Changes in the urinary proteome in rats with regular swimming exercise
- Published
- Accepted
- Received
- Academic Editor
- Young Jin Lee
- Subject Areas
- Animal Behavior, Biochemistry, Molecular Biology, Kinesiology, Urology
- Keywords
- Urine, Proteome, Swimming, Exercise
- Copyright
- © 2021 Meng et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Changes in the urinary proteome in rats with regular swimming exercise. PeerJ 9:e12406 https://doi.org/10.7717/peerj.12406
Abstract
Purpose
Urine can sensitively reflect early pathophysiological changes in the body. The purpose of this study was to explore the changes of urine proteome in rats with regular swimming exercise.
Methods
In this study, experimental rats were subjected to daily moderate-intensity swimming exercise for 7 weeks. Urine samples were collected at weeks 2, 5, and 7 and were analyzed by using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS).
Results
Unsupervised clustering analysis of all urinary proteins identified at week 2 showed that the swimming group was distinctively different from the control group. Compared to the control group, a total of 112, 61 and 44 differential proteins were identified in the swimming group at weeks 2, 5 and 7, respectively. Randomized grouping statistical analysis showed that more than 85% of the differential proteins identified in this study were caused by swimming exercise rather than random allocation. According to the Human Protein Atlas, the differential proteins that have human orthologs were strongly expressed in the liver, kidney and intestine. Functional annotation analysis revealed that these differential proteins were involved in glucose metabolism and immunity-related pathways.
Conclusion
Our results revealed that the urinary proteome could reflect significant changes after regular swimming exercise. These findings may provide an approach to monitor the effects of exercise of the body.
Introduction
Urine is a good source for biomarker discovery. Without homeostatic mechanisms, urine can sensitively reflect early pathophysiological changes in the body, and these changes might be useful disease biomarkers (Gao, 2013). Since the composition of urine is affected by various factors, such as age, sex, and diet (Gao, 2014; Guo et al., 2015; Wu & Gao, 2015; Li & Gao, 2016), animal models are an effective tool to minimize external influencing factors due to their similar genetic backgrounds and the same living environment. Thus, disease animal models can establish relationships between the disease and the corresponding changes in the urine proteome. Our laboratory found that changes in urinary proteins occurred before pathologic or clinical manifestations appeared in various types of animal models, such as subcutaneous tumor model (Wu, Guo & Gao, 2017), Alzheimer’s disease model (Zhang et al., 2018b), chronic pancreatitis model (Zhang, Li & Gao, 2018), liver fibrosis model (Zhang et al., 2018a), and myocarditis model (Zhao et al., 2018). Recent studies have shown that the urine proteome has potential for differential diagnosis. For example, early urinary proteins were different when the same tumor cells were grown in different organs (Wu, Guo & Gao, 2017; Wei et al., 2019; Zhang et al., 2019; Wang et al., 2020; Zhang, Gao & Gao, 2020) and when different cells were injected into the same organ (Zhang et al., 2018a; Zhang et al., 2018b; Zhang et al., 2021). Furthermore, several clinical studies performed urine proteomics to discover diagnostic biomarkers, such as for gastric cancer (Shimura et al., 2020) and familial Parkinson’s disease (Winter et al., 2021).
Physical exercise as a pathophysiological process that can improve health conditions and has a positive role in numerous chronic conditions (Pate et al., 1995; Haskell et al., 2007; Seo et al., 2014), including cancer and coronary heart diseases (Stewart et al., 2017; Hojman et al., 2018). Many studies have shown that exercise has a profound effect on the immune system. Furthermore, it has been demonstrated that physical exercise exerts a positive impact on the nervous system, learning and memory (Inoue et al., 2015; Faria et al., 2016; Faria et al., 2018). Urinary proteomics of athletes after training and competition were analyzed in previous studies (Kohler et al., 2009; McCullough et al., 2011). To the best of our knowledge, there are very few studies on global urinary proteomes after daily exercise. Swimming is a popular physical activity and an effective option for maintaining and improving cardiovascular health. Recent studies have shown that swimming is beneficial for mental health and cognitive ability (Hillman, Castelli & Buck, 2005; Da Silva et al., 2020). Rats have the innate ability to swim and are the first choice for swimming models (Souza et al., 2009). The purpose of this study was to explore the changes of urine proteome in rats with regular swimming exercise.
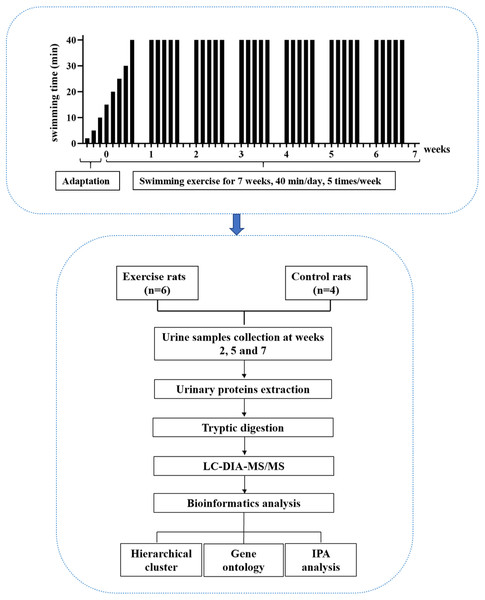
In this study, experimental rats were subjected to daily moderate-intensity swimming exercise for 40 min per day for 7 weeks. Urine samples were collected at weeks 2, 5, and 7. The urine proteome was analyzed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). The experimental design and workflow of the proteomics analysis in this study are shown in Fig. 1.
Figure 1: The experimental design and workflow of the proteomics analysis in this study.
The experimental rats were subjected to daily moderate-intensity swimming exercise for 7 weeks. Urine samples were collected at weeks 2, 5, and 7 during swimming exercise. Urine proteins were identified by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS).Materials & Methods
Experimental animals
Male SD rats (seven days old) were supplied by the Department of Neurobiology, School of Basic Medical Sciences, Peking University. All animals were housed with free access to a standard laboratory diet and water with a 12-h light-dark cycle under standard conditions (indoor temperature 22 ± 1 °C, humidity 65–70%). The experiment was approved by the Institute of Basic Medical College (ID: ACUC-A02-2014-007). The study was performed according to guidelines developed by the Institutional Animal Care and Use Committee of Peking Medical College. After the experiment, all the animals were euthanized by intraperitoneal injection of barbiturates.
Swimming exercise
A large pool (diameter: 1,500 mm, height: 500 mm) served as the swimming pool. The water temperature was maintained at 36 °C. For the adaptation phase, rats swam for increasing amounts of time, from 2 min to 10 min over three days. For the exercise phase, the intensity of exercise in the first week gradually increased from 15 min to 40 min, and the intensity at 40 min lasted for 6 weeks, which is considered to be moderate exercise (Seo et al., 2014). The animals were quickly and gently dried after each training session. The rats (n = 10) were randomly divided into the following two groups: experimental rats (n = 6) and control rats (n = 4). In the experimental group, the rats underwent the 7-week swimming exercise program. The control rats did not swim.
Urine collection and sample preparation
Urine samples were collected from the experimental and control groups at weeks 2, 5 and 7 during the swimming exercise. The animals were individually placed in metabolic cages for 10 h to collect urine samples without any treatment. After collection, the urine samples were stored at −80 °C. The urine samples (n = 30) were centrifuged at 12,000 g for 40 min at 4 °C to remove cell debris. The supernatants were precipitated with three volumes of ethanol at −20 °C overnight and then centrifuged at 12,000 g for 30 min. Then, lysis buffer (8 mol/L urea, 2 mol/L thiourea, 50 mmol/L Tris, and 25 mmol/L DTT) was used to dissolve the pellets. The protein concentration of the urine samples was measured by the Bradford assay.
Tryptic digestion
Urinary proteins (100 µg of each sample) were digested with trypsin (Trypsin Gold, Mass Spec Grade, Promega, Fitchburg, WI, USA) using filter-aided sample preparation (FASP) methods (Wisniewski et al., 2009). These peptide mixtures were desalted using Oasis HLB cartridges (Waters, Milford, MA) and dried by vacuum evaporation (Thermo Fisher Scientific, Bremen, Germany). The digested peptides (n = 30) were redissolved in 0.1% formic acid to a concentration of 0.5 µg/µL. The iRT reagent (Biognosys, Switzerland) was spiked at a concentration of 1:10 v/v into all samples for calibration of the retention time of the extracted peptide peaks. For analysis, 1 µg of peptides from an individual sample was analyzed by LC-DIA-MS/MS.
Reversed-phase fractionation spin column separation
A total of 90 µg of pooled peptides was generated from 6 µl from each sample and then separated by a high-pH reversed-phase peptide fractionation kit (Thermo Pierce, Waltham, MA, USA) according to the manufacturer’s instructions. A step gradient of increasing acetonitrile concentrations (5, 7.5, 10, 12.5, 15, 17.5, 20 and 50%) was applied to the columns to elute the peptides. Ten different fractionated samples (including the flow-through fraction, wash fraction, and eight step gradient sample fractions) were collected and dried by vacuum evaporation. The ten fractions were resuspended in 20 µl of 0.1% formic acid, and 1 µg of each of the fractions was analyzed by LC-DDA-MS/MS.
LC-MS/MS analysis
A total of 30 peptide samples were analyzed in an EASY-nLC 1200 chromatography system (Thermo Fisher Scientific) and an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). The samples were loaded onto a trapping column (75 µm × 2 cm, 3 µm, C18, 100 Å) and separated by a reverse-phase analysis column (75 µm × 25 cm, 2 µm, C18, 100 Å). The eluted gradient was 4%–35% buffer B (0.1% formic acid in 80% acetonitrile) at a flow rate of 300 nL/min for 90 min.
To generate the spectral library, 1 µg of each of ten fractions was analyzed in DDA mode. The parameters were set as follows: the full scan ranged from 350 to 1500 m/z with a resolution of 120,000; MS/MS scans were acquired with a resolution of 30,000; the cycle time was set to 3 s; the HCD energy was set to 30%; the autogain control (AGC) target was set to 4e5; and the maximum injection time was set to 50 ms. In DIA mode, 1 µg of each sample was analyzed. The variable isolation window of the DIA method with 36 windows was set (Table S1). The parameters were set as follows: the full scan ranged from 350 to 1,500 m/z with a resolution of 60,000; the DIA scan was acquired from 200 to 2,000 m/z with a resolution of 30,000; the HCD energy was set to 32%; the AGC target was set to 1e6; and the maximum injection time was set to 100 ms. During the samples analysis, a mixture from each sample was analyzed after every six samples for quality control (QC).
Data analysis
The DDA data of ten fractions were searched against the Swiss-Prot rat database (released in 2017, including 7,992 sequences) appended with the iRT peptide sequence using Proteome Discoverer software (version 2.1, Thermo Scientific). The search parameters were set as follows: two missed trypsin cleavage sites were allowed; the parent ion mass tolerances were set to 10 ppm; the fragment ion mass tolerances were set to 0.02 Da; the carbamidomethyl of cysteine was set as a fixed modification; and the oxidation of methionine was set as a variable modification. The false discovery rate (FDR) of the proteins was less than 1%. A total of 873 protein groups, 4098 peptide groups and 37555 peptide spectrum matches were identified. The search results were used to set the DIA method. The DDA raw files were processed using Spectronaut’s Pulsar database (Biognosys, Switzerland) with the default parameters to generate the spectral library. The DIA raw files were processed using Spectronaut for analysis with the default setting. All of the results were filtered by a Q value cutoff of 0.01 (corresponding to an FDR of 1%). Peptide intensity was calculated by summing the peak areas of their respective fragment ions of MS2, and the protein intensity was calculated by summing the intensities of their respective peptides.
Statistical analysis
The k-nearest neighbor (K-NN) method was used to fill the missing values of protein abundance (Armitage et al., 2015). Comparisons between experimental and control groups were performed by one-way ANOVA. The differential proteins at weeks 2, 5 and 7 were screened by the following criteria: fold change ≥ 1.5 or ≤ 0.67; and P < 0.05 by independent sample t-test. Group differences resulting in p < 0.05 were considered statistically significant.
Functional annotation of the differential proteins
DAVID 6.8 (https://david.ncifcrf.gov/) was used to perform the functional annotation of the differential proteins between the experimental and control groups. The canonical pathways were analyzed with IPA software (Ingenuity Systems, Mountain View, CA, USA).
Results
Urine proteome changes in the swimming exercise rats
In this study, thirty urine samples from three time points (weeks 2, 5, and 7) from six experimental rats and four control rats were used for LC-DIA-MS/MS quantitation. A total of 729 proteins and 5,265 peptides were identified in all urine samples. A quality control sample of a mixture from each sample was analyzed after every six samples. A total of 518 proteins were identified that had a coefficient of variation (CV) of the QC samples below 30%, and all of the identification and quantification details are listed in Table S2.
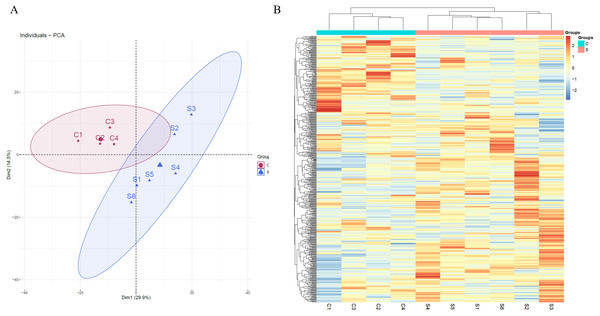
Unsupervised clustering analysis of all of proteins identified at three time points was performed (Fig. S1). We found that the samples at week 2 were clustered together, indicating that swimming exercise has a great impact on urine after 2 weeks. It is speculated that the clustering effect of the samples at weeks 5 and 7 was poor because the body had adapted to long-term exercise. To further characterize the effects of 2 weeks of swimming exercise, all urinary proteins from 10 urine samples between the two groups at week 2 were analyzed by principal component analysis (PCA). As shown in Fig. 2A, the swimming exercise rats were differentiated from the control rats. Meanwhile, unsupervised clustering analysis of all urinary proteins from 10 urine samples between two groups at week 2 was performed. As shown in Fig. 2B, the proteomics profiles of the swimming group were distinctively different from those of the control group. These results demonstrated that the urinary proteome changed significantly after swimming exercise.
Figure 2: Proteomic analysis of the urine samples of swimming exercise rats.
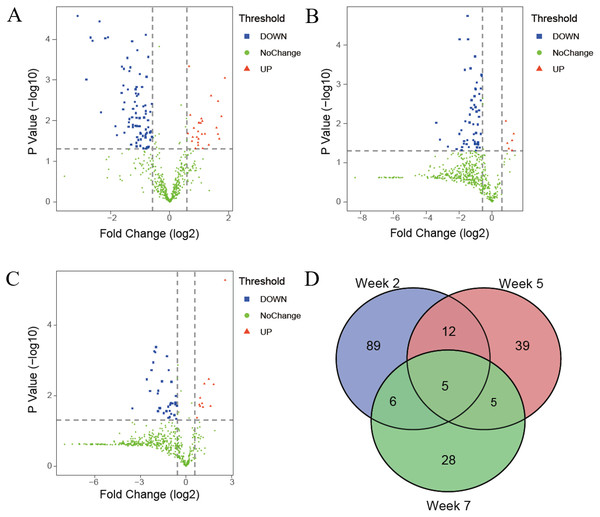
(A) PCA analysis of all proteins from experimental and control urine proteome at week 2. (B) Cluster analysis of all the proteins from experimental and control urine proteome at week 2.The differential proteins were screened with a p value < 0.05 by two-sided, unpaired t-test and a fold change ≥ 1.5 compared with controls. Compared to the control group, 112 differential proteins were identified after 2 weeks of swimming exercise, among which 28 proteins were upregulated and 84 proteins were downregulated (Fig. 3A); 61 differential proteins were identified after 5 weeks of swimming exercise, among which 6 proteins were upregulated and 55 proteins were downregulated (Fig. 3B); and 44 differential proteins were identified after 7 weeks of swimming exercise, among which 11 proteins were upregulated and 33 proteins were downregulated (Fig. 3C). The details of these differential proteins are presented in Table 1. Among these differential proteins, 171 proteins had human orthologs. The overlap of these differential proteins is shown by the Venn diagram in Fig. 3D. Five proteins were commonly identified at three time points (Fig. 3D), including Ig gamma-1 chain C region, hemopexin, transthyretin, cathepsin D and chondroitin sulfate proteoglycan 4.
Figure 3: Differential proteins identified between experimental and control group.
(A) Volcano plots showing P values (−log10) versus protein ratios between experimental and control rats (log2) at week 2. (B) Volcano plots showing P values (−log10) versus protein ratios between experimental and control rats (log2) at week 5. (C) Volcano plots showing P values (−log10) versus protein ratios between experimental and control rats (log2) at week 7. (D) Overlap evaluation of differential proteins at three time points.| Uniprot ID | Protein names | Human ortholog | P-value | Fold change | ||
|---|---|---|---|---|---|---|
| Week 2 | Week 5 | Week 7 | ||||
| P20059 | Hemopexin | P02790 | 0.039679 | 2.501 | 1.896 | 3.514 |
| P24268 | Cathepsin D | P07339 | 0.000272 | 0.605 | 0.420 | 0.453 |
| P02767 | Transthyretin | P02766 | 0.049310 | 0.575 | 0.316 | 0.618 |
| P20759 | Ig gamma-1 chain C region | P01859 | 0.003593 | 0.346 | 0.322 | 0.483 |
| Q00657 | Chondroitin sulfate proteoglycan 4 | Q6UVK1 | 0.000090 | 0.235 | 0.254 | 0.419 |
| Q63556 | Serine protease inhibitor A3M | P01011 | 0.021564 | 3.103 | 0.479 | |
| P27590 | Uromodulin | P07911 | 0.002444 | 2.642 | 0.453 | |
| Q6IFW6 | Keratin, type I cytoskeletal 10 | P13645 | 0.048471 | 2.172 | 2.019 | |
| O70534 | Protein delta homolog 1 | P80370 | 0.048375 | 1.828 | 0.463 | |
| P82450 | Sialate O-acetylesterase | Q9HAT2 | 0.007297 | 1.623 | 0.647 | |
| P47820 | Angiotensin-converting enzyme | P12821 | 0.018595 | 0.642 | 0.303 | |
| Q6AYS7 | Aminoacylase-1A | Q03154 | 0.023841 | 0.532 | 2.322 | |
| P02651 | Apolipoprotein A-IV | P06727 | 0.029010 | 0.508 | 0.362 | |
| P13635 | Ceruloplasmin | P00450 | 0.002251 | 0.479 | 0.501 | |
| Q64319 | Neutral and basic amino acid transport protein rBAT | Q07837 | 0.025707 | 0.465 | 2.484 | |
| B5DFC9 | Nidogen-2 | Q14112 | 0.005150 | 0.423 | 0.392 | |
| P20761 | Ig gamma-2B chain C region | NO | 0.001365 | 0.338 | 1.785 | |
| P00689 | Pancreatic alpha-amylase | NO | 0.015215 | 1.763 | 5.916 | |
| P29975 | Aquaporin-1 | P29972 | 0.016488 | 0.569 | 0.334 | |
| P52759 | 2-iminobutanoate/2-iminopropanoate deaminase | P52758 | 0.006852 | 0.569 | 0.589 | |
| P08721 | Osteopontin | P10451 | 0.048467 | 0.541 | 0.247 | |
| O70513 | Galectin-3-binding protein | Q08380 | 0.000185 | 0.468 | 0.507 | |
| P01015 | Angiotensinogen | P01019 | 0.003462 | 0.440 | 2.158 | |
| P14046 | Alpha-1-inhibitor 3 | NO | 0.040909 | 0.529 | 0.446 | |
| P10960 | Prosaposin | P07602 | 0.001299 | 0.491 | 0.449 | |
| Q99041 | Protein-glutamine gamma-glutamyltransferase 4 | P49221 | 0.028307 | 0.306 | 0.225 | |
| Q4V885 | Collectin-12 | Q5KU26 | 0.014972 | 0.273 | 0.305 | |
| P05544 | Serine protease inhibitor A3L | P01011 | 0.000894 | 3.651 | ||
| P28648 | CD63 antigen | P08962 | 0.007863 | 3.366 | ||
| P84039 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 5 | Q9UJA9 | 0.028089 | 3.202 | ||
| P05545 | Serine protease inhibitor A3K | P01011 | 0.003344 | 3.111 | ||
| O89117 | Beta-defensin 1 | P60022 | 0.015008 | 2.999 | ||
| P32038 | Complement factor D | P00746 | 0.021309 | 2.288 | ||
| P15950 | Glandular kallikrein-3, submandibular | NO | 0.010257 | 2.152 | ||
| P20909 | Collagen alpha-1 | P12107 | 0.021608 | 2.129 | ||
| Q63514 | C4b-binding protein alpha chain | P04003 | 0.008936 | 2.104 | ||
| D3ZTX0 | Transmembrane emp24 domain-containing protein 7 | Q9Y3B3 | 0.011501 | 2.073 | ||
| P49134 | Integrin beta-1 | P05556 | 0.011293 | 1.988 | ||
| Q76HN1 | Hyaluronidase-1 | Q12794 | 0.011380 | 1.982 | ||
| Q6RY07 | Acidic mammalian chitinase | Q9BZP6 | 0.028264 | 1.965 | ||
| P05539 | Collagen alpha-1 | P02458 | 0.040399 | 1.950 | ||
| Q6AXR4 | Beta-hexosaminidase subunit beta | P07686 | 0.018512 | 1.949 | ||
| Q9R1T3 | Cathepsin Z | Q9UBR2 | 0.034738 | 1.942 | ||
| Q5XIL0 | E3 ubiquitin-protein ligase RNF167 | Q9H6Y7 | 0.025554 | 1.891 | ||
| P48199 | C-reactive protein | P02741 | 0.025494 | 1.716 | ||
| P08649 | Complement C4 | P0C0L4 | 0.032173 | 1.686 | ||
| P50430 | Arylsulfatase B | P15848 | 0.000462 | 1.575 | ||
| Q6AYP5 | Cell adhesion molecule 1 | Q9BY67 | 0.020851 | 1.534 | ||
| Q00238 | Intercellular adhesion molecule 1 | P05362 | 0.025199 | 0.662 | ||
| Q920H8 | Hephaestin | Q9BQS7 | 0.003917 | 0.659 | ||
| B0BND0 | Glycerophosphocholine cholinephosphodiesterase ENPP6 | Q6UWR7 | 0.030004 | 0.636 | ||
| Q6Q0N1 | Cytosolic non-specific dipeptidase | Q96KP4 | 0.006146 | 0.636 | ||
| P29598 | Urokinase-type plasminogen activator | P00749 | 0.019588 | 0.630 | ||
| Q8R5M3 | Leucine-rich repeat-containing protein 15 | Q8TF66 | 0.043831 | 0.608 | ||
| Q62638 | Golgi apparatus protein 1 | Q92896 | 0.014649 | 0.607 | ||
| P31044 | Phosphatidylethanolamine-binding protein 1 | NO | 0.046012 | 0.606 | ||
| Q9QX79 | Fetuin-B | Q9UGM5 | 0.009853 | 0.602 | ||
| Q5U367 | Multifunctional procollagen lysine hydroxylase and glycosyltransferase LH3 | O60568 | 0.020139 | 0.586 | ||
| Q5U2Q3 | Ester hydrolase C11orf54 homolog | Q9H0W9 | 0.045189 | 0.584 | ||
| P35704 | Peroxiredoxin-2 | P32119 | 0.000841 | 0.581 | ||
| P46413 | Glutathione synthetase | P48637 | 0.021466 | 0.573 | ||
| Q4FZV0 | Beta-mannosidase | O00462 | 0.000078 | 0.571 | ||
| Q99MA2 | Xaa-Pro aminopeptidase 2 | O43895 | 0.005837 | 0.569 | ||
| Q63530 | Phosphotriesterase-related protein | Q96BW5 | 0.026074 | 0.567 | ||
| P48500 | Triosephosphate isomerase | P60174 | 0.004231 | 0.566 | ||
| P19804 | Nucleoside diphosphate kinase B | P22392 | 0.023348 | 0.564 | ||
| P27139 | Carbonic anhydrase 2 | P00918 | 0.008439 | 0.548 | ||
| P51647 | Retinal dehydrogenase 1 | P00352 | 0.004435 | 0.538 | ||
| P04639 | Apolipoprotein A-I | P02647 | 0.001486 | 0.524 | ||
| P51635 | Aldo-keto reductase family 1 member A1 | P14550 | 0.021157 | 0.509 | ||
| P69897 | Tubulin beta-5 chain | P07437 | 0.019827 | 0.504 | ||
| P53813 | Vitamin K-dependent protein S | P07225 | 0.021142 | 0.503 | ||
| P62963 | Profilin-1 | P07737 | 0.042120 | 0.501 | ||
| P08650 | Complement C5 | NO | 0.000712 | 0.496 | ||
| Q9QXQ0 | Alpha-actinin-4 | O43707 | 0.013840 | 0.494 | ||
| O55004 | Ribonuclease 4 | P34096 | 0.008646 | 0.492 | ||
| P85971 | 6-phosphogluconolactonase | O95336 | 0.013814 | 0.488 | ||
| P19112 | Fructose-1,6-bisphosphatase 1 | P09467 | 0.029793 | 0.480 | ||
| Q62930 | Complement component C9 | P02748 | 0.004371 | 0.464 | ||
| P60711 | Actin, cytoplasmic 1 | P60709 | 0.002492 | 0.462 | ||
| P22282 | Cystatin-related protein 1 | NO | 0.043085 | 0.460 | ||
| P42123 | L-lactate dehydrogenase B chain | P07195 | 0.001504 | 0.460 | ||
| P08289 | Alkaline phosphatase, tissue-nonspecific isozyme | P05186 | 0.013575 | 0.460 | ||
| P05964 | Protein S100-A6 | P06703 | 0.008557 | 0.459 | ||
| P00884 | Fructose-bisphosphate aldolase B | P05062 | 0.035549 | 0.457 | ||
| P50399 | Rab GDP dissociation inhibitor beta | P50395 | 0.006541 | 0.453 | ||
| P02770 | Albumin | P02768 | 0.002136 | 0.451 | ||
| Q63716 | Peroxiredoxin-1 | Q06830 | 0.001691 | 0.449 | ||
| Q06496 | Sodium-dependent phosphate transport protein 2A | Q06495 | 0.013307 | 0.448 | ||
| P41562 | Isocitrate dehydrogenase [NADP] cytoplasmic | O75874 | 0.014681 | 0.422 | ||
| P01041 | Cystatin-B | P04080 | 0.012801 | 0.421 | ||
| P04642 | L-lactate dehydrogenase A chain | P00338 | 0.000113 | 0.415 | ||
| Q9Z339 | Glutathione S-transferase omega-1 | P78417 | 0.008351 | 0.415 | ||
| D4ACX8 | Protocadherin-16 | Q96JQ0 | 0.039161 | 0.408 | ||
| Q9WTW7 | Solute carrier family 23 member 1 | Q9UHI7 | 0.038746 | 0.402 | ||
| P17475 | Alpha-1-antiproteinase | P01009 | 0.000491 | 0.396 | ||
| P50115 | Protein S100-A8 | P05109 | 0.009132 | 0.389 | ||
| Q6P734 | Plasma protease C1 inhibitor | P05155 | 0.000603 | 0.388 | ||
| P34080 | Aquaporin-2 | P41181 | 0.010950 | 0.383 | ||
| P20762 | Ig gamma-2C chain C region | NO | 0.014244 | 0.382 | ||
| Q5FVQ0 | Metal cation symporter ZIP8 | Q9C0K1 | 0.001720 | 0.376 | ||
| P15978 | Class I histocompatibility antigen, Non-RT1.A alpha-1 chain | P01891 | 0.000287 | 0.370 | ||
| P34058 | Heat shock protein HSP 90-beta | P08238 | 0.019878 | 0.344 | ||
| P09006 | Serine protease inhibitor A3N | P01011 | 0.000536 | 0.341 | ||
| P04276 | Vitamin D-binding protein | P02774 | 0.005304 | 0.335 | ||
| Q66HG4 | Galactose mutarotase | Q96C23 | 0.000467 | 0.326 | ||
| Q63772 | Growth arrest-specific protein 6 | Q14393 | 0.002087 | 0.323 | ||
| P50116 | Protein S100-A9 | P06702 | 0.022965 | 0.290 | ||
| P00697 | Lysozyme C-1 | P61626 | 0.013674 | 0.279 | ||
| P12346 | Serotransferrin | P02787 | 0.000095 | 0.220 | ||
| P01026 | Complement C3 | P01024 | 0.006223 | 0.200 | ||
| P06866 | Haptoglobin | P00739 | 0.000036 | 0.193 | ||
| Q64268 | Heparin cofactor 2 | P05546 | 0.000109 | 0.164 | ||
| Q9EQV9 | Carboxypeptidase B2 | Q96IY4 | 0.000091 | 0.156 | ||
| Q63313 | Lipopolysaccharide-binding protein | P18428 | 0.000977 | 0.141 | ||
| P15399 | Probasin | NO | 0.000027 | 0.115 | ||
| Q6IFV1 | Keratin, type I cytoskeletal 14 | P02533 | 0.048522 | 2.314 | ||
| Q642A7 | Protein FAM151A | Q8WW52 | 0.000571 | 0.641 | ||
| P23680 | Serum amyloid P-component | P02743 | 0.001278 | 0.622 | ||
| P70490 | Lactadherin | Q08431 | 0.002590 | 0.619 | ||
| P15083 | Polymeric immunoglobulin receptor | P01833 | 0.018502 | 0.617 | ||
| P16391 | RT1 class I histocompatibility antigen, AA alpha chain | NO | 0.041460 | 0.590 | ||
| P36373 | Glandular kallikrein-7, submandibular/renal | P06870 | 0.001770 | 0.582 | ||
| Q05820 | Putative lysozyme C-2 | P61626 | 0.006730 | 0.575 | ||
| Q63041 | Alpha-1-macroglobulin | NO | 0.004656 | 0.574 | ||
| P26051 | CD44 antigen | P16070 | 0.034146 | 0.560 | ||
| P61972 | Nuclear transport factor 2 | P61970 | 0.029915 | 0.550 | ||
| Q63621 | Interleukin-1 receptor accessory protein | Q9NPH3 | 0.031691 | 0.546 | ||
| P97829 | Leukocyte surface antigen CD47 | Q08722 | 0.010609 | 0.543 | ||
| Q6AYE5 | Out at first protein homolog | Q86UD1 | 0.033694 | 0.541 | ||
| P13221 | Aspartate aminotransferase, cytoplasmic | P17174 | 0.000894 | 0.521 | ||
| P54759 | Ephrin type-A receptor 7 | Q15375 | 0.004971 | 0.518 | ||
| P16573 | Carcinoembryonic antigen-related cell adhesion molecule 1 | P13688 | 0.017399 | 0.506 | ||
| P36376 | Glandular kallikrein-12, submandibular/renal | P06870 | 0.049426 | 0.500 | ||
| Q9WUK5 | Inhibin beta C chain | P55103 | 0.003809 | 0.497 | ||
| Q9EPB1 | Dipeptidyl peptidase 2 | Q9UHL4 | 0.000194 | 0.486 | ||
| Q9R0D6 | Transcobalamin-2 | P20062 | 0.033115 | 0.484 | ||
| P13852 | Major prion protein | P04156 | 0.033316 | 0.479 | ||
| P43303 | Interleukin-1 receptor type 2 | P27930 | 0.002016 | 0.474 | ||
| Q9R0T4 | Cadherin-1 | P12830 | 0.003331 | 0.473 | ||
| Q794F9 | 4F2 cell–surface antigen heavy chain | P08195 | 0.026565 | 0.450 | ||
| Q64604 | Receptor-type tyrosine-protein phosphatase F | P10586 | 0.004355 | 0.444 | ||
| Q6IUU3 | Sulfhydryl oxidase 1 | O00391 | 0.003978 | 0.412 | ||
| Q7TPB4 | CD276 antigen | Q5ZPR3 | 0.000390 | 0.403 | ||
| P11232 | Thioredoxin | P10599 | 0.009423 | 0.386 | ||
| P98158 | Low-density lipoprotein receptor-related protein 2 | P98164 | 0.040045 | 0.373 | ||
| P53369 | 7,8-dihydro-8-oxoguanine triphosphatase | P36639 | 0.000018 | 0.354 | ||
| P35859 | Insulin-like growth factor-binding protein complex acid labile subunit | P35858 | 0.026722 | 0.351 | ||
| P07154 | Procathepsin L | P07711 | 0.033555 | 0.341 | ||
| Q91XT9 | Neutral ceramidase | Q9NR71 | 0.039975 | 0.281 | ||
| Q0PMD2 | Anthrax toxin receptor 1 | Q9H6X2 | 0.048291 | 0.276 | ||
| Q30 kJ2 | Beta-defensin 50 | 0.018575 | 0.251 | |||
| P52796 | Ephrin-B1 | P98172 | 0.045230 | 0.224 | ||
| P32736 | Opioid-binding protein/cell adhesion molecule | Q14982 | 0.038970 | 0.156 | ||
| P97580 | Beta-microseminoprotein | P08118 | 0.025914 | 0.110 | ||
| P06760 | Beta-glucuronidase | P08236 | 0.009520 | 0.095 | 0.208 | |
| P20760 | Ig gamma-2A chain C region | P01859 | 0.019815 | 3.050 | ||
| P42854 | Regenerating islet-derived protein 3-gamma | NO | 0.003409 | 2.815 | ||
| P01836 | Ig kappa chain C region, A allele | P01834 | 0.004604 | 2.299 | ||
| P20611 | Lysosomal acid phosphatase | P11117 | 0.016619 | 2.073 | ||
| P01835 | Ig kappa chain C region, B allele | P01834 | 0.011652 | 1.900 | ||
| Q920A6 | Retinoid-inducible serine carboxypeptidase | Q9HB40 | 0.020110 | 1.885 | ||
| Q5XI43 | Matrix remodeling-associated protein 8 | Q9BRK3 | 0.018083 | 1.828 | ||
| Q6P7S1 | Acid ceramidase | Q13510 | 0.042780 | 1.643 | ||
| Q499T2 | Gamma-interferon-inducible lysosomal thiol reductase | P13284 | 0.019485 | 0.660 | ||
| P08592 | Amyloid-beta A4 protein | P05067 | 0.045641 | 0.637 | ||
| P07897 | Aggrecan core protein | P16112 | 0.010204 | 0.629 | ||
| Q9JHY1 | Junctional adhesion molecule A | Q9Y624 | 0.016015 | 0.615 | ||
| P04906 | Glutathione S-transferase P | P09211 | 0.016839 | 0.541 | ||
| Q6MG71 | Choline transporter-like protein 4 | Q53GD3 | 0.004089 | 0.515 | ||
| P0CG51 | Polyubiquitin-B [Cleaved into: Ubiquitin] | P0CG47 | 0.016453 | 0.504 | ||
| P02650 | Apolipoprotein E | P02649 | 0.040693 | 0.477 | ||
| Q6TUD4 | Protein YIPF3 | Q9GZM5 | 0.027382 | 0.472 | ||
| Q05695 | Neural cell adhesion molecule L1 | P32004 | 0.027649 | 0.411 | ||
| P10247 | H-2 class II histocompatibility antigen gamma chain | P04233 | 0.031337 | 0.362 | ||
| Q9JJ19 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | O14745 | 0.022332 | 0.297 | ||
| Q62632 | Follistatin-related protein 1 | Q12841 | 0.007274 | 0.282 | ||
| Q5M871 | Fas apoptotic inhibitory molecule 3 | O60667 | 0.027711 | 0.279 | ||
| Q9WUC4 | Copper transport protein ATOX1 | O00244 | 0.008730 | 0.275 | ||
| Q06880 | Neuroblastoma suppressor of tumorigenicity 1 | P41271 | 0.000421 | 0.254 | ||
| Q63467 | Trefoil factor 1 | P04155 | 0.000552 | 0.234 | ||
| P07171 | Calbindin | P05937 | 0.007365 | 0.193 | ||
| Q09030 | Trefoil factor 2 | Q03403 | 0.003471 | 0.168 | ||
| P97574 | Stanniocalcin-1 | P52823 | 0.023304 | 0.087 | ||
Tissue distribution of the human orthologs of the differential proteins
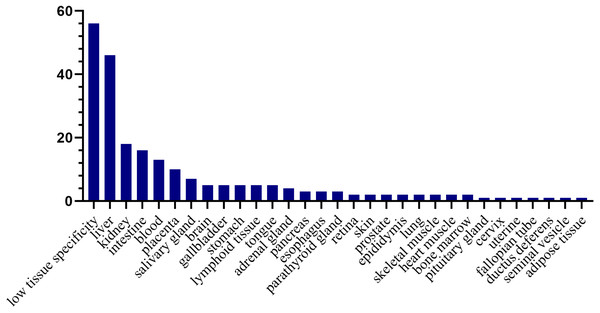
To investigate the expression levels of the differential proteins in different tissues and organs, 171 differential proteins that had human orthologs were searched from the Human Protein Atlas. According to the Tissue Atlas, 31 tissues were identified (Fig. 4). The differential proteins were strongly expressed in the liver, kidney, intestine, and blood, indicating that these organs may be affected after swimming exercise. Swimming exercise can recruit a large volume of muscle mass. Notably, two proteins, triosephosphate isomerase (TIPSS) and aspartate aminotransferase (AATC), were strongly expressed in skeletal muscle, indicating that moderate-intensity swimming exercise might have an effect on the muscles of rats.
Figure 4: Tissue distribution of the human orthologs of differential proteins.
X-axis represents human tissues; Y- axis represents the number of differential proteins.Randomized grouping statistical analysis
Considering that omics data are large but the sample size is limited, the differences between the two groups may be randomly generated. To confirm whether the differential proteins were indeed due to swimming exercise, we performed a randomized grouping statistical analysis. We randomly allocated the proteomic data of 10 samples (6 for experimental and 4 for control samples) at each time point and screened for the differential proteins with the same criteria. Then, the average number of differential proteins in all random combinations was calculated, which was the false positive in the actual grouping. There were 210 random allocations at each time point, and the average number of differential proteins in all random combinations at each time point was 15, 5 and 6. The results showed that the false-positive rates were 13.4%, 5% and 13.6% at weeks 2, 5 and 7, respectively. Therefore, most of the differential proteins identified at each time point in this study were caused by swimming exercise rather than random allocation. The details are presented in Table S3. These results suggested that the sample size of this study was sufficient to prove the significant difference in the urine proteome between the swimming group and the control group.
Functional annotation analysis of the differential proteins
Functional annotation of differential proteins at weeks 2, 5 and 7 was performed by DAVID (Huang, Sherman & Lempicki, 2009). The differential proteins identified at three time points were classified into three categories: biological process, cellular component and molecular function.
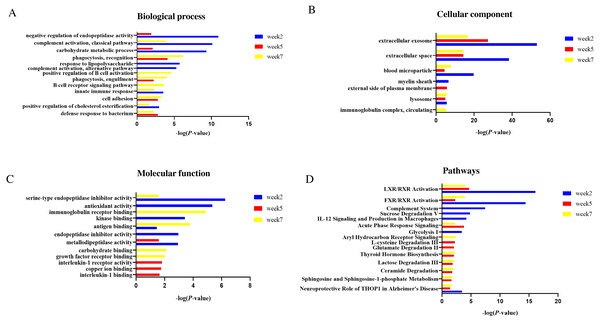
In the biological process category (Fig. 5A), negative regulation of endopeptidase activity and carbohydrate metabolic process were overrepresented at weeks 2 and 5; complement activation, classical pathway, innate immune response and positive regulation of cholesterol esterification were overrepresented at weeks 2 and 7. Response to lipopolysaccharide and positive regulation of cholesterol esterification were only overrepresented at week 2; B cell receptor signaling pathway and positive regulation of B cell activation were only overrepresented at week 7.
Figure 5: Functional enrichment analysis of differential proteins in this study.
(A) Biological process (B) Cellular component (C) Molecular function (D) Canonical pathways.In the cellular component category (Fig. 5B), the majority of these differential proteins were from extracellular exosome and extracellular space. In the molecular function category (Fig. 5C), metallodipeptidase activity was overrepresented at weeks 2 and 5; serine-type endopeptidase inhibitor activity and antigen binding were overrepresented at weeks 2 and 7.
To characterize the canonical pathways involved with these differential proteins, IPA software was used for analysis. As shown in Fig. 5D, LXR/RXR activation and FXR/RXR activation were enriched at three time points. Sphingosine and sphingosine-1-phosphate metabolism, ceramide degradation, lactose degradation III, and thyroid hormone biosynthesis were enriched at weeks 5 and 7. Complement system, sucrose degradation V, IL-12 signaling and production in macrophages, and glycolysis I were enriched at week 2. Glutamate degradation II was enriched at week 5. Aryl hydrocarbon Receptor signaling was enriched at week 7.
Discussion
In this study, daily moderate-intensity swimming exercise rat model was established. Compared to the control group, a total of 112, 61 and 44 differential proteins were identified after 2, 5 and 7 weeks of swimming exercise, respectively. Randomized grouping statistical analysis showed that more than 85% of the differential proteins identified in this study were caused by swimming exercise rather than random allocation.
By biological process analysis, we found that some processes of differential proteins were consistent with previous researches. For example, some immune-related processes were enriched after swimming exercise. Exercise has a profound effect on immune system function, and studies have shown that regular moderate intensity exercise is beneficial for immunity (Pedersen & Hoffman-Goetz, 2000; Simpson et al., 2015). Furthermore, we found that positive regulation of cholesterol esterification was enriched after swimming exercise in this study. Regular physical exercise provides a wide range of cardiovascular benefits as a nonpharmacological treatment and promotes cholesterol esterification and transport from peripheral tissues to the liver (Simko & Kelley, 1979; Mann, Beedie & Jimenez, 2014).
Additionally, some pathways were previously reported to be associated with physical exercise. For example, sphingosine-1-phosphate (S1P) plays an important role in skeletal muscle pathophysiology, and S1P metabolism was found to be regulated by exercise (Hodun, Chabowski & Baranowski, 2021). The S1P content in plasma and its receptors in skeletal muscles were reported to be increased in the skeletal muscle of rats after resistance training (Banitalebi et al., 2013). Sphingosine and sphingosine-1-phosphate metabolism were enriched in the urine after swimming exercise in this study. Additionally, carbohydrates are the most efficient fuel for working muscles. The first metabolic pathways of carbohydrate metabolism are skeletal muscle glycogenolysis and glycolysis, and circulating glucose becomes an important energy source. Lactate was also reported to play a primary role as either a direct or indirect energy source for contracting skeletal muscle. We found that some glucose metabolism-related pathways were enriched in urine. Furthermore, glutamate has been implicated in exhaustive or vigorous exercise (Guezennec et al., 1998), and a study showed that glutamate increased significantly in the visual cortex following exercise (Maddock et al., 2016). In this study, we found that glutamate degradation II was enriched in urine following moderate-intensity exercise. Overall, the urine proteome can reflect changes associated with physical exercise.
This study was a preliminary study with a limited number of rats, and the differential proteins identified in this study require further verification in a large number of human urine samples. Urine proteomes after different lengths of exercise were different, suggesting that urine proteomics may distinguish long-term and short-term responses to exercise. Additionally, this is a starting point for further studies of urinary proteome after different types and intensities of exercise to monitor the amount of exercise and to develop an optimal exercise plan. Physical exercise may be an influencing factor in urine proteomics research. When using human urine samples to discover disease biomarkers, physical exercise-related effects can be excluded in future studies.
Conclusions
Our results revealed that the urinary proteome could reflect significant changes after swimming exercise. These findings may provide an approach to monitor the effects of exercise of the body.