Multiple virulence factors regulated by AlgU contribute to the pathogenicity of Pseudomonas savastanoi pv. glycinea in soybean
- Published
- Accepted
- Received
- Academic Editor
- Heng Yin
- Subject Areas
- Agricultural Science, Microbiology, Molecular Biology, Plant Science
- Keywords
- AlgU, Pseudomonas savastanoi pv. glycinea, Pathogenicity, Soybean, Coronatine, Extracellular polysaccharide
- Copyright
- © 2021 Nguyen et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Multiple virulence factors regulated by AlgU contribute to the pathogenicity of Pseudomonas savastanoi pv. glycinea in soybean. PeerJ 9:e12405 https://doi.org/10.7717/peerj.12405
Abstract
Pseudomonas savastanoi pv. glycinea (Psg) causes bacterial blight of soybean. To identify candidate virulence factors, transposon-mediated mutational analysis of Psg was carried out. We syringe-inoculated soybean leaves with Psg transposon mutants and identified 28 mutants which showed reduced virulence from 1,000 mutants screened. Next, we spray-inoculated soybean leaves with these mutants and demonstrated that the algU mutant showed significantly reduced virulence together with reduced bacterial populations in planta. Expression profiles comparison between the Psg wild-type (WT) and algU mutant in HSC broth revealed that expression of coronatine (COR)-related genes (including cmaA and corR) were down-regulated in the algU mutant compared with Psg WT. Moreover, we also showed that COR production were reduced in the algU mutant compared with WT. We also demonstrated that algD, which is related to alginate biosynthesis, showed reduced expression and biofilm formation was significantly suppressed in the algU mutant. Furthermore, hrpL also showed less expression in the algU mutant. These results indicate that AlgU plays a critical role in promoting Psg pathogenesis by regulating multiple virulence factors.
Introduction
Pseudomonas savastanoi pv. glycinea (Psg) causes bacterial blight of soybean. The disease is characterized by circular necrotic lesions on leaves surrounded by a chlorotic halo (Ignjatov et al., 2007). In P. syringae, P. cannabina, and P. savastanoi, the phytotoxin Coronatine (COR) is important in inducing chlorosis, and contributes to bacterial growth and lesion formation (Bender et al., 1987; Budde & Ullrich, 2000; Peñaloza-Vázquez et al., 2000; Uppalapati et al., 2005; Qi et al., 2011; Sakata et al., 2021). In Psg PG4180, COR synthesis genes reside on a 90 kb plasmid designated p4180A (Bender, Young & Mitchell, 1991), with a 32 kb COR gene cluster which consists of two distinct regions encoding coronafacic acid (CFA) and coronamic acid (CMA) (Bender, 1999). Psg produces COR not only for the biological fitness of pathogens in planta (Ullrich et al., 1993) but also in vitro (Hoitink & Sinden, 1970; Palmer & Bender, 1993). Many studies showed the roles of COR in chlorosis, promoting lesion formation, and suspension of both stomatal and salicylic acid (SA)-dependent defenses (Peñaloza-Vázquez et al., 2000; Kloek et al., 2001; Zhao et al., 2003; Brooks, Bender & Kunkel, 2005; Melotto et al., 2006; Uppalapati et al., 2007). COR contributes to P. syringae pv. tomato (Pst) DC3000 virulence by suppressing the host defense response (Uppalapati et al., 2007). COR suppresses pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), especially stomatal-based defense in the early Pst DC3000 infection stage in Arabidopsis thaliana and tomato (Melotto et al., 2006; Ishiga et al., 2018).
Besides COR, the type three secretion system (T3SS) also plays a critical role in P. syringae virulence. The T3SS, is encoded by the hrp (hypersensitive response and pathogenicity) cluster, and transfers type three effectors (T3Es) into plant cells to suppress PTI, contribute to pathogenesis, and enhance disease symptoms and bacterial multiplication (Brooks, Bender & Kunkel, 2005; Lam et al., 2014). Furthermore, Psg also produces pectolytic enzymes which allow the pathogen to invade and multiply in the intercellular spaces of host tissues. This is the physiological capability of Psg in using plant polysaccharides and providing flexibility for its pathogen activity (Haefelet & Lindow, 1987).
AlgU, an extracytoplasmic function (ECF) sigma factor, is also important in supporting P. syringae growth and disease development (Markel et al., 2016). AlgU regulates between 800 to 1,000 genes (Yu et al., 2014), and importantly contributes to virulence gene regulation as well as flagellin repression (Schreiber & Desveaux, 2011; Markel et al., 2016; Bao et al., 2020). Pst DC3000 AlgU is not only able to regulate gene expression associated with T3SEs and the phytotoxin COR, but also alginate biosynthesis (Ishiga et al., 2018). Moreover, AlgU (previously called AlgT) also induces transcription of the algT-mucAB gene cluster and the algD operon, which are responsible for alginate biosynthesis in Psg PG4180 (Schenk et al., 2006). Although AlgU has been extensively studied in several P. syringae pathovars, its roles in Psg pathogenicity have not been elucidated yet.
To identify genes related to P. syringae pathogenicity, researchers carried out a screen for P. syringae mutants with reduced virulence. Pst DC3000 Tn5 mutants with reduced virulence on A. thaliana found the crucial functions of COR in virulence (Brooks et al., 2004). Sakata et al. (2019) also screened for P. cannabina pv. alisalensis (Pcal) KB211 Tn5 mutants with reduced virulence on cabbage plants using a dip-inoculation method, and identified multiple virulence factors including the T3SS, membrane transporters, transcription factors, and amino acid metabolism genes. Thus, it is tempting to speculate that each P. syringae pathovar has developed its own virulence factors. However, a screening study to identify Psg virulence factors has not been conducted previously.
To investigate Psg virulence factors, we constructed a bacterial mutant library based on transposon insertion in Psg, and screened for mutants with less or no chlorosis on soybean leaves after syringe-inoculation. We successfully identified several virulence factors including COR, T3Es, and AlgU. Expression profiles revealed that AlgU promotes virulence in host plants by up-regulation of COR-related gene expression. We also showed that algU mutant showed reduced COR production and biofilm formation compared to WT. Our results provide evidence that AlgU plays a critical role in promoting Psg pathogenesis.
Materials & methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas strains were routinely cultured on King’s B (KB; King, Ward & Raney, 1954) medium or mannitol-glutamate (MG; Keane, Kerr & New, 1970) medium at 28 °C. Escherichia coli (E. coli) cultures were grown on Luria-Bertani (LB; Sambrook, Fritsch & Maniatis, 1989) medium at 37 °C. The bacterial cell densities at 600 nm (OD600) were measured using a Biowave CO8000 Cell Density Meter (Funakoshi, Tokyo, Japan) as described in Sakata et al. (2021).
| Bacterial strain or plasmid | Locus | Relevant characteristics | Reference or source |
|---|---|---|---|
| E. coli strain | |||
| DH5α | F − λ − φ80dLacZΔM15Δ (lacZYA-argF) U169 recA1 endA1 hsdR17 (rK − mK+) supE44 thi-1gyrA relA1 |
Takara, Kyoto, Japan | |
| S17-1 | Thi pro hsdR-hsdM + recA (chr::RP4-2-Tc::Km::Tn7) | Schäfer et al. (1994) | |
| P. savastanoi pv. glycinea | |||
| Psg | Psg wild-type | MAFF301684 | |
| VTB8 | PsgB076_27735 | cfa6::mTn5, Nalr, Kmr | This study |
| VTC23 | PsgB076_27795 | cmaA::mTn5, Nalr, Kmr | This study |
| VTD16 | PsgB076_27730 | cfa5::mTn5, Nalr, Kmr | This study |
| VTD28 | PsgB076_27725 | cfa4::mTn5, Nalr, Kmr | This study |
| VTD29 | PsgB076_27740 | cfa7::mTn5, Nalr, Kmr | This study |
| VTD30 | PsgB076_27720 | cfa3::mTn5, Nalr, Kmr | This study |
| VTE13 | PsgB076_27750 | cfa8::mTn5, Nalr, Kmr | This study |
| VTE17 | PsgB076_27740 | cfa7::mTn5, Nalr, Kmr | This study |
| VTF3 | PsgB076_27705 | cfa2::mTn5, Nalr, Kmr | This study |
| VTG22 | PsgB076_27735 | cfa6::mTn5, Nalr, Kmr | This study |
| VTG29 | PsgB076_27740 | cfa7::mTn5, Nalr, Kmr | This study |
| VTG41 | PsgB076_27725 | cfa5::mTn5, Nalr, Kmr | This study |
| VTH39 | PsgB076_27735 | cfa6::mTn5, Nalr, Kmr | This study |
| VTI15 | PsgB076_27740 | cfa7::mTn5, Nalr, Kmr | This study |
| VTR4 | PsgB076_27740 | cfa7::mTn5, Nalr, Kmr | This study |
| VTT14 | PsgB076_27805 | cmaC::mTn5, Nalr, Kmr | This study |
| VTT22 | PsgB076_27730 | cfa5::mTn5, Nalr, Kmr | This study |
| VTB2 | PsgB076_28870 | Type III effector protein XopAD::mTn5, Nalr, Kmr | This study |
| VTB52 | PsgB076_09885 | Heat shock protein 90 (Hsp90)::mTn5, Nalr, Kmr | This study |
| VTH40 | PsgB076_15537 | MFS transporter::mTn5, Nalr, Kmr | This study |
| VTM22 | PsgB076_27425 | Hypothetical protein::mTn5, Nalr, Kmr | This study |
| VTO15 | PsgB076_01129 | ABC transporter permease::mTn5, Nalr, Kmr | This study |
| VTO41 | PsgB076_06035 | Sigma factor algU::mTn5, Nalr, Kmr | This study |
| VTP20 | PsgB076_26940 | DNA-binding protein::mTn5, Nalr, Kmr | This study |
| VTE41 | PsgB076_09720 | Unknown, Nalr, Kmr | This study |
| VTF6 | PsgB076_10315 | Unknown, Nalr, Kmr | This study |
| VTO37 | PsgB076_19532 | Unknown, Nalr, Kmr | This study |
| VTT8 | PsgB076_26618 | Unknown, Nalr, Kmr | This study |
|
algU mutant (VTO41) + pDSKG-algU |
algU mutant complemented with pDSKG-algU, Nalr, Genr |
This study | |
| Plasmid | |||
| pBSLC1 | Transposon vector constructed by ligation of pBSL118 and pHSG396 at EcoRI site, Ampr, Kmr, Cmr | Sawada et al. (2018) | |
| pDSK519 | Broad-host-range cloning vector, Kenr | Keen et al. (1988) | |
| pDSKG | Broad-host-range cloning vector, Genr | This study | |
| pDSKG-algU | The vector containing algU gene inserted into pDSKG, Genr | This study | |
Note:
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Genr, gentamicin; Kmr, kanamycin resistance; Nalr, nalidixic acid resistance.
Bacterial in vitro growth measurements
Wild-type, the algU mutant, and the algU-complemented strain were grown at 28 °C on LB medium. The bacterial suspensions were standardized to an OD600 of 0.05 with LB, and bacterial growth was measured at OD600 for 6, 9, and 12 h.
Plant material and inoculation procedures
Soybean plants (Glycine max), cultivar “Enrei”, were grown in a growth chamber at 22 °C, with approximately 60% humidity, and a supplementary light intensity of 200–350 μmol/(m2 s) for a 14 h photoperiod. All soybean plants used for virulence studies were 3 or 4-week-old.
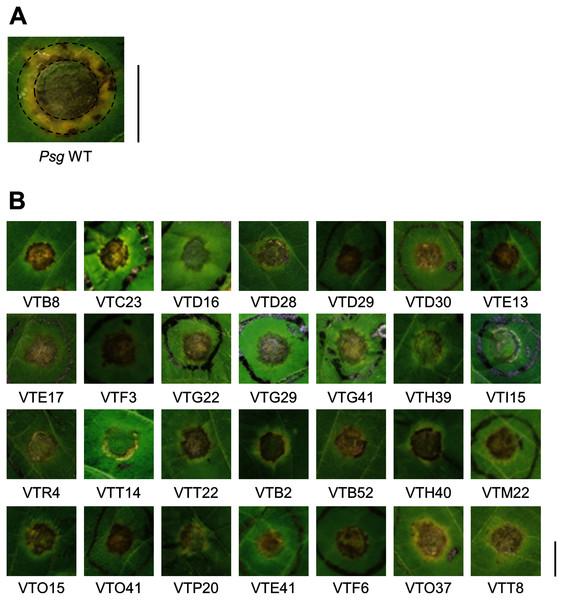
Psg carrying Tn5 transposon was syringe-infiltrated into soybean leaves at an OD600 of 0.1 (5 × 107 CFU/ml) containing 0.02% Silwet L-77 (OSi Specialties Inc., Danbury, CT, USA). The disease symptoms were fully developed at 6 days post inoculation (dpi) (Fig. 1A). The mutants which showed different disease symptoms or virulence reduction in comparison to Psg WT were selected.
Figure 1: Disease symptoms on soybean leaves syringe-inoculated with Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT) and mutants.
(A) Disease symptoms on soybean leaves syringe-inoculated with 5 × 107 colony forming units (CFU)/ml of the Psg WT at six dpi. (B) Disease symptoms on soybean leaves syringe-inoculated with 5 × 107 colony forming units (CFU)/ml of the Psg mutants at six dpi. Scale bars shows 1 cm.For spray inoculation, bacterial suspensions were applied to observe disease symptoms on whole soybean plants as described previously (Uppalapati et al., 2007). Plants were sprayed with a bacterial suspension at an OD600 of 0.2 (1 × 108 CFU/ml) in sterile distilled water containing 0.025% Silwet L-77 until runoff. After inoculation, plants were transferred to growth chambers at 28 °C with approximately 90% to 100% humidity for 24 h in the dark before maintaining plants at approximately 70% humidity.
For syringe inoculation, bacteria were suspended at a final concentration at an OD600 of 0.1 (5 × 107 CFU/ml), 0.01 (5 × 106 CFU/ml), and 0.001 (5 × 105 CFU/ml), and infiltrated with a one-ml blunt syringe into leaves. The plants were then incubated at 70–80% humidity for the rest of the experimental period. Leaves were removed and photographed at five dpi.
To measure bacterial growth in soybean leaves after spray inoculation, individual second leaf pairs were selected at six dpi, weighed and surface-sterilized in 5% H2O2 for 3 min, and then rinsed three times with sterile water. The leaves were then homogenized, and appropriate dilutions were plated on KB medium containing the appropriate antibiotics. The bacterial colony forming units (CFU) were normalized as CFU/g using the total inoculated leaf mass. The population at 0 dpi was estimated using leaves harvested 1 h post inoculation (hpi) without surface-sterilization. For syringe-inoculation, leaf discs were harvested using a 3.5 mm-diameter cork-borer from syringe-infiltrated leaf zone. The leaves were then homogenized, and appropriate dilutions were plated on KB medium containing the appropriate antibiotics. The bacterial colony forming units (CFU) were normalized as CFU/cm2 using the leaf square meters. The bacterial populations were evaluated in at least three independent experiments.
Transposon-mediated mutagenesis and identification of mutated genes
Transposon-mediated mutagenesis was carried out, as described previously (Sakata et al., 2019). Briefly, pBSLC1 (Sawada et al., 2018) carrying mini-Tn5 transposon were transferred into Psg to build a mutant library. We developed more than 1,000 individual Psg mutant lines. After the inoculation assay, we identified the mutated genes by rescuing the transposon insertion sites into an E. coli plasmid and sequencing (Sakata et al., 2019).
Complementation of the algU mutant
The algU-complemented strain was constructed as described in Ishiga et al. (2018). Briefly, the pDSKG vector was made from pDSK519 vector (Keen et al., 1988) by replacing kanamycin cassette to gentamycin. The algU and promoter region were transferred into the pDSKG vector to generate pDSKG-algU. The pDSKG-algU construct was introduced into the algU mutant by electrophoresis to generate the complemented strain.
Real-time quantitative RT-PCR
For Psg gene expression profiles, data were collected as previously described in Sakata et al. (2021). Specifically, bacteria were grown in HS medium optimized for COR production (HSC; Palmer & Bender, 1993) for 3 and 48 h. Bacterial RNA was extracted using the ReliaPrep RNA Cell Miniprep System Kit (Promega, WI, USA) according to the manufacture’s protocol. Two micrograms of total RNA were treated with gDNA Remover (TOYOBO, Osaka, Japan) to eliminate genomic DNA, and the DNase-treated RNA was reverse transcribed using the ReverTra Ace qPCR RT Master Mix (TOYOBO). The cDNA (1:10) was then used for RT-qPCR using the primers shown in Table S1 with THUNDERBIRD SYBR qPCR Mix (TOYOBO) on a Thermal Cycler Dice Real Time System (TaKaRa). Psg outer membrane lipoprotein I (oprI) was used to normalize gene expression.
COR quantification by HPLC
Psg WT, the algU mutant, and the algU-complemented strain were cultured in HSC for 7 days. Culture supernatant was obtained by centrifugation (12,000 × g for 5 min). Cell pellets were dried at 65 °C and weighed. The 500 µl of supernatants were extracted twice with 500 µl of ethyl acetate and 25 µl of HCl, and the organic phase was transferred to a new microcentrifuge tube. The sample was dried by centrifugal evaporator at 55 °C, and the dried sample was dissolved with 0.05% trifluoroacetic acid (TFA)/acetonitrile (9:1, v/v). The culture supernatant was analyzed by HPLC with a Shimadzu LC20A system equipped with a Symmetry C8 column (4.6 × 250 mm; Waters Corporation, MA, USA) as described previously (Sakata et al., 2021).
Hypertrophy-inducing activity assay on potato tuber tissue
Potato tubers were cut from the central tuber portion to ensure samples of high uniformity. After washing in tap water for 5 min, each disc was washed with sterile distilled water several times. Potato tuber discs were inoculated using toothpicks by placing the tip in Psg WT, COR-defective mutants (cfa6 and cmaA), the algU mutant, and the algU-complemented strain on a KB medium plate, and then placing the toothpick on the potato tuber disc. The discs were then placed at 23 °C incubator (darkness) for 5 days. Photographs were taken at five dpi.
Biofilm formation assay
Biofilm formation was assayed as described previously (Shao et al., 2019). Briefly, the bacterial strains were incubated overnight in LB broth and resuspended in fresh LB broth to an OD600 of 0.1. Bacterial suspensions (120 μl) were put into 96-well plates and incubated at 28 °C for 24, 48, 72, and 96 h. The bacterial solutions were discarded and washed three times with distilled water. The biofilm forming bacteria were treated with 150 μl of 0.1% crystal violet (CV; Fujifilm, Tokyo, Japan) for 20 min without shaking. The dye was discarded and washed twice with distilled water. The plate was dried completely, subsequently the biofilm was eluted with 150 μl of 100% ethanol, and the CV were dissolved completely. Finally, the eluted biofilm sample’s absorbance was measured at OD595.
Results
Identification and characterization of reduced virulence mutants
To identify Psg virulence genes, we screened 1,000 transposon insertion mutants for reduced disease symptoms on soybean leaves using the syringe-infiltration method. Disease symptoms caused by Psg WT showed a small water-soaked lesion surrounded by regions of chlorosis (Fig. 1A). A total of 28 mutants showed no or less chlorosis at six dpi (Fig. 1B). Seventeen mutants out of 28 had transposon insertions in genes encoding COR biosynthesis-related genes (Table 1). Soybean leaves inoculated with COR biosynthesis mutants (VTD29, VTE13, VTI15, and VTR4), the algU mutant (VTO41), and an unknown-function mutant (VTF6) showed no chlorosis (Fig. 1B).
Reduced disease symptoms and bacterial growth in soybean
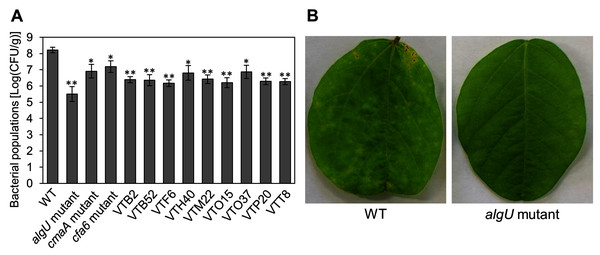
We identified the 28 mutants which showed reduced disease symptoms compared to Psg WT by syringe-inoculation (Fig. 1B). We further investigated whether these mutants also showed reduced virulence via spray-inoculation, and selected two mutants related to COR biosynthesis (cmaA and cfa6), and others. Soybean leaves inoculated with all mutants showed significantly reduced bacterial populations than those of Psg WT (Fig. 2A). Among all mutants, the algU mutant (VTO41) showed dramatically reduced bacterial populations and disease symptoms at six dpi (Figs. 2A and 2B). To confirm whether the altered algU mutant phenotype originates from a corresponding mutation, an algU-complemented strain was generated. Psg WT and the algU-complemented strain showed the same bacterial population levels as well as disease symptom development in soybean (Figs. S1A and S1B). We also confirmed that algU is apparently dispensable for Psg growth in rich LB medium, since no growth difference was observed among WT, algU mutant, and algU-complemented strain (Fig. S2).
Figure 2: Bacterial populations and disease symptoms in soybean leaves.
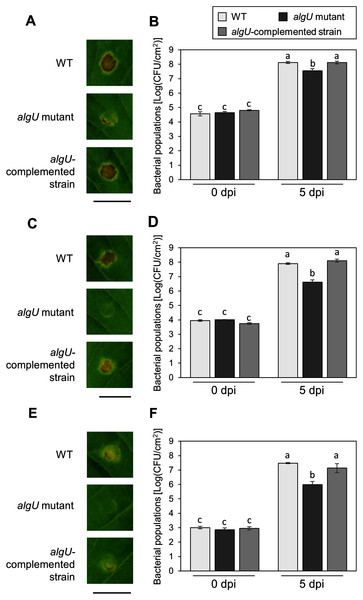
(A) Bacterial populations in leaves spray-inoculated with Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT) and mutants (1 × 108 colony forming units (CFU)/ml). Bacterial populations in leaves were estimated at 6 days post inoculation (dpi) by dilution plating on selective medium as described in the methods. Vertical bars indicate standard error for three independent experiments. Asterisks indicate a significant difference from WT in a t test (*P < 0.05; **P < 0.01). (B) Disease symptoms in leaves spray-inoculated with the Psg WT and the algU mutant (1 × 108 colony forming units (CFU)/ml) at six dpi.To further investigate the algU contribution to Psg virulence, we conducted syringe infiltration with WT, algU mutant, and algU-complemented strain. As a result, the algU mutant showed reduced symptoms and bacterial populations at all inoculum levels we tested (Figs. 3A–3F). Taken together, these results indicate that AlgU contributes to growth both on leaf surface and in apoplast, and to causing disease.
Figure 3: Disease symptoms and bacterial populations in soybean leaves after syringe inoculation.
Disease symptom and bacterial populations in leaves syringe-inoculated with Pseudomonas syringae pv. glycinea (Psg) wild-type (WT), the algU mutant, and the algU-complemented strain at 5 × 107 colony forming units (CFU)/ml (A, B), 5 × 106 CFU/ml (C, D), and 5 × 105 CFU/ml (E, F), respectively. Photographs were taken at 5 days post inoculation (dpi). Scale bars shows 1 cm. Bacterial populations in leaves were estimated at five dpi by dilution plating on selective medium as described in the methods. Vertical bars indicate standard error for three independent experiments. The different letters indicate significant statistical differences (P < 0.05, Turkey’s HSD test).AlgU regulates the expression of Psg virulence genes in HSC medium
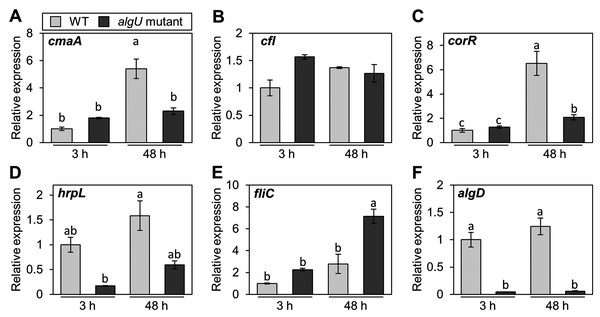
Pst DC3000 AlgU positively regulates virulence gene transcription (Markel et al., 2016; Ishiga et al., 2018). To investigate whether Psg AlgU also regulates virulence genes, we analyzed virulence gene expression profiles in HSC medium. COR biosynthesis-related genes including cmaA and corR, in the algU mutant showed reduced expression at 48 h after incubation (Figs. 4A and 4C). Moreover, hrpL, encoding HrpL (an alternative sigma factor recognizing the hrp box in the promoter of T3SS genes), also showed significantly less expression in the algU mutant at both 3 and 48 h after incubation compared to Psg WT (Fig. 4D). These results indicate that AlgU positively regulates COR biosynthesis-related genes and hrpL.
Figure 4: Gene expression profiles involved in the virulence of Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT) and algU mutant in liquid HSC broth.
Psg WT and algU mutant were grown in HSC broth for 3 and 48 h, adjusted to an OD600 of 0.1, and grown again in fresh HSC broth for 3 h. Gene expression was normalized using the housekeeping gene Psg outer membrane lipoprotein I (oprI) by real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR) with gene-specific primer sets (Table S1). (A) cmaA, (B) cfl, (C) corR, (D) hrpL, (E) fliC, and (F) algD. Vertical bars indicate the standard error for three biological replicates. The different letters (a–c) indicate a significantly statistical difference (P < 0.05, Turkey’s HSD test).To investigate whether AlgU can coordinate gene expression involved in Psg motility, we determined the expression profile of fliC (encoding flagellin, relating to flagellar mobility). At 3 h after incubation, there was no difference in the flagellar-encoding gene expression between the algU mutant and Psg WT. However, after 48 h incubation, relative fliC expression was greater in the algU mutant compared to Psg WT (Fig. 4E). Additionally, algD expression was down regulated in the algU mutant (Fig. 4F), indicating that AlgU positively regulates alginate biosynthesis-related genes.
AlgU contributes to COR biosynthesis and biofilm formation in Psg
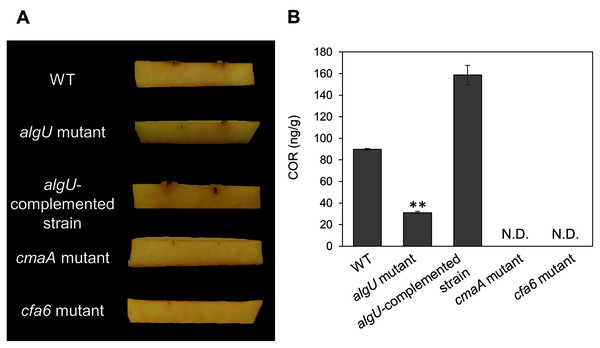
We demonstrated that COR biosynthesis-related genes in the algU mutant showed reduced expression in HSC medium (Figs. 4A–4D). To investigate whether AlgU contributes to COR production, we first conducted a hypertrophy-inducting activity test on potato tuber tissues for COR detection (Sakai et al., 1979; Völksch, Bublitz & Fritsche, 1989). Potato tuber tissues inoculated with Psg WT showed hypertrophy response, but those inoculated with COR-defective mutants (cmaA and cfa6) showed no response (Fig. 5A). The algU mutant-inoculated potato tuber tissues showed less hypertrophy response compared to those inoculated with Psg WT and the reduction was restored in the algU-complemented strain (Fig. 5A). Furthermore, we also quantified COR production by using HPLC. Psg WT produced around 90 ng/g of COR in HSC medium (Fig. 5B). However, the algU mutant produced only around one third as much COR compared to Psg WT (Fig. 5B). We also confirmed that the algU-complemented strain recovered COR production more than WT (Fig. 5B). Taken together, these results suggest that AlgU contributes to COR biosynthesis in Psg.
Figure 5: COR quantification of Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT), the algU mutant, and the algU-complemented strain.
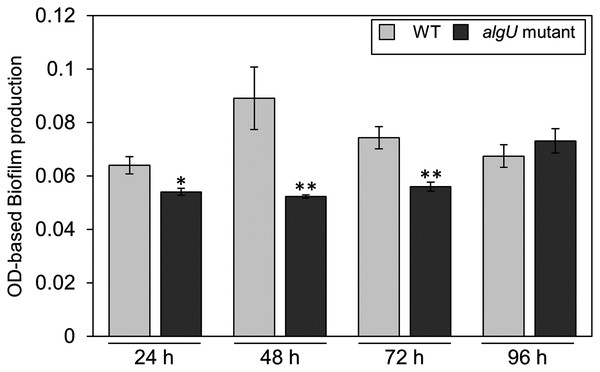
(A) Observation of hypertrophy-inducting activity on potato tuber tissue inoculated with Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT), COR-defective mutants (cmaA and cfa6), the algU mutant, and the algU-complemented strain. Potato tuber discs were inoculated using toothpicks by placing the tips in the Psg WT, cmaA, cfa6, algU mutant, and the algU-complemented strain on a KB medium plate and then placing the toothpick on the potato tuber disc. Photographs were taken at five dpi. (B) COR quantification of Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT) and the algU mutant grown in liquid HS broth by HPLC. Psg WT, the algU mutant, and the algU-complemented strain were cultured in HSC broth for 7 days. HPLC analysis was conducted by a Shimadzu LC20A system equipped with a Symmetry C8 column. COR in the culture supernatant was identified, as compared with authentic COR as the standard. Vertical bars indicate the standard error for three biological replicates. Asterisks indicate a significant difference from WT in a t test (**P < 0.01). N.D. indicates not detected by HPLC.Since we also demonstrated that alginate biosynthesis-related genes algD showed reduced expression in algU mutant (Fig. 4F), we next investigated the biofilm formation ability in Psg WT and the algU mutant. The algU mutant showed a reduction in biofilm formation at 24, 48, and 72 hpi (Fig. 6). These results suggest that AlgU also contributes to biofilm formation in Psg.
Figure 6: Biofilm biomass measurement of Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT) and the algU mutant with crystal violet (CV) grown in LB broth at 28 °C for 24, 48, 72, and 96 h.
The biofilm eluted sample’s absorbance was measured at OD595. Vertical bars indicate the standard error for three biological replicates. Asterisks indicate a significant difference from WT in a t test (*P < 0.05, **P < 0.01).Discussion
We attempted to identify Psg virulence factors that are crucial in soybean pathogenicity. We screened 1,000 Psg mutants by syringe-infiltration and identified 28 mutants with reduced virulence (Fig. 1B). Several important virulence factors contribute to Psg virulence including COR, the T3SS, and AlgU (Table 1). Sigma factor AlgU regulates not only algD, but also other virulence genes including hrpL and COR (Figs. 4A–4F). Our study provides new insights into AlgU function as a global regulatory hub for Psg pathogenicity by regulating the expression of multiple virulence genes.
Our screening identified that 17 out of 28 reduced virulence mutants were related to COR biosynthesis genes. These COR biosynthesis mutants were mostly disrupted by Tn5 on the cfa and cma operons (Table 1). The cfa and cma operons encode enzymes related to CFA and CMA biosynthesis, respectively, the two elements that are ligated together to form COR (Bender, 1999). Together, these results indicate that COR is an important Psg virulence factor.
The algU mutant showed reduced virulence in plants both spray-and syringe-inoculated (Figs. 2, S1, 3), indicating that AlgU contributes to Psg multiplication both on leaf surface and in apoplast, and causing disease. Our results indicate that AlgU regulate several virulence factors. Firstly, gene expression related to COR biosynthesis, such as cmaA and corR (but not cfl) were suppressed in the algU mutant (Figs. 4A–4C). Moreover, COR production in the algU mutant also less than that of Psg WT (Figs. 5A and 5B), suggesting that AlgU contributes to COR production in Psg. Consistent with our results, Ishiga et al. (2018) demonstrated that gene expression related to COR biosynthesis was suppressed during Pst DC3000 algU mutant infection. Furthermore, AlgU also contributes to Pst DC3000 virulence by regulating COR production to overcome stomatal-based defense (Ishiga et al., 2018). Together, these results suggest that AlgU suppresses stomatal-based defense in the early Psg infection stage with soybean plants. Moreover, COR contributes to virulence by overcoming apoplastic defense as well as stomatal-based defense in Pcal (Sakata et al., 2021). Further study on COR contribution in Psg virulence will be needed to understand AlgU-mediated COR regulation.
Secondary, the algD expression profile in the algU mutant was significantly reduced in comparison with Psg WT, indicating that AlgU is important in regulating alginate biosynthesis gene expression (Fig. 4F). This result was consistent with a previous report in Pst DC3000 (Ishiga et al., 2018), in which algD expression was significantly suppressed in an algU mutant. Further, alginate plays a crucial role in epiphytic fitness and survival, and contributes to P. syringae virulence (Yu et al., 1999). Alginate is one of the exopolysaccharides (EPSs), which are the major components of biofilms, in Psg (Osman, Fett & Fishman, 1986; Sutherland, 2001). We demonstrated that biofilm formation was significantly decreased in the algU mutant compared with Psg WT (Fig. 6). Psg PG4180 AlgU is important in virulence and bacterial growth in host plants, but it is not dependent on alginate production (Schenk, Weingart & Ullrich, 2008; Yu et al., 2014). Additionally, AlgU, but not AlgD plays a crucial role in Pst DC3000 virulence (Markel et al., 2016). Together, it is tempting to speculate that alginate function in virulence differs in each P. syringae pathovar. Thus, further study is needed to understand AlgU regulating genes involved in biofilm formation and alginate function in Psg virulence.
Thirdly, expression profiles also revealed hrpL transcripts were suppressed in the algU mutant compared with Psg WT (Fig. 4D). Pst DC3000 AlgU functions to regulate hrpL expression (Ishiga et al., 2018). Markel et al. (2016) also demonstrated that AlgU plays an important role in virulence by regulating the expression of T3Es and hrpL. In P. syringae, both the T3SS and T3Es genes are in turn encoded by the hrp gene cluster, while the sigma factor HrpL directly regulates both hrc and hop genes (Lam et al., 2014). Although many studies were caried out to elucidate the functions and mode of actions of the T3SS and its T3Es, AlgU regulation on the T3SS system is still unknown in P. syringae infection processes. Therefore, further precise characterization of AlgU-mediated T3SS regulation will be needed to understand global gene expression networks during Psg infection.
Lastly, fliC transcripts in the algU mutant were increased compared with those of Psg WT (Fig. 4E). In Pst DC3000, AlgU not only downregulates flagellar and chemotaxis genes in vitro (Markel et al., 2016), but also negatively regulates fliC expression during infection (Bao et al., 2020). fliC encodes the flagellin protein including the flg22 epitope which triggers PTI (Felix et al., 1999; Zipfel et al., 2004; Parys et al., 2021; Colaianni et al., 2021). Recent studies reported the important role of AlgU in de-flagellation during the P. syringae-plant interaction to reduce PTI activation, and promote bacterial fitness in its host (Bao et al., 2020). Likewise, AlgU also plays an important role in de-flagellation of P. syringae pv. maculicula ES4236, in which transposon inactivation of AlgW led to decreased AlgU activity and increased the flagella expression, as well as reduced bacterial growth in planta (Schreiber & Desveaux, 2011). Therefore, it is tempting to speculate that high levels of flagellin protein production in the algU mutant activate PTI.
Conclusions
Our findings indicate that multiple virulence factors regulated by AlgU, including COR biosynthesis, biofilm formation, and T3SS, contributes to Psg virulence in soybean. Our findings help to expand understanding of AlgU roles in Psg virulence. Further studies on AlgU regulated mechanisms will be needed to fully understand Psg virulence.
Supplemental Information
Disease symptoms and bacterial population dynamics in soybean leaves spray-inoculated with 1 × 108 colony forming units (CFU)/ml of the Pseudomonas savastanoi pv. glycinea (Psg) wild-type (WT), algU mutant, and algU.
(A) Disease symptoms in leaves spray-inoculated with 1 × 108 colony forming units (CFU)/ml of the Psg WT, algU mutant, and algU-complemented line (algU mutant + pDSKG-algU) at six dpi. (B) Bacterial populations in leaves were estimated at 2, 4, and 6 dpi by dilution plating on selective medium as described in the methods. Vertical bars indicate the standard error for three independent experiments. The different letters indicate significant statistical differences (P < 0.05, Turkey’s HSD test).
Pseudomonas savastanoi pv.glycinea WT, the algU mutant and the algU-complemented line growth in LB medium.
All strains were adjusted to an OD600 of 0.05 in LB medium and incubated with shaking at 28 °C.Bacterial growth was quantified at 6, 9, and 12 h. Vertical bars indicate the standard error for three biological replicates. Different letters indicate a significant difference among treatments based on a Tukey’s HSD test (P < 0.05).