Apis andreniformis associated Actinomycetes show antimicrobial activity against black rot pathogen (Xanthomonas campestris pv. campestris)
- Published
- Accepted
- Received
- Academic Editor
- Joseph Gillespie
- Subject Areas
- Agricultural Science, Microbiology, Molecular Biology, Zoology
- Keywords
- Honey bee, Apis andreniformis, Insect associated actinomycetes, Streptomyces, Rare actinomycetes, Plant pathogenic bacteria, Xanthomonas campestris pv. campestris
- Copyright
- © 2021 Promnuan et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Apis andreniformis associated Actinomycetes show antimicrobial activity against black rot pathogen (Xanthomonas campestris pv. campestris) PeerJ 9:e12097 https://doi.org/10.7717/peerj.12097
Abstract
This study aimed to investigate cultivable actinomycetes associated with rare honey bee species in Thailand and their antagonistic activity against plant pathogenic bacteria. Actinomycetes were selectively isolated from the black dwarf honey bee (Apis andreniformis). A total of 64 actinomycete isolates were obtained with Streptomyces as the predominant genus (84.4%) followed by Micromonospora (7.8%), Nonomuraea (4.7%) and Actinomadura (3.1%). All isolates were screened for antimicrobial activity against Xanthomonas campestris pv. campestris, Pectobacterium carotovorum and Pseudomonas syringae pv. sesame. Three isolates inhibited the growth of X. campestris pv. campestris during in vitro screening. The crude extracts of two isolates (ASC3-2 and ASC5-7P) had a minimum inhibitory concentration (MIC) of 128 mg L−1against X. campestris pv. campestris. For isolate ACZ2-27, its crude extract showed stronger inhibitory effect with a lower MIC value of 64 mg L−1 against X. campestris pv. campestris. These three active isolates were identified as members of the genus Streptomyces based on their 16S rRNA gene sequences. Phylogenetic analysis based on the maximum likelihood algorithm showed that isolate ACZ2-27, ASC3-2 and ASC5-7P were closely related to Streptomyces misionensis NBRC 13063T (99.71%), Streptomyces cacaoi subsp. cacaoi NBRC 12748T (100%) and Streptomyces puniceus NBRC 12811T (100%), respectively. In addition, representative isolates from non-Streptomyces groups were identified by 16S rRNA gene sequence analysis. High similarities were found with members of the genera Actinomadura, Micromonospora and Nonomuraea. Our study provides evidence of actinomycetes associated with the black dwarf honey bee including members of rare genera. Antimicrobial potential of these insect associated Streptomyces was also demonstrated especially the antibacterial activity against phytopathogenic bacteria.
Introduction
Apis andreniformis or black dwarf honey bee is one of the rare native bees found in Thailand and both tropical and near subtropical regions of Southeast Asia (Hepburn & Radloff, 2011). They build a single, exposed comb on the small branches of shrubs, banana, bamboos or small trees (Wongsiri et al., 1996). Actinomycetes are Gram-positive bacteria which produce two types of branching mycelia, namely aerial and substrate mycelium. They are well known as prolific producer of bioactive compounds, especially members of the Streptomyces species. Actinomycetes are widely distributed in nature both in terrestrial and aquatic environments (Goodfellow & Williams, 1983). They are recently found to be associated with several insects such as ants (Currie et al., 1999; Currie et al., 2003; Cafaro & Currie, 2005; Oh et al., 2009; Van Arnam et al., 2016; Chevrette et al., 2019), leafcutter bees (Inglis, Sinler & Goette, 1993), wasp (Kroiss et al., 2010; Matarrita-Carranza et al., 2021), southern pine beetle (Scott et al., 2008), honey bees and stingless bees (Promnuan, Kudo & Chantawannakul, 2009; Promnuan, Kudo & Chantawannakul, 2011; Promnuan, Promsai & Meelai, 2020).
There are few studies on actinomycetes associated with honey bee (Apis) species in Thailand. Actinomycetes belonging to the genera Streptomyces, Nonomurea and Nocardiopsis were isolated from three species of honey bees (Apis mellifera, Apis cereana and Apis florea) collected from northern Thailand. Furthermore, some of these isolates were able to inhibit the growth of Paenibacillus larvae and Melisococcus plutonius in vitro (Promnuan, Kudo & Chantawannakul, 2009). A novel actinobacterial species, Actinomadura apis was isolated from an A. mellifera hive in Chiang Mai province, Thailand (Promnuan, Kudo & Chantawannakul, 2011). In addition, Streptomyces spp. isolated from giant honey bee (Apis dorsata) showed the ability to inhibit the growth of Xanthomonas oryzae pv. oryzae, Xanthomonas campestris pv. campestris, Ralstonia solanacearum and Pectobacterium carotovorum (Promnuan, Promsai & Meelai, 2020). These studies indicated that honey bees harbored actinomycetes with potential to produce novel antimicrobial compounds to combat bacterial diseases in agriculture. Nevertheless, currently the discovery rate of new antibiotics from actinomycetes has been declining especially those from common habitats. Therefore, the focus of search and discovery programs for novel antibiotics from actinomycetes has shifted toward unexplored habitats (Bundale et al., 2019; Rangseekaew & Pathom-aree, 2019). In addition, rare actinomycetes are of interest for drug discovery programs as they are producers of many antibiotics in the market including rifamycins (Amycolatopsis mediterranei) and erythromycin (Saccharopolyspora erythraea). During the last two decades, known antibiotics produced by rare actinomycetes were increased up to 25–30% (Ding et al., 2019). The Gram-negative bacterium, X. campestris pv. campestris, is known to cause significant losses in many crop plants from diseases such as tomato speck, rice and pomegranate bacterial blight, citrus canker and brassica black rot (Yan et al., 2019). This bacterium can cause disease in a large number of species in the Brassicaceae, including Brassica and Arabidopsis. Black rot is a seed-borne disease and typical symptoms include V-shaped yellow lesions starting from the leaf margins and blackening of the veins (Vicente & Holub, 2013). The use of bactericidal compounds to control phytopathogenic bacteria such as antibiotics and copper could cause serious problems to human health and environment such as antibiotic resistance and toxicity. Furthermore, some emerging strains have shown strong resistance to the antibiotics (Satish, Raveesha & Jandrdhana, 2002; Sabir et al., 2017; Mougou & Boughalleb-M’hamdi, 2018; Wu et al., 2019). For these reasons, other control measures have been developed and reported including antagonistic bacteria (Bacillus spp. and Pseudomonas spp.) (Wulff et al., 2002; Mishra & Arora, 2012), antimicrobial compounds from plant extracts (Satish, Raveesha & Jandrdhana, 2002; Kaur et al., 2016) and essential oils (Sabir et al., 2017; Amini et al., 2018).
Actinomycetes isolated from the black dwarf honey bee (A. andreniformis) with antagonistic activity against the phytobacterial pathogen have never been studied and reported. Therefore, this study focused on the isolation of cultivable actinomycetes from A. andreniformis collected from Chiang Mai province, Thailand and their antimicrobial activity against phytopathogenic bacteria was investigated.
Materials & Methods
Sample collection
This study was approved by the Institutional Animal Care and the Use Committee (IACUC), Silpakorn University (Ethic number: 8603.16/0328). A. andreniformis combs were obtained from the local villages in Mae-rim district, Chiang Mai province, Thailand during October 2013–January 2014. The permission was received from the farm owners (Mr. Ton Tatiya and Mr. Ma Madamun). Six combs were stored in sterilized containers and transferred back to the laboratory. The samples (adult bees, pupae, honey and pollen) were kept in sterile plastic tubes and stored at −20 °C until further processing.
Isolation and characterization of actinomycetes from A. andreniformis
Actinomycetes were isolated from six combs of A. andreniformis. Five adult bees and pupae were taken from each of the six collected combs, surface sterilized and ground before the isolation process. Isolation of actinomycetes from one milliliter of honey and one gram of pollen was achieved using a standard 10-fold dilution spread plate method on glycerol-asparagine (ISP5) (Pridham & Lyons, 1961), starch casein nitrate agar (Küster & Williams, 1964), Czapek’s agar (Waksman, 1950) and nutrient agar supplemented with 25 µg mL−1 of nalidixic acid and nystatin. All plates were incubated at 30 °C for 14–21 days. Presumptive actinomycetes colonies were purified and maintained on yeast extract-malt extract agar (ISP2) (Shirling & Gottlieb, 1966) slants and kept in 4 °C. Sixty-four actinomycetes were grouped based on their morphological characteristics in particular the colour of substrate mycelium, aerial spore mass and diffusible pigments.
Phytopathogenic bacteria
The bacterial phytopathogens, X. campestris pv. campestris and P. carotovorum were obtained from the Department of Agriculture, Ministry of Agriculture and Cooperative, Thailand. Pseudomonas syringae pv. sesame TISTR 901 was obtained from Thailand Institute of Scientific and Technological Research (TISTR), Thailand. These phytopathogenic bacteria were activated and maintained on nutrient agar and yeast extract-malt extract (ISP2) agar for 24–48 h, at 30 °C before use.
Screening for antagonistic activity of actinomycetes against plant pathogenic bacteria
All actinomycete isolates were tested for their activity against three plant pathogenic bacteria (X. campestris pv. campestris, P. carotovorum and Ps. syringae pv. sesame TISTR 901) using a modified cross streak method as described by Promnuan, Promsai & Meelai (2020) on glucose yeast extract (GYE) and ISP2 agar plate. The inhibition zones were recorded after 24 h. Each experiment was performed in triplicate.
Extraction of bioactive compounds from potent actinomycete isolates
Selected actinomycete isolates that showed potent activity against the growth of X. campestris pv. campestris from the cross-streaking method were grown on ISP2 agar plates (for ACZ2-27) and GYE agar plates (for ASC3-2 and ASC5-7P) and incubated at 30 °C. After 14 days incubation, the agar media of each isolate was extracted using ethyl acetate followed the method as described by Promnuan, Promsai & Meelai (2020). Briefly, small pieces of agar medium (approximately 0.5 cm × 0.5 cm) were added to 200 ml of ethyl acetate and shaken at 150 rpm, 30 °C for 48 h. The extracts were filtered through Whatman No. 1 filter paper and concentrated in a rotary evaporator at 40 °C. One microliter of sterile dimethyl sulfoxide (DMSO) was added into the dried extracts and stored at −20 °C.
Determination of minimum inhibitory concentration (MIC) of antagonistic actinomycete isolates
The MIC of ethyl acetate extract was determined using modified broth microdilution method in 96-well microtiter plate as described by Wiegand, Hilpert & Hancock (2008). The concentration of the test organism, X. campestris pv. campestris was adjusted to 0.5 McFarland standard. The initial concentration of the crude extract was adjusted to 1,280 mg L−1 using sterile DMSO. The crude extract was prepared as two-fold dilution series using ISP2 broth in a 96-well microtiter plate. The concentration of the crude extract was ranged from 0.25 to 128 mg L−1. Sterile DMSO was used as a negative control. After incubation at 30 °C for 24 h, the suspension from each well was inoculated onto an ISP2 agar plate using streak plate technique. Plates were incubated at 30 °C for 24 h. The lowest concentration of the extracts that showed no bacterial growth on the ISP2 agar plate was recorded as the MIC value. This experiment was conducted in triplicate.
Identification of actinomycetes using 16S rRNA gene
The representative isolates of each morphological group and the actinomycetes that showed strong activity against X. campestris pv. campestris were grown in ISP2 broth, incubated at 30 °C for 7 days on the rotary shaker (120 rev min−1). The cells were collected by centrifugation (91,000 g) and washed three times using sterile distilled water. Genomic DNA extraction and PCR amplification of the 16S rRNA gene were carried out as described by Nakajima et al. (1999) using primers 20F (5′-AGTTTGATCCTGGCTC) and 1540R (5′-AAGGAGGTGATCCAGCC). The PCR product was purified using Invitrogen™ PureLink™ PCR Purification Kit (Thermo Fisher Scientific, USA). The 16S rRNA genes were sequenced by 1st BASE, Singapore using the Sanger method. BLAST analysis of actinomycete isolates was done using the EzBioCloud database (https://www.ezbiocloud.net/) (Yoon et al., 2017). Multiple alignment of the closely related type strain sequences obtained from the GenBank database were carried out using Clustal_W in BioEdit Sequence Alignment Editor 7.2.5 (Hall, 1999). The maximum likelihood (ML) trees were constructed using MEGA X version 10.1.8 (Kumar et al., 2018) based on a comparison of 1,299-1,386 nucleotides present in all the strains used after elimination of gaps and ambiguous nucleotides from the sequences. Streptomyces thermocarboxydus DSM 44293T was used as an outgroup. Bootstrap analyses based on 1,000 resamplings was used to determine the confidence values for branches of the maximum likelihood (ML) tree (Felsenstein, 1985). The percentage of sequence similarity were calculated using the pairwise alignments function in BioEdit 7.2.5.
Results
Isolation and characterization of the actinomycetes from A. andreniformis
Adult bees, pupae, honey and pollen were obtained from six combs of the black dwarf honey bee (A. andreniformis). A total of sixty-four actinomycetes were isolated from different media as summarized in Table 1. Most actinomycetes were obtained from adult bees (65.6%) followed by honey (20.3%), pollen (12.5%) and pupae (1.6%). Based on the morphological characteristics, all actinomycete isolates were assigned to four groups (Table 2). Streptomyces (group IV) was the predominant genus with 84.4% followed by Micromonospora (group I) (7.8%), Nonomuraea (group III) (4.7%) and Actinomadura (group II) (3.1%). Streptomyces (ACZ2-27 and ASC3-2) and all rare genera were recovered from the adult honey bee (Table 2).
Antimicrobial activity of actinomycetes against phytobacterial pathogens
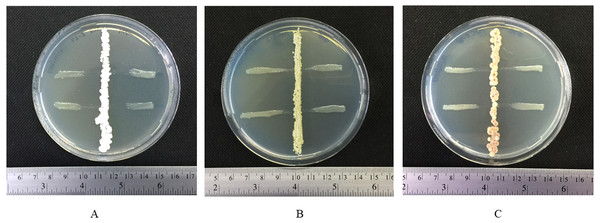
Three isolates from group IV (ACZ2-27, ASC3-2 and ASC5-7P) showed strong inhibition of the growth of a phytobacterial pathogen. Actinomycete isolate ACZ2-27 inhibited the growth of X. campestris pv. campestris on an ISP2 agar plate (Fig. 1A) with an inhibition zone diameter of 10.75 ± 0.83 mm. The actinomycete isolates ASC3-2 and ASC5-7P inhibited the growth of X. campestris pv. campestris on a GYE agar plate (Figs. 1B, 1C) with an inhibition zone diameter of 6.00 ± 0.71 and 9.0 ± 0.82 mm, respectively. The MIC levels of ethyl acetate extracts of the three isolates were determined against the growth of X. campestris pv. campestris using the 96-well microtiter assay. The MIC value of the crude extract of ACZ2-27 against X. campestris pv. campestris was 64 mg L−1 and the MIC values of the crude extracts of ASC3-2 and ASC5-7P were 128 mg L−1. These results provide the first evidence of antibacterial extracts from black dwarf honey bee associated actinomycetes against the black rot pathogen, X. campestris pv. campestris.
| Isolation medium | |||||
|---|---|---|---|---|---|
| Sample source | ISP5* | SC* | CZ* | NA* | Total (%) |
| Adults | 17 | 14 | 6 | 5 | 42 (65.6%) |
| Pupae | 1 | 0 | 0 | 0 | 1 (1.6%) |
| Pollen | 2 | 5 | 1 | 0 | 8 (12.5%) |
| Honey | 9 | 4 | 0 | 0 | 13 (20.3%) |
| Total (%) | 29 (45.3%) | 23 (35.9%) | 7 (10.9%) | 5 (7.8%) | 64 (100%) |
Notes:
| Group | Morphological characteristics | Sources | Occurrence (%) | Representative isolates | Accession no. | Genus |
|---|---|---|---|---|---|---|
| I | wrinkled colony with brown or orange colour | Adult Adult |
7.8 | AGA3-9 AGA3-53 |
LC546088
LC546089 |
Micromonospora |
| II | convex and rigid colony with cream colour | Adult | 3.1 | AGA3-58 | LC546090 | Actinomadura |
| III | wrinkled colony with cream colour | Adult | 4.7 | ASC2-5 | LC546091 | Nonomuraea |
| IV | powdery colonies and aerial spore mass with white or grey | Adult Adult Pollen |
84.4 | ACZ2-27 ASC3-2 ASC5-7P |
LC500236
LC506284 LC506285 |
Streptomyces |
Figure 1: Inhibitory effect of actinomycetes against growth of X. campestris pv. campestris.
Fig. 1 Inhibitory effect of actinomycetes against growth of X. campestris pv. campestris: (A) ACZ2-27 on ISP2 agar plate and (B) ASC3-2 and (C) ASC5-7P on GYE agar plates.Identification of actinomycetes using 16S rRNA gene
Three actinomycete isolates which showed strong inhibition against the growth of X. campestris pv. campestris were identified based on the 16S rRNA gene sequence analysis. The 16S rRNA gene sequences of strains ACZ2-27 (LC500236), ASC3-2 (LC506284) and ASC5-7P (LC506285) were analyzed by BLAST in the EzBioCloud database. All strains exhibited high similarities with members of the genus Streptomyces. The 16S rRNA gene sequences of all three isolates were compared with the corresponding sequences of the most closely related strains of the genus Streptomyces. The maximum likelihood tree (Fig. 2) revealed that strain ACZ2-27, ASC3-2 and ASC5-7P were closely related to S. misionensis NBRC 13063T, S. cacaoi subsp. cacaoi NBRC 12748T and S. puniceus NBRC 12811T. The 16S rRNA gene sequence similarity percentage between each isolate and their closely related type strains was calculated using the pairwise alignment option in BioEdit program version 7.2.5. The results revealed that isolates ACZ2-27, ASC3-2 and ASC5-7P were closely related to S. misionensis NBRC 13063T (99.71%), S. cacaoi subsp. cacaoi NBRC 12748T (100%) and S. puniceus NBRC 12811T (100%), respectively. This is the first report of actinomycete species isolated from the black dwarf honey bee (A. andreniformis) that showed the ability to produce antimicrobial metabolites that inhibited the growth of the phytobacterial pathogen X. campestris pv. campestris.
Figure 2: The Maximum likelihood tree of antagonistic Streptomyces strains ACZ2-27, ASC3-2 and ASC5-7P.
The Maximum likelihood tree based on 16S rRNA gene sequences (1,296 nucleotides) showing the phylogenetic position of antagonistic Streptomyces strains ACZ2-27, ASC3-2 and ASC5-7P relative to the type strains of other related Streptomyces species. Streptomyces thermocarboxydus DSM 44293T was used as an outgroup. The number at each node is the bootstrap support value (%) based on 1,000 replicates (only values >50% are shown). The bar shows 0.005 substitutions per nucleotide position.In addition, the representative isolates of group I (AGA3-9, AGA3-53), group II (AGA3-58) and group III (ASC2-5) were identified by 16S rRNA gene sequence analysis. The 16S rRNA gene sequences of strains AGA3-9 (LC546088), AGA3-53 (LC546089), AGA3-58 (LC546090) and ASC2-5 (LC546091) were analyzed by BLAST using the EzBioCloud database. The representative isolates from group I-III exhibited high similarity with members of the genera Micromonospora, Actinomadura and Nonomuraea, respectively (Table 2). The 16S rRNA gene sequence similarity between each representative isolate and their closely related type strains showed that group I (AGA3-9 and AGA3-53) were most closely related to Micromonospora chalcea DSM 43026T (99.34%) and M. halotolerans CR18T (99.70%), respectively. The representative isolates of group II (AGA3-58) and group III (ASC2-5) were closely related with Actinomadura meyerii DSM 44715T (99.34%) and Nonomuraea salmonea DSM 43678T (99.92%), respectively. The maximum likelihood tree also confirmed the placement of these actinomycetes to their respective genera (Fig. 3). This is the first evidence of rare actinomycetes associated with the black dwarf honey bee A. andreniformis.
Figure 3: The Maximum likelihood tree of the representative strains of non-Streptomyces isolates.
The Maximum likelihood tree based on 16S rRNA gene sequences (1,297 nucleotides) showing the phylogenetic position of the representative strains of non-Streptomyces isolates and their nearest neighbors. Streptomyces thermocarboxydus DSM 44293T was used as an outgroup. The number at each node is the bootstrap support value (%) based on 1,000 replicates (only values >50% are shown). The bar shows 0.01 substitutions per nucleotide position.Discussion
In this study, Streptomyces were found as the majority of the obtained isolates. In addition, members of rare actinomycete genera Micromonospora, Nonomuraea and Actinomadura were recovered from the black dwarf honey bee. All of these isolates were recovered from the adult honey bee except for isolate ASC5-7P which was obtained from the pollen. Actinomycetes belonging to the genera Streptomyces, Nonomurea and Nocardiopsis have been previously isolated from three species of honey bees (A. mellifera, A. cereana and A. florea) collected from northern Thailand (Promnuan, Kudo & Chantawannakul, 2009). Recently, Streptomyces spp. were also isolated from giant honey bee (A. dorsata) (Promnuan, Promsai & Meelai, 2020). These observations suggested that members of the genus Streptomyces may be commonly isolated from at least 5 species of honey bee in Thailand (A. mellifera, A. cereana, A. florea, A. dorsata and A. andreniformis). The evidence of rare actinomycetes associated with the black dwarf honey bee, A. andreniformis, implied that this honey bee species may harbor diverse actinomycete populations. It is also interesting to note that members of the genus Micromonospora were isolated from honey bee for the first time. Though, the function of these associated actinomycetes in A. andreniformis is still unknown, it is tempting to suggest that this relationship is at least neutral to the bees as all the bee samples used for isolation were healthy.
Actinomycetes are known for the production of bioactive compounds with antimicrobial activity against plant pathogens such as bacteria and fungi (Viaene et al., 2016). Several actinomycete species obtained from various habitats have been reported for secondary metabolites against phytopathogens. Streptomyces spp. obtained from marine samples in Egypt showed variable antimicrobial activity, secretion of numerous hydrolytic enzymes, in vitro and in vivo nematicidal activity against root-knot nematodes and supported plant growth (Rashad et al., 2015). S. violaceusniger strain A5 isolated from chitin-rich partially decomposed molted snake skin, showed strong inhibitory activity against Xanthomonas axonopodis pv. punicae, the causative agent of oily spot disease in pomegranate, with a MIC in the range of 0.625–1.25 mg mL−1 (Chavan et al., 2016). Endophytic Streptomyces spp. (AB131-1 and AB131-2) reduced the infection of X. oryzae pv. oryzae a causal agent of bacterial leaf blight (BLB disease) and improved the growth of rice seedlings in a pot experiment in the greenhouse (Hastuti et al., 2012). Streptomyces isolated from compost-amended soil had antimicrobial activity against Agrobacterium tumefaciens (Cuesta et al., 2010). S. caeruleatus isolated from the Cassia fistula rhizosphere soil showed strong activity against X. campestris pv. glycine, the soybean pathogen (Mingma et al., 2014). It is evident from these studies and our results that Streptomyces are effective in control of plant pathogens including Xanthomonas species.
Several publications support the view that insects provide new sources of actinomycetes that may produce novel natural products with antimicrobial properties (Chevrette et al., 2019). Streptomyces spp. have been isolated from A. andreniformis and showed high activity in reducing the egg hatch rate, increasing the infective second-stage juvenile mortality rate of the root-knot nematode (Meloidogyne incognita) and reducing root gall of chili in a pot experiment (Santisuk et al., 2018). This study and our results indicated that the black dwarf honey bee (A. andreniformis) is an interesting new source for screening of actinomycetes that may produce novel natural products for agriculture. Streptomyces spp., Nonomurea spp. and Nocardiopsis spp. were isolated from honey bees (A. mellifera, A. cereana and A. florea) and showed antibacterial activity against the growth of Paenibacillus larvae and Melisococcus plutonius that cause American foulbrood and European foulbrood disease in honey bee (Promnuan, Kudo & Chantawannakul, 2009). The actinobacteria (Pseudonocardia spp.) associated with fungus-growing ants (Apterostigma dentigerum) produced dentigerumycin which inhibited the growth of parasitic fungus (Escovopsis sp.) (Oh et al., 2009). Poulsen et al. (2011) reported the novel macrocyclic lactam, sceliphrolactam compounds from Streptomyces symbionts with mud dauber wasps (Chalybion californicum and Sceliphron caementarium). Streptomyces spp., Micromonospora spp. and Actinoplanes spp. obtained from the paper wasp Polistes dominulus nests showed antibacterial activity against the growth of some pathogenic bacteria (Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, Serratia marcescens and Bacillus subtilis) (Madden et al., 2013). A novel compound cyphomycin was isolated from Streptomyces (ISID311) isolated from the microbiome of the fungus-growing ant which active against multidrug resistant fungal pathogens (Chevrette et al., 2019). Matarrita-Carranza et al. (2021) reported the genome sequence and the potential for antibiotic production of Streptomyces sp. M54, actinomycetes associated with the eusocial wasp (Polybia plebeja). This actinobacterium produces antimicrobial compounds that are active against Hirsutella citriformis, a natural fungal enemy of its host, and the human pathogens Staphylococcus aureus and Candida albicans. Recently, three actinomycetes (S. ramulosus, S. axinellae and S. drozdowiczii) isolated from giant honey bee (A. dorsata) combs showed the ability to inhibited the growth of black rot pathogen (X. campestris pv. campestris) with MIC value 32 mg L−1 (Promnuan, Promsai & Meelai, 2020). These data indicated that actinomycetes associated with insects represent a valuable source for new antimicrobial compounds.
Conclusions
Black rot disease, which is caused by X.campestris pv. campestris, can cause significant losses of the Brassicaceae in Asian countries. In this study, we provide evidence that actinomycetes including members of rare genera are associated with the black dwarf honey bee (A. andreniformis) especially in adult stage. The organic extract of isolate ACZ2-27 showed the lowest MIC of 64 mg L−1 against the growth of black rot pathogen (X. campestris pv. campestris). This isolate was obtained from an adult bee and is phylogenetically closely related to S. misionensis. This is the first report of actinomycetes isolated from black dwarf honey bee which showed growth inhibitory activity against phytopathogenic bacterium pathogen, X. campestris pv. campestris. Black dwarf honey bee associated actinomycetes may represent an interesting source for the search of bioactive compounds for biotechnological usage, especially in agriculture.