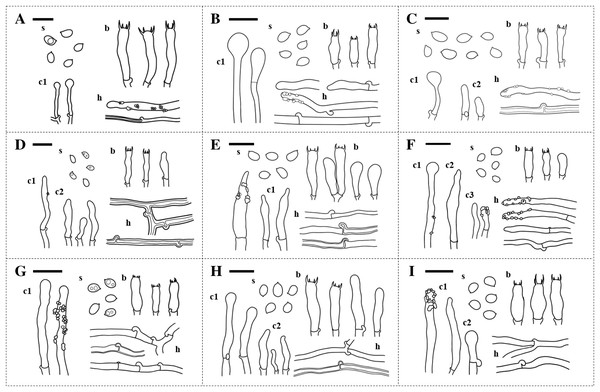
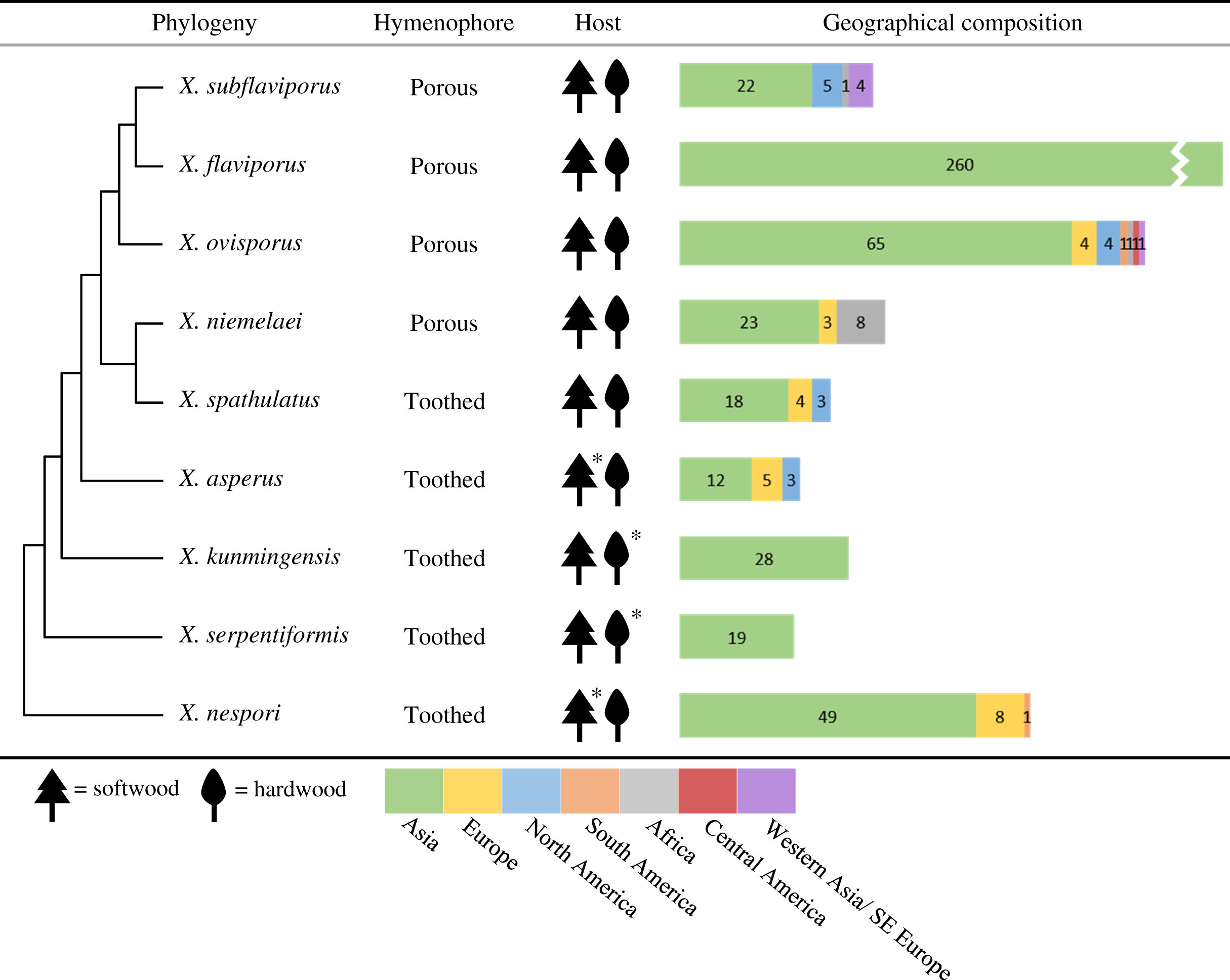
The specimens were divided into nine different groups according to morphology. Features including hymenophore types, pore densities, and basidiospore sizes of Xylodon specimens from Korea were analyzed. The basidiomes of all species were resupinate and adnate on wood. There were two hymenophore shapes, raduloid (toothed) and poroid (Fig. 1). Five groups had raduloid hymenophores, and the other four groups were poroid. Through the detailed morphological features, two raduloid groups were identified as X. asperus and X. spathulatus, and two poroid groups were identified as X. niemelaei and X. ovisporus. The unidentified raduloid and poroid groups of species were comparably similar in macromorphology, but were divided by several micromorphological characteristics (Table 3; Fig. 2). A taxonomic key to each species is provided below, and detailed morphological characteristics are provided in Table 3. Along with morphological characteristics, the preferential host types of specimens from Korea were compared—host preference of the nine Xylodon species was divided into either angiosperms, gymnosperms, or both, and each species corresponded to the respective reference (Tables 1 and 4). All poroid groups and a raduloid species, X. spathulatus, had no host preference. The remaining two raduloid groups favored angiosperms, and the other two, including X. asperus, preferred gymnosperms.
Figure 1: Basidiomes of Xylodon species in Korea.
(A) X. asperus (SFC20170209-11). (B) X. flaviporus (SFC20180410-17). (C) X. kunmingensis (SFC20160114-24). (D) X. nespori (SFC20150523-08). (E) X. niemelaei (KUC20160721B-26). (F) X. ovisporus (SFC2010718-08). (G) X. serpentiformis (KUC20121019-31). (H) X. spathulatus (SFC20180818-36). (I) X. subflaviporus (SFC20170426-14). Scale bars are 1 mm. ‘(P)’ refers to poroid type and ‘(T)’ refers to toothed type.Figure 2: Microscopic characteristics of Xylodon species in Korea.
(A) X. asperus. (B) X. flaviporus. (C) X. kunmingensis. (D) X. nespori. (E) X. niemelaei. (F) X. ovisporus. (G) X. serpentiformis. (H) X. spathulatus. (I) X. subflaviporus. Scale bars are 10 µm. ‘s’ indicates basidiospores, ‘b’ indicates basidia, ‘c1,’ ‘c2,’ and ‘c3,’ indicate different types of cystidia, and ‘h’ indicates hyphae.| Characteristics | X. asperusa | X. flaviporusb | X. kunmingensisc | X. nesporid | X. niemelaeie | X. ovisporusb | X. serpentiformisf | X. spathulatusd | X. subflaviporusg | |
|---|---|---|---|---|---|---|---|---|---|---|
| Hymenophore type | raduloid | poroid | odontioid | grandinioid | poroid and arachnoid | poroid | odontioid | raduloid | poroid | |
| Hymenophore color | cream to buff | cream to buff or pinkish buff | cream | cream | cream to buff | cream to buff or pinkish buff | cream | cream | cream to buff | |
| Hyphal system | monomitic | pseudodimitic | monomitic | monomitic | monomitic | pseudodimitic | pseudodimitic | monomitic | pseudodimitic | |
| Clamp connections | present | present | present | present | present | present | present | present | present | |
| Cystidia | capitate, subulate | acicular, apically-encrusted, capitate | capitate | encrusted, subcapitate | capitate, short | acicular, apically-encrusted, capitate | encrusted, tubular | capitate, cylindrical, subulate | acicular, apically-encrusted, capitate | |
| Basidia | shape | suburniform | suburniform | utriform | subclavate to suburniform | subclavate to suburniform | suburniform | suburniform | subclavate to suburniform | suburniform |
| length/µm | 9.7–18.8 | 9.7–18.8 |
15.0–22.0 (23.0–27.0) |
15.0–20.0 |
13.0–18.0 (17.0–23.0) |
8.0–15.6 |
11.3–14.7 (15.0–17.0) |
14.0–21.0 | 11.3–15.4 | |
| width/µm | 3.5–5.6 | 3.5–5.6 | 3.0–5.3 | 3.6–4.1 | 4.0–5.0 | 3.1–5.1 |
2.8–3.9 (4.0–5.0) |
3.5–5.0 |
4.3–6.4 (4.0–5.0) |
|
| Basidio-spores | ornamentation | ellipsoid | ellipsoid | narrowly ellipsoid | narrowly ellipsoid | ellipsoid | ellipsoid | ellipsoid | ellipsoid | ellipsoid |
| length (l)/µm | 4.2–5.2 | 4.2–5.2 | 5.0–6.3 |
4.4–5.1 (5.0–6.0) |
5.0–6.2 | 3.5–4.4 | 4.8–5.8 | 4.8–5.8 | 3.9–4.8 | |
| width (w)/µm | 3.0–4.0 | 3.0–4.0 | 2.5–3.4 | 2.1–2.6 | 3.2–3.7 | 2.6–3.3 | 3.3–4.3 | 3.5–4.5 | 2.8–3.5 | |
| mean (l × w) | 4.6 × 3.2 | 4.6 × 3.2 | 5.7 × 3.2 | 4.6 × 2.4 | 4.6 × 3.5 | 3.9 × 2.9 | 5.3 × 3.8 | 5.3 × 3.9 | 4.4 × 3.2 | |
| Q value |
1.4 (1.2–1.3) |
1.4 | 1.8 | 1.9 | 1.6 | 1.3 | 1.4 | 1.4 | 1.4 | |
Notes:
Measurements in bold are deviant from the references, which are given in parentheses.
 |
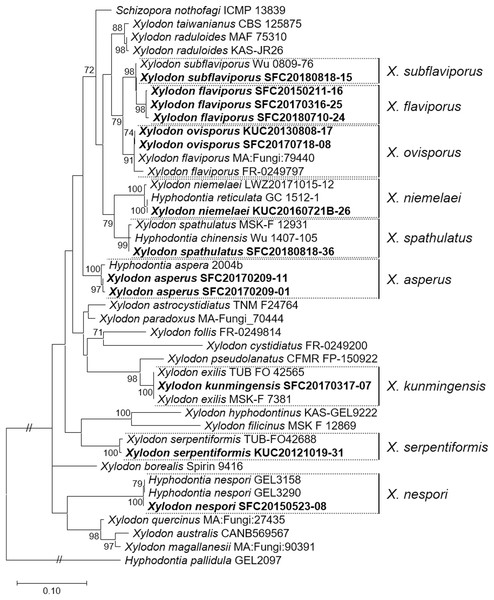
Five to ten representative specimens from each morphological group were selected for sequencing to verify their identities and distinguish enigmatic species. Nine different taxa were confirmed through the analyses of ITS and nrLSU, corresponding to the morphological groupings: five raduoid species (X. asperus, X. kunmingensis, X. nespori, X. serpentiformis, and X. spathulatus) and four poroid species (X. flaviporus, X. niemelaei, X. ovisporus, and X. subflaviporus). In the public database, type sequences for ITS were available for three of the nine species: X. kunmingensis (MK404532, and NR_163312 as X. exilis), X. spathulatus (as X. bubalinus, NR_154097), and X. subflaviporus (NG_068781); they were included in the phylogenetic analyses to obtain a more reliable conclusion (Fig. 3).
Figure 3: Maximum likelihood (ML) tree of Xylodon species constructed based on ITS and nrLSU regions.
Hyphodontia pallidula (DQ340317) was used as an outgroup. Sequences from Korea are indicated in bold. Bootstrap values >70 are shown.The concatenated sequences for the maximum likelihood (ML) tree consisted of 578 bases of the ITS region and 517 bases of the nrLSU region. No reference sequence was available for X. flaviporus in the nrLSU region from other countries. Xylodon nespori, X. ovisporus, X. serpentiformis, and X. subflaviporus sequences from Korea were marginally different from those of the other countries. For instance, X. subflaviporus (SFC20180818-15) was different from the holotype (Wu 0809-76) only in two separate regions, one base each in the ITS and nrLSU regions. Neighbor-joining (NJ) tree of ITS for X. asperus was constructed to present the division of clades by the geographical origin, and for X. niemelaei and X. spathulatus to display the related species names that are parallel or highly similar in sequence (Fig. S1).
Sequence validation
A total of 646 GenBank ITS and nrLSU sequences corresponded to the nine Xylodon species in Korea. For ITS, there were 553 sequences with 404 (73.1%) labeled with misleading names and 93 nrLSU sequences with 57 (61.3%) sequences labeled with misleading names (Tables S1 and S2). The misleading names included misidentified, synonymous names, and labels such as “Hyphodontia sp.,” “Xylodon sp.,” “Fungal sp.,” and “Uncultured fungus”.
Xylodon flaviporus, X. ovisporus, and X. subflaviporus had ITS sequences misidentified as one another, along with sequences identified as Hyphodontia tropica (Table S1). Xylodon flaviporus had the most ITS sequences (n = 260) in GenBank, but only nine were rightly annotated. The remaining 251 sequences were misidentified as X. ovisporus (n = 63) and H. tropica (n = 188). Xylodon ovisporus had the second largest number of ITS sequences (n = 77) in GenBank, with nine annotated correctly. The rest of the sequences were either misidentified as X. flaviporus (n = 63) under genera Hyphodontia, Schizopora, and Xylodon or as unspecified (n = 5). Of the 58 ITS sequences defined as X. nespori, 54 were correctly labeled, one was mislabeled as X. magallanesii, and three were ambiguously labeled (‘uncultured Hyphodontia’ and ‘uncultured fungi’). Most ITS sequences of X. asperus and X. kunmingensis were correctly annotated as to their current name or synonyms. For X. niemelaei, 19 ITS sequences were assigned as X. niemelaei or H. niemelaei. Three other Xylodon species names showed high sequence similarity with X. niemelaei. Similarly, ITS sequences defined as X. spathulatus included X. bubalinus, X. chinensis, and ambiguously labeled sequences (‘Xylodon sp.’ and ‘Hyphodontia sp.’). Only for X. serpentiformis, all GenBank references were labeled accurately, except for the query ITS sequence from Korea (KJ668517), which was annotated as ‘Hyphodontia sp. 1’.
For nrLSU, X. kunmingensis and X. serpentiformis had no mislabeled sequences, and sequences for the remaining species were revised for their identity (Table S2). Xylodon asperus and X. nespori had sequences labeled as Hyphodontia species. The nrLSU sequences of X. flaviporus and X. ovisporus had sequences misidentified as one another, similar to the trend seen for ITS. The complexity of X. niemelaei and X. spathulatus was also reflected in the nrLSU sequences.
For all nine Xylodon species, public ITS sequences from Asia were present (Table 4); and three of the nine species, X. flaviporus, X. kunmingensis, and X. serpentiformis, had sequences only from Asia. The other six species were from various parts of the world. Xylodon ovisporus sequences were reported from most regions, including Africa, the Americas, Asia, and Europe. Xylodon asperus and X. spathulatus sequences were from Asia, Europe, and North America. Xylodon nespori sequences were from Asia, Europe, and South America. Xylodon niemelaei sequences were from Africa, Asia, and Europe. Xylodon subflaviporus sequences were from Africa, Asia, and North America. For sequences that had no description of the country of origin, it was presumed that they belonged to the country where the authors or depositors were affiliated.
Taxonomic key to Xylodon in Korea
1 Hymenophore raduloid2
1* Hymenophore poroid6
2 Aculei length up to 2 mmX. asperus
2* Aculei length <1 mm3
3 Aculei length up to 0.8 mm; 1–2 aculei per mmX. spathulatus
3* >10 aculei per mm4
4 Up to 25 aculei per mmX. serpentiformis
4* 10< aculei <20 per mm5
5 Hymenophore cream to sand; aculei length up to 200 µm long; up to 12 aculei per mm; basidiospores 4.4–5.1 × 2.1–2.6 µmX. nespori
5* Hymenophore cream to buff; aculei length <200 µm; up to 15 aculei per mm; distorted capitate cystidia; bigger basidiospores of 5.0–6.3 × 2.5–3.4 µmX. kunmingensis
6 Hymenophore pores arachnoidX. niemelaei
6* Hymenophore pores not arachnoid7
7 >4 pores per mmX. ovisporus
7* <4 pores per mm8
8 Hymenophore cream to buff or pinkish buff; basidiospores 4.2–5.2 × 3.0–4.0 µmX. flaviporus
8* Hymenophore cream to buff; more lacerated pore dissepiments; smaller basidiospores of 3.9–4.8 × 2.8–3.5 µmX. subflaviporus
Discussion
Update on the taxonomy of Xylodon in Korea
Through the integration of recent taxonomic revisions of the genus Xylodon, nine species were confirmed to occur in Korea. Three Xylodon species were reported new to the country: X. kunmingensis, X. serpentiformis, and X. subflaviporus. The remaining six species have previously been reported to reside in Korea, some as Hyphodontia or Schizopora. For example, X. asperus has been reported as H. aspera, as a synonym of H. granulosa (Lee & Jung, 2005), and X. niemelaei has been reported by its synonym, H. reticulata (Kwon et al., 2018).
Most of the Xylodon specimens collected in Korea were X. flaviporus, X. ovisporus, and X. subflaviporus. They were also abundant nationwide. The lack of clear distinction between these species brought confusion in Korea for a long time. In terms of morphological characteristics, basidiospore dimensions vary among these three species (Table 3; Riebesehl & Langer, 2017; Chen, Wu & Chen, 2018). Their pore sizes are also comparable, with X. ovisporus specimens having relatively dense and smaller pores (>4 pores per mm) than those of X. flaviporus and X. subflaviporus (<4 pores per mm). Furthermore, the pores of X. subflaviporus were found to be uneven in shape, with more lacerated pore dissepiments. In Korea, many X. flaviporus, X. ovisporus, and X. subflaviporus specimens have been recorded as Schizopora paradoxa (now Xylodon paradoxus; Lee et al., 1992; Lim & Jung, 2001). However, our study showed that X. paradoxus does not reside in Korea, which was consistent with the results in Fernández-López et al. (2018), where X. paradoxus was phylogenetically proven to reside only in Europe.
Both Xylodon exilis and X. kunmingensis have been reported as new to science in 2019 but were recognized as conspecific, with X. kunmingensis having priority (Wang et al., 2021). Wang et al. (2021) have also assessed the monophyletic clade of X. niemelaei with its neighboring species, X. apacheriensis, X. reticulatus, and X. rhizomorphus, which was also supported in the present study (Fig. S1A). Similarly, X. spathulatus has been recognized to be conspecific to X. bubalinus and X. chinensis (Riebesehl et al., 2019). Xylodon spathulatus specimens from Korea have previously been reported as X. chinensis based on NCBI BLAST results (Lupala et al., 2019), but after a thorough examination of the specimens, we recognized that they correspond to the descriptions of X. spathulatus (Eriksson & Ryvarden, 1976), and support the morphological (Table 3) and phylogenetic (Fig. 3; Fig. S1B) synonymy with X. bubalinus (Wang & Chen, 2017) and X. chinensis (Chen, Wu & Chen, 2017).
The morphological characteristics of nine Xylodon species in Korea corresponded to the descriptions of their respective references (Table 3). However, measurements of basidia and basidiospores of some species were slightly different. The basidia of the specimens from Korea were generally smaller than those from other regions: Xylodon kunmingensis from South China (Shi et al., 2019), X. niemelaei from Taiwan (Wu, 1990), X. serpentiformis from Taiwan (Langer et al., 1992), and X. spathulatus from the Northern Europe (Eriksson & Ryvarden, 1976; Table 4); only the basidia of X. subflaviporus specimens in Korea were broader than those of the reference specimens from China (Chen, Wu & Chen, 2018). For basidiospores, the basidiospore length of X. nespori was shorter than that of the reference from Northern Europe (Eriksson & Ryvarden, 1976). The basidiospores of X. asperus from Korea were broader than that of Hyphodontia aspera (= X. asperus) from Finland (Kotiranta & Saarenoksa, 2000). These micromorphological variations between specimens may result from the geographical distance or the difference in landscape or habitat. More care is required when studying wood-degrading resupinate fungi such as Xylodon, as morphological discrepancies are often only observed at a finer scale. The variations were also reflected in the phylogeny. Xylodon asperus sequences from Korea were grouped in a clade with most sequences from Asia (Fig. S1C).
The number of sequences for each species was variable worldwide (Table 4), indicating an unevenness of species dispersal or biases of the locations where research was actively conducted. The ITS sequences of X. flaviporus were only submitted from Asia, but X. flaviporus has been reported to reside in Europe and South America through morphological analyses (Cooke, 1886; Keizer, 1990; Drechsler-Santos, Groposo & Loguercio-Leite, 2008). Similarly, all ITS sequences of X. serpentiformis were from Asia, but nrLSU sequence for the same species was available from Germany (AJ406465). As there is such inconsistency in specimen analyses, sequence-based information should not be solely relied upon when identifying or differentiating species.
White-rotting fungi have different mechanisms to degrade hardwood, which is more intractable than softwood (MacDonald & Master, 2012; Couturier et al., 2015). The difference in mechanisms may contribute to the division of species by host type. Most Xylodon species in Korea have been found to grow on dead or decayed wood of both angiosperms and gymnosperms (Eriksson & Ryvarden, 1976; Kotiranta & Saarenoksa, 2000; Chen, Wu & Chen, 2017, 2018). Most angiosperm hosts were oak trees (Quercus spp., Fagaceae), and most gymnosperm hosts were pine trees (Pinus spp., Pinaceae). Some species have been noted to have a preferential host type. Xylodon kunmingensis and X. serpentiformis were noted to grow more frequently on deciduous woods (Langer et al., 1992), which was also reflected in our study (Table 1). Xylodon asperus and X. nespori have been reported to mostly grow on conifers (Eriksson & Ryvarden, 1976; Kotiranta & Saarenoksa, 2000). This was also found in our study despite the small number of specimens collected, where the two species were found to grow on red pine (Pinus densiflora Siebold & Zucc.). Xylodon nespori has been reported in Korea as a fungus that co-occurs with the ant species Pristomyrmex punctatus (Lupala et al., 2019). Wood-decay fungi take several measures to disperse basidiospores, including the use of insect vectors. The host preference of insects increases the chance of wood-decaying fungal basidiospore transfer to suitable host types (Stenlid & Gustafsson, 2001). Therefore, interpretation of the preferential host type of each Xylodon species could be used to understand the interactions of Xylodon species with other organisms.
Validity of Xylodon sequences in GenBank
Public databases are increasingly being used in multifarious ways by researchers from various biological fields (Duck et al., 2016). Researchers have raised concerns about the use of misidentified sequences in succeeding studies (Stavrou et al., 2018; Fort et al., 2021). A substantial number of Xylodon ITS and nrLSU sequences were misidentified or not updated in taxonomy in GenBank (Tables S1 and S2). There were more ITS sequences available in GenBank than nrLSU, possibly owing to the ITS region being a universal DNA barcode marker for fungi. This resulted in more ITS sequences labeled with misleading names. Initial misperceptions of sequences led to an accumulation of mislabeled sequences that were deposited based on identity designated through a BLAST search. For Xylodon species, many sequences were still labeled as Hyphodontia spp., even after the transfer of some species from Hyphodontia to Xylodon (Hjortstam & Ryvarden, 2009; Riebesehl & Langer, 2017).
In GenBank, Xylodon flaviporus and X. subflaviporus had many ITS sequences labeled as Hyphodontia tropica (Table S1). We recognized the H. tropica sequences as part of the genus Xylodon, as of Yurchenko & Wu (2016), who integrated H. tropica (nom. inval.) into X. ovisporus. However, our study re-identified most of the H. tropica sequences as X. flaviporus. Thorough evaluation of the morphology and phylogeny of X. flaviporus and X. ovisporus specimens in the present study revealed that many GenBank ITS sequences of X. flaviporus and X. ovisporus were identified as one another (Table S1). The two species in this study were identified based on the morphological characteristics (Table 3), which agreed with previous descriptions (Wu, 2001; Yurchenko & Wu, 2016; Riebesehl & Langer, 2017; Chen, Wu & Chen, 2018). However, some of these previous studies did not analyze phylogeny, whereas few studies identified species solely based on sequence alignment at the beginning of GenBank sequence uploads of the two species (Paulus et al., 2000; Buzina et al., 2003). These factors led to misalignments between morphology and sequences of specimens, and this further led to an accumulation of misidentified sequences in GenBank.
Many correctly identified sequences of Xylodon species have been described as Hyphodontia, or Schizopora, possibly due to the shortage of updates upon taxonomic revisions by the authors responsible for sequence uploads. As amendments to GenBank submissions are managed by the submitters (Benson et al., 2004), it is essential for all taxonomists to consistently stay aware of the changes made in the classification and taxonomy of organisms and request essential revisions to NCBI (Schoch et al., 2020). Miscellaneous Xylodon sequences that were inaccurate or repetitious in the genus classification exemplify the need for comprehensive research on the taxonomic history of the genus, consistent updates, and morphological observations to prevent the submission and buildup of misleading sequences. The validated sequence information of Xylodon species in this study will reduce the errors for sequence-based identification and contribute to the subsequent research based on accurate identification.
Conclusion
We report nine Xylodon species in Korea based on taxonomic descriptions and molecular analyses. Some of the nine species were previously grouped under the genera Hyphodontia or Schizopora. Through the integration of the upgrade in taxonomy, species that were previously placed in Hyphodontia and Schizopora were synonymized under the genus Xylodon, resulting in nine Xylodon species being present in the country today. We provide a taxonomic key and assess the host preference and global distributions of the nine species described here for species delimitation. We also validated the reference sequences on a public database in the hopes of avoiding future misidentification of sequences and preventing further accumulation of mislabeled sequences in GenBank.
Supplemental Information
Neighbor-joining tree of Xylodon species.
(A) X. niemelaei clade. (B) X. spathulatus clade. (C) X. asperus clade.