Pseudolebinthus lunipterus sp. nov.: a striking deaf and mute new cricket from Malawi (Orthoptera, Gryllidae, Eneopterinae)
- Published
- Accepted
- Received
- Academic Editor
- Joseph Gillespie
- Subject Areas
- Entomology, Evolutionary Studies, Taxonomy
- Keywords
- New species, Acoustic communication, Malawi, Africa, Xenogryllini, Mitogenome, Molecular phylogeny, Cricket, Evolution of communication, Taxonomy
- Copyright
- © 2020 Salazar et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Pseudolebinthus lunipterus sp. nov.: a striking deaf and mute new cricket from Malawi (Orthoptera, Gryllidae, Eneopterinae) PeerJ 8:e8204 https://doi.org/10.7717/peerj.8204
Abstract
This article presents an intriguing new cricket species of the tribe Xenogryllini discovered in Northern Malawi. This is the first case of mute and deaf species in the subfamily Eneopterinae; it shows no stridulatory apparatus on short male forewings and no tympana on either side of fore tibiae in both sexes. We introduce the new species and its complete mitogenome and assess phylogenetic relationships based on molecular data obtained from next-generation sequencing genome skimming method. Phylogenetic analyses place the new species within the genus Pseudolebinthus in Xenogryllini, as the sister species of Pseudolebinthus gorochovi Robillard. We describe Pseudolebinthus lunipterus sp. nov., provide illustrations of main morphology, male and female genitalia, photographs of living specimens and information about habitat and update the identification key for species of genus Pseudolebinthus. We discuss the differences between the new species and related taxa and the striking loss of acoustic communication in this cricket.
Introduction
Crickets have long been studied for their capacity to communicate with sound (Horch et al., 2017). Their mechanism of sound production by wing stridulation and their hearing system has been extensively studied during the last 50 years (Michelsen, 1998; Bennet-Clark, 1989; Gerhardt & Huber, 2002). What is less known is that many lineages within the cricket clade have independently lost their capacity to produce sound, and sometimes their hearing capacity too. Numerous examples of mute lineages are spread in the taxonomic literature about crickets (Otte & Alexander, 1983; Otte, 1992; Wang et al., 2018), some species being completely wingless and some retaining the wings while losing stridulatory structures at different degrees (Zuk, Rotenberry & Tinghitella, 2006; Pascoal et al., 2014). Recent studies have demonstrated that the loss of sound production structures on male forewings (FWs) could occur convergently and very rapidly in populations of Teleogryllus oceanicus (Le Guillou) as a result to strong selective pressures by a parasitoid fly (Zuk, Rotenberry & Tinghitella, 2006; Pascoal et al., 2014).
In many mute lineages of crickets, auditory tympana are retained after the tegminal stridulatory mechanism is lost (Otte, 1992), which could be linked with avoidance of bat predation. Species still able to fly but in which males have lost the stridulum usually retain the tympana (Otte & Alexander, 1983; Otte, Alexander & Cade, 1987). Species becoming both mute and deaf are relatively less common, even among the diversity of situations presented by crickets.
The subfamily Eneopterinae has been well studied for its diversity of traits related to acoustic communication (Robillard & Desutter-Grandcolas, 2004a, 2004b; Robillard, Grandcolas & Desutter-Grandcolas, 2007; ter Hofstede et al., 2015). Among this diversity, eneopterines include several lineages which have lost capacity to produce sound independently (Robillard & Desutter-Grandcolas, 2004a), either by losing stridulatory structures while retaining long FWs (genus Swezwilderia Chopard, 1929 in tribe Lebinthini), or by becoming apterous (genus Paranisitra Chopard, 1925 in tribe Nisitrini), but no example of complete loss of hearing was documented yet.
In this study, we describe the species Pseudolebinthus lunipterus sp. nov., a new eneopterine from Northern Malawi being both mute and deaf (Fig. 1). The new species is the first member of Eneopterinae showing no stridulatory apparatus on short male FWs and no tympana on either side of fore tibiae in both sexes. We provide illustrations about main morphology, male and female genitalia, photographs of living specimens and information about its natural habitat. We describe its complete mitogenome and assess phylogenetic relationships based on molecular data obtained by next-generation sequencing using the genome skimming method (Straub et al., 2012). We discuss the differences between the new species and related taxa, their phylogenetic relationships and the possible origins of muteness and deafness of these crickets.
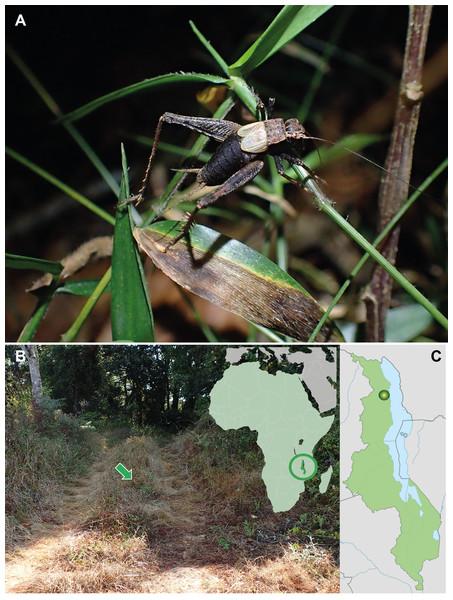
Figure 1: Pseudolebinthus lunipterus sp. nov.
(A) Male habitus on low vegetation at night; (B) natural habitat indicated by a green arrow (left) and location of Malawi on simplified map of Africa (left); (C) type locality in Malawi. Photo Tony Robillard.Materials and Methods
Material and taxonomy
The new collected material comes from a field expedition in Malawi in September and October 2018 under the collection and export permit number EAD-12-07-087-18-20a from The Forestry Research Institute of Malawi (FRIM) and from the personal collection of R.J.M. Specimens are deposited in the collections of Muséum national d’Histoire naturelle, Paris (MNHN).
The electronic version of this article in Portable Document Format will represent a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank Life Science Identifiers (LSIDs) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/. The LSID for this publication is: urn:lsid:zoobank.org:pub:DA17C29A-B265-4819-A9E6-55FCB137E3F0. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central and CLOCKSS.
Description follows terminologies as proposed by Robillard et al. (2014). Observations of external morphological characters and dissection of male and female genitalia were performed using a Leica stereomicroscopes MZ16. Terminologies for male FW venation follow Ragge (1955) and Robillard & Desutter-Grandcolas (2004a). Male and female genitalia were dissected from freshly killed specimens. Male genitalia were dissected by making a small slit between paraproct and subgenital plate. Female copulatory papilla was dissected out by cutting the membrane between ovipositor and subgenital plate. Dissected genitalia were cleared in 10% cold KOH solution and preserved in glass vials containing glycerine. Imaging of male and female genitalia were made using Canon EOS 60D Digital SLR camera on a Nikon stereomicroscope SMZ1500. To highlight the structural components of male and female genitalia, water solution containing a drop of JBL Punktol was used. To fix orientation and stabilization of genitalia for photography, a clear and viscous Power Plast Hand Sanitizer was used following Su (2016). The dry mounted adults were photographed with a Canon EOS 6D Digital SLR camera.
Abbreviations used in taxonomic descriptions and figures
General morphology: FI, FII, FIII, fore, median, hind femur; FW, forewing; TI, TII, TIII, fore, median, hind tibia; Tarsomere I/II/III-1: basal segment of fore, median and hind leg tarsomere. Tegminal venation: 1A–4A, first to fourth anal veins; CuA, anterior cubitus; CuP, posterior cubitus; M, median vein; R, radial vein; Sc, subcostal vein and its projections.
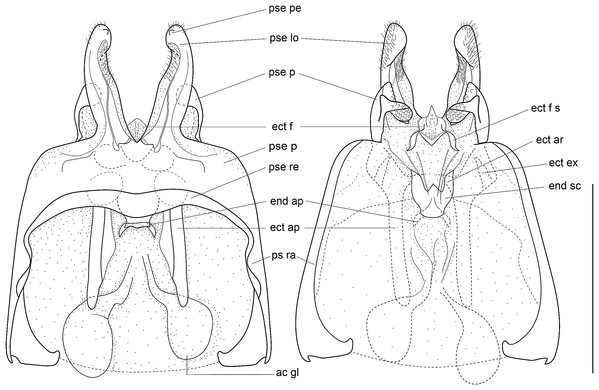
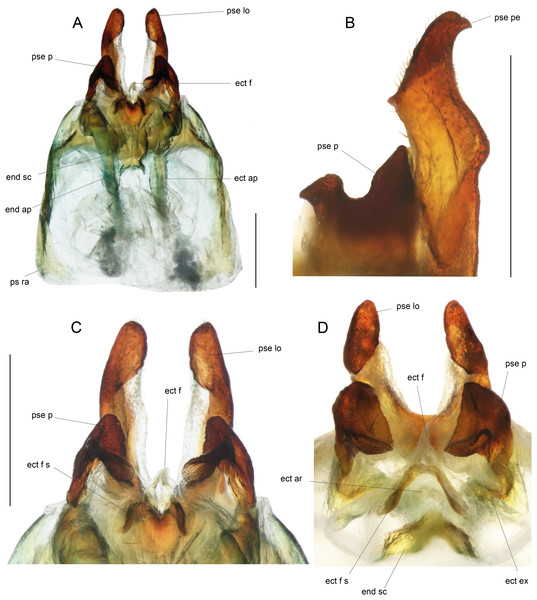
Male genitalia: ac gl, accessory gland; ect ar, ectophallic arc; ect ex, ectophallic lateral expansion; ect f, ectophallic fold; ect f s, sclerite of ectophallic fold; ect ap, ectophallic apodeme; end sc, endophallic sclerite; end ap, endophallic apodeme; pse, pseudepiphallus; pse pe, pseudepiphallic posterior expansion; pse lo, pseudepiphallic lophi; pse pa, pseudepiphallic paramere; pse ra, pseudepiphallic rami; pse re, pseudepiphallic basal reinforcement.
Measurements: (in mm, except for spine numbers) BL, body length in dorsal view, from fastigium to apex of abdomen; FIIIL, length of FIII; FIIIW, width of FIII; TIIIL, length of TIII; FWL, forewing length; FWW, forewing width (at the level of maximal width at about 1/3 of FWL); Ias, inner spines on TIII dorsal side above the spurs; Ibs, inner spines on TIII dorsal side between the spurs; Oas, outer spines on TIII dorsal side above the spurs; Obs, outer spines on TIII dorsal side between the spurs; OL, ovipositor length; PronL, pronotum length; PronW, pronotum width; TaIIIs, spines of third hind tarsomere, not including the apical spines: Ids, inner dorsal spines; Ods, outer dorsal spines; Ols, outer spines on lateral side of TaIII.
Laboratory methods
DNA extraction, PCR amplification and bank preparations were carried out at Service de Systématique Moléculaire of the MNHN. We extracted DNA from ethanol-preserved median legs for four newly collected specimens (two P. lunipterus sp. nov. and two Pseudolebinthus gorochovi Robillard, 2018) (see Table 1 for details about specimen vouchers). Total genomic DNA was extracted using a DNeasy Blood and Tisue Kit (Qiagen Inc., Venlo, Netherlands and Germany) following the manufacturer’s instructions. For each newly generated extract, we amplified the mitochondrial gene maker 12S (12S rRNA gene, amplicon ~400 bp) with the protocols described in Nattier et al. (2012) with the following primers and annealing temperatures: 12SF 5′-TACTATGTTACGACTTAT′3′, 12sr 5′-AAACTAGGATTAGATACCC-3′ at 48 °C (Kambhampati, 1995). PCR products were sequenced with the Ion Torrent PGM platform in MNHN. Assembling and annotations were executed with Geneious Prime 2019.1.3 (Biomatter Ltd., Auckland, New Zealand, Oceania, www.geneious.com, Kearse et al., 2012).
| Species | Voucher/lab code (type status) | 16S | 12S | COI | COII | Cytb | 18S | 28S | H3 |
|---|---|---|---|---|---|---|---|---|---|
| Acheta domesticus Linnaeus (1758) | MNHN-EO-ENSIF3523/Adom | AF248698 | ADZ97611 | JX897403 | JX897439 | AF248682 | AD18SITS1 | JX897465 | KR903150 |
| Agnotecous meridionalis Desutter-Grandcolas (2006) | MNHN-ENSIF-2772/AmeIP | JX897349 | JX897401 | JX897420 | NA | JX897311 | JX897579 | JX897488 | JX897553 |
| Agnotecous meridionalis Desutter-Grandcolas (2006) | MNHN-EO-ENSIF-2771/AmePB | JX897350 | JX897402 | JX897410 | JX897442 | JX897313 | JX897597 | JX897489 | JX897550 |
| Cardiodactylus novaeguineae Haan (1844) | MNHN-ENSIF2038/C3CnoNC | JF972520 | JF972504 | MH662977 | MH662880 | JF972488 | JF972535 | MH663420 | NA |
| Cardiodactylus novaeguineae Haan (1844) | MNHN-EO-ENSIF2030/C2CnoPe | JF972521 | JF972506 | KU705563 | KU705551 | JF972490 | JF972537 | KR903500 | KR903151 |
| Eneoptera guyanensis Chopard (1931) | MNHN-EO-ENSIF2741/Egu | AY905301 | AY905272 | JX897404 | KU705553 | AY905355 | AY905331 | KU705581 | JX897547 |
| Eurepini sp. | MNHN-EO-ENSIF3155/Eursp | KR903674 | KR903834 | KU705565 | KU705554 | KR903331 | KR904028 | KR903503 | KR903153 |
| Gryllus bimaculatus De Geer (1773) | MNHN-EO-ENSIF3524/3404/Gbi | AF248685 | AY905292 | NA | KU705555 | AF248659 | AF514509 | KR903002 | KR903154 |
| Indigryllus kudremu sp. nov. | ZSI/X3Xsp2 (AT) | KY595509 | KY595483 | KY646248 | MK761340 | AY905377 | AY905345 | KY605247 | KY646293 |
| Lebinthus bitaeniatus Stål (1877) | MNHN-EO-ENSIF4393/L18LbiP1 | MK761250 | MK761274 | MK761331 | MK761341 | MK761353 | MK761293 | MK761313 | MK761370 |
| Lebinthus bitaeniatus Stål (1877) | MNHN-EO-ENSIF4394/L28LbiP2 | MK761252 | MK761275 | MK761332 | NA | MK761354 | MK761294 | MK761314 | MK761371 |
| Lebinthus luae Robillard & Tan (2013) | MNHN-EO-ENSIF2740/L8LbiS1 (PT) | JF972524 | KR904017 | KU705567 | KU705557 | JF972493 | KR904199 | KR903665 | KR903321 |
| Lebinthus luae Robillard & Tan (2013) | MNHN/L10LbiS3 | MK761253 | MK761276 | MK761333 | MK761342 | MK761355 | MK761295 | MK761315 | MK761372 |
| Microbinthus santoensis Robillard (2009) | MNHN-EO-ENSIF2484/L7LsaPe | KU705528 | KU708011 | KU705569 | NA | KU705535 | KU705543 | KU705585 | KU705601 |
| Microbinthus santoensis Robillard (2009) | MNHN-EO-ENSIF2437/LsaV (PT) | JF972527 | JF972511 | JX897405 | JX897441 | JF972495 | JF972542 | JX897467 | JX897548 |
| Nisitrus vittatus Haan (1842) | MNHN-EO-ENSIF2742/NviS | MH575026 | MH575158 | KU705572 | NA | MH662741 | AY905340 | KR903667 | JX897546 |
| Pseudolebinthus gorochovi Robillard (2018) | ZIN/X17PsMal1 (HT) | KY595508 | KY595472 | KY646231 | NA | NA | KY595511 | KY605231 | MK761373 |
| Pseudolebinthus gorochovi Robillard (2018) | MNHN-EO-ENSIF10732/X27 | NA | MN583263 | NA | NA | NA | NA | NA | NA |
| Pseudolebinthus gorochovi Robillard (2018) | MNHN-EO-ENSIF10744/X31 | NA | MN583262 | NA | NA | NA | NA | NA | NA |
| Pseudolebinthus lunipterus sp. nov. | MNHN-EO-ENSIF10720/X28 (PT) | MN414243 | MN414243 | MN414243 | MN414243 | MN414243 | MN583259 | MN583260 | MN583264 |
| Pseudolebinthus lunipterus sp. nov. | MNHN-EO-ENSIF10718/X34 (PT) | NA | MN583261 | NA | NA | NA | NA | NA | NA |
| Xenogryllus eneopteroides Bolívar (1890) | MNHN-EO-ENSIF3159/XenAC | KR903829 | KR904023 | KY646249 | NA | KR903490 | KR904205 | KR903670 | KR903328 |
| Xenogryllus eneopteroides Bolívar (1890) | MNHN-EO-ENSIF3442/XenCI | MK761256 | MK761279 | NA | NA | NA | MK761298 | NA | MK761375 |
| Xenogryllus eneopteroides Bolívar (1890) | MNHN-EO-ENSIF3442/XenGA | MK761257 | MK761280 | NA | NA | NA | MK761299 | MK761318 | MK761376 |
| Xenogryllus maichauensis Gorochov (1992) | ZFMK/XtrTh | MK761258 | NA | NA | NA | MK761357 | NA | NA | MK761377 |
| Xenogryllus marmoratus Haan (1844) | MNHN-EO-ENSIF3161/Xma2 | KR903830 | KR904024 | NA | MK761343 | KR903491 | KR904206 | NA | KR903329 |
| Xenogryllus marmoratus Haan (1844) | MNHN-EO-ENSIF1599/XmaCh1 | KY595510 | KY595484 | NA | MK761344 | KY646274 | KY595518 | KY605248 | KY646292 |
| Xenogryllus marmoratus Haan (1844) | MNHN-EO-ENSIF1594/XmaCh2 | MK761261 | MK761283 | NA | MK761345 | MK761360 | MK761302 | MK761320 | MK761379 |
| Xenogryllus marmoratus Haan (1844) | MNHN-EO-ENSIF3562/XmaCh3 | MK761262 | MK761284 | NA | MK761346 | MK761361 | MK761303 | MK761321 | MK761380 |
| Xenogryllus mozambicus Robillard (2019) | MNHN-EO-ENSIF1515/XenMoz (PT) | MK761263 | MK761285 | MK761336 | NA | MK761362 | MK761304 | NA | MK761381 |
| Xenogryllus transversus Walker (1869) | IISERM/Xtr715 | MK761264 | MK761286 | NA | NA | MK761363 | MK761305 | MK761322 | MK761382 |
| Xenogryllus transversus Walker (1869) | IISERM/Xtr765 | MK761265 | NA | MK761337 | NA | MK761364 | MK761306 | MK761323 | MK761383 |
| Xenogryllus transversus Walker (1869) | IISERM/Xtr766 | MK761266 | NA | NA | NA | MK761365 | NA | MK761324 | MK761384 |
| Xenogryllus transversus Walker (1869) | MNHN-EO-ENSIF87/XtrIn | JF972530 | NA | KY646247 | MK761347 | JF972499 | KY595519 | KY605246 | KY646294 |
| Xenogryllus ululiu Gorochov (1990) | ZIN/X18XulV2 | MK761268 | MK761287 | NA | MK761348 | NA | MK761308 | MK761326 | MK761386 |
| Xenogryllus ululiu Gorochov (1990) | ZIN/X19XulSi | MK761269 | MK761288 | NA | MK761349 | MK761367 | MK761309 | MK761327 | MK761387 |
| Xenogryllus ululiu Gorochov (1990) | MNHN-EO-ENSIF4385/X20XulCam1 | MK761270 | MK761289 | NA | MK761350 | NA | MK761310 | MK761328 | MK761388 |
| Xenogryllus ululiu Gorochov (1990) | ZIN/X21XulCam2 | MK761271 | MK761290 | NA | MK761351 | NA | MK761311 | MK761329 | MK761389 |
| Xenogryllus ululiu Gorochov (1990) | ZFMK/Xulth | MK761272 | MK761291 | MK761339 | NA | MK761368 | NA | NA | NA |
Note:
Abbreviation of museums: IISERM, Indian Institute of Science Education and Research Mohali, Punjab, India; MNHN, Muséum national d’Histoire naturelle, Paris, France; ZFMK, Zoologisches Forschungsinstitut und Museum Alexander Koenig, Bonn, Germany; ZIN, Zoological Institute, Russian Academy of Sciences, S. Petersburg, Russia, and ZSI, Zoological Survey of India, Kolkata, India. Other abbreviations: HT, Holotype; PT, Paratype.
One extract of the new species P. lunipterus (X28, MNHN-EO-ENSIF10720) was used for library preparation in a Genome Skimming approach (Straub et al., 2012). We assessed total DNA with a Qubit™ dsDNA High-Sensitivity Assay Kit (Life Technologies, Paisley, UK) with a Fluorescence Microplate Reader in 1.0 µL of sample. Prior to library preparation, we fragmented the DNA by sonication using BioRuptor® UCD-200 (Life Technologies and Invitrogen, Carlsbad, CA, USA) using 50 µL DNA sample and 50 µL TE buffer 0.1X. The molecular weight of the fragmented DNA sample was analyzed in agarose gel electrophoreses (3.0 µL DNA sample plus BG 1.0 µL; gel agarose 1% in TAE buffer (Tris-acetate-EDTA) 1.0X; migration buffer TAE 0.5X; migration time 20 min) before and after the sonification of the DNA. We then used the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA, USA; dsDNA protocol) with a modified version of the protocol based on Meyer & Kircher (2010). After library preparation, total DNA was quantified with a Qubit™ dsDNA (HS) Assay Kit using Qubit™ Fluorometer (Life Technologies, Carlsbad, CA, USA) in 1.0 µL of sample. Libraries were then analyzed with a Bioanalyzer 2100 DNA 1000 series II chip (Agilent Technologies, Santa Clara, CA, USA) (High Sensitivity DNA Assay). Pooled libraries were sequenced as paired-end reads (150 bp) on an Illumina HiSeq 3000 HWI-J0015 at the Genome and Transcriptome Platform of Toulouse (Genotoul, Toulouse, France).
Sequence analyses and mitogenome annotation
Sequencing reads from both paired-end libraries were imported in Geneious Prime 2019.1.3, then filtered and trimmed by quality using the BBDuk plugin (minimum quality score of 30 and minimum length of reads of 30 bp). Quality and length distribution of the sequences were inspected using FastQC v. 0.11.8 (Andrews, 2010) under the open-source application Galaxy (http://galaxyproject.org/) (Afgan et al., 2018). We then extracted sequences of interest from the total read using the Map to reference option in Geneious (Custom sensibility, fine tuning: iterate up to 10 times; Maximum Mismatches Per Read 30). The Mitochondrial genomes of Xenogryllus marmoratus (Haan, 1842) (Ma, Zhang & Li, 2019; GenBank Accession MK033622) and Cardiodactylus muiri Otte, 2007 (Dong et al., 2017; GenBank Accession MG680938) were used as references. After removing the reference sequence from the resulting contig, a step of De Novo assemble (Sensibility: High sensibility/Medium) was performed in Geneious. The longest resultant contig (Number of reads 14,817; Sequence length 16,075 bp) was chosen as a seed and mapped with the filtered reads again (Custom sensibility, fine-tuning: iterate up to 25 times; max. Mismatches Per Read 10).
The consensus sequence (Threshold: Highest quality; Assign quality: Highest) was then circularized and annotated with Geneious with X. marmoratus as reference. Genome annotation based on sequence similarity was performed independently using Geneious and with MITOS (Bernt et al., 2013), available at http://mitos.bioinf.uni-leipzig.de/index.py using the invertebrate mitogenome genetic code.
A similar process was used to extract sequences of three nuclear genes: histone H3 (H3, ~330 bp) and the sequences of the non-protein-coding genes corresponding to nuclear small ribosomal subunit (18S rRNA, 18S, ~650 bp) and of the nuclear large ribosomal subunit (28S rRNA, 28S, ~400 bp). Nuclear genes were assembled using references from the species P. gorochovi from (Jaiswara, Dong & Robillard, 2019). All the newly generated sequences are available on GenBank (Table 1).
Phylogenetic analysis
The cricket tribe Xenogryllini is composed of three genera and 13 valid species (including the new species): Xenogryllus Bolívar (eight species), Pseudolebinthus Robillard (four species) and Indigryllus Robillard & Jaiswara (one species). To infer the phylogenetic position of the new species, we refer to the recent molecular phylogeny of Xenogryllini (Jaiswara, Dong & Robillard, 2019) which included eight Xenogryllini species representing the three genera and eight species representing all four other tribes of the Eneopterinae subfamily, plus two more distant species belonging to the subfamily Gryllinae. We used DNA markers from eight genes, five from the mitochondrial and three from the nuclear genome based on Jaiswara, Dong & Robillard (2019) and previous studies (Robillard & Desutter-Grandcolas, 2006; Nattier et al., 2012). The mitochondrial markers were partial sequences of the small subunit rRNA gene (12S, amplicon ~400 bp), the large subunit rRNA gene (16S, ~500 bp), of the cytochrome b gene (Cytb, ~400 bp), and of the cytochrome c oxidase subunit 1 (CO1, ~750 bp) and subunit 2 (CO2, ~400 bp). Nuclear markers were partial sequences of protein coding histone H3 gene (H3, ~330 bp), and partial sequences of two non-protein-coding genes corresponding to nuclear ribosomal subunits 18S rRNA (18S, ~650 bp) and 28S rRNA (28S, ~400 bp).
Newly generated sequences were added to the previous data set. We extracted the mitochondrial markers from the mitogenome of P. lunipterus sp. nov. See Table 1 for detailed information about taxon and molecular sampling.
The sequences were aligned with MAFFT version 7 online (Katoh & Standley, 2013). The aligned sequences of all eight markers were further concatenated in Sequencematrix (Vaidya, Lohman & Meier, 2011). The concatenated dataset was analyzed using maximum likelihood (ML) using IQ-TREE 1.6. 2 web portal (Nguyen et al., 2015) (http://iqtree.cibiv.univie.ac.at/; Kalyaanamoorthy et al., 2017; Trifinopoulos et al., 2016) with data partitioned by gene marker and the following options: Edge-unlinked partitions, Substitution model Auto. Clade support was assessed by conducting 1,000 bootstrap replicates (standard bootstrap). Nodes supported by bootstrap support values (BS) ≥ 70% were considered strongly supported.
Results
Mitogenome of P. lunipterus
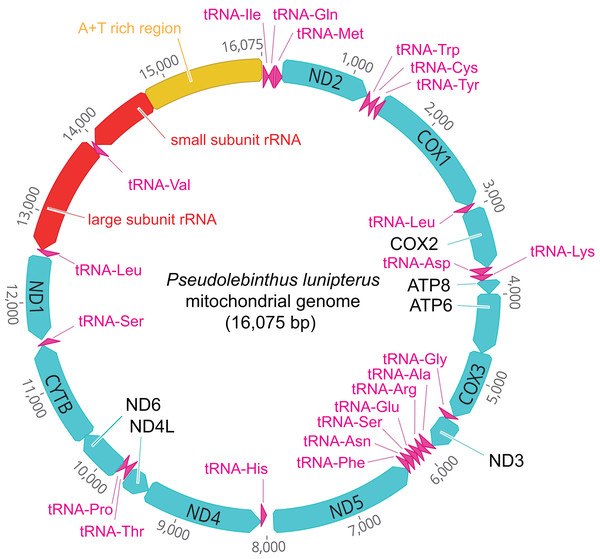
The mitogenome of P. lunipterus is 16,075 bp in length and has a typical circular structure (Fig. 2). The nucleotide composition of this genome has a GC content of 24.2%. The identity and position of 13 PCGs, 22 tRNA and 2 rRNA genes is detailed in Table 2.
Figure 2: Map of the mitochondrial genome of Pseudolebinthus lunipterus sp. nov.
The 13 PCGs are shown in blue, the 22 tRNA in purple, the 2 rRNA genes in red and the A + T rich region in orange. The direction of transcription is indicated by an arrow.| Gene | Location | Length (bp) | Strand | Start codon | Stop codon | Anticodon sequence |
|---|---|---|---|---|---|---|
| tRNA-Ile | 1–63 | 63 | + | GAT | ||
| tRNA-Gln | 61–129 | 69 | − | TTG | ||
| tRNA-Met | 147–215 | 69 | + | CAT | ||
| ND2 | 217–1,233 | 1,017 | + | ATT | TAA | |
| tRNA-Trp | 1,232–1,296 | 65 | + | TCA | ||
| tRNA-Cys | 1,289–1,351 | 63 | − | GCA | ||
| tRNA-Tyr | 1,367–1,430 | 64 | − | GTA | ||
| COX1 | 1,424–2,971 | 1,548 | + | ATC | TAA | |
| tRNA-Leu2 | 2,966–3,030 | 65 | + | TAA | ||
| COX2 | 3,031–3,706 | 676 | + | ATG | TAA1 | |
| tRNA-Lys | 3,706–3,775 | 70 | + | CTT | ||
| tRNA-Asp | 3,776–3,843 | 68 | + | GTC | ||
| ATP8 | 3,845–4,003 | 159 | + | ATT | TAG | |
| ATP6 | 3,997–4,677 | 678 | + | ATG | TAA | |
| COX3 | 4,681–5,467 | 787 | + | ATG | TAA1 | |
| tRNA-Gly | 5,467–5,530 | 64 | + | TCC | ||
| ND3 | 5,532–5,885 | 354 | + | ATA | TAA | |
| tRNA-Ala | 5,886–5,949 | 64 | + | TGC | ||
| tRNA-Arg | 5,950–6,012 | 63 | + | TCG | ||
| tRNA-Glu | 6,007–6,070 | 64 | − | TTC | ||
| tRNA-Ser1 | 6,071–6,137 | 67 | − | GCT | ||
| tRNA-Asn | 6,138–6,203 | 66 | − | GTT | ||
| tRNA-Phe | 6,211–6,278 | 68 | − | GAA | ||
| ND5 | 6,251–7,927 | 1,677 | − | CAT | TTA | |
| tRNA-His | 8,002–8,064 | 63 | − | GTG | ||
| ND4 | 8,066–9,409 | 1,342 | − | CAT | TTA | |
| ND4L | 9,403–9,687 | 285 | − | CAT | TTA | |
| tRNA-Thr | 9,704–9,767 | 64 | + | TGT | ||
| tRNA-Pro | 9,768–9,833 | 66 | − | TGG | ||
| ND6 | 9,837–10,361 | 525 | + | ATT | TAA | |
| CYTB | 10,361–11,500 | 1,140 | + | ATG | TAA | |
| tRNA-Ser2 | 11,499–11,563 | 65 | + | TGA | ||
| ND1 | 11,564–12,518 | 952 | − | TAT | TAA1 | |
| tRNA-Leu1 | 12,521–12,587 | 67 | − | TAG | ||
| Large subunit rRNA | 12,599–13,895 | 1,297 | − | |||
| tRNA-Val | 13,877–13,944 | 68 | − | TAC | ||
| Small subunit rRNA | 13,944–14,733 | 790 | − | |||
| A+T rich region | 14,734–16,075 | 1,342 |
Note:
Direction of transcription: +, forward; −, reverse. Exponent numerals in gene column differentiate each of the two leucine- and serine-specifying tRNAs (Leu1 and Leu2, Ser1 and Ser2). 1TAA stop codon is completed by the addition of 3′A residues to mRNA.
Phylogenetic relationships
The alignment of all eight markers consists of 3,684 aligned base pairs (bp) for 34 terminals: 416 bp for 12S, 521 bp for 16S, 707 bp for CO1, 335 bp for CO2, 346 bp for Cytb, 328 bp for H3, 652 bp for 18S and 379 bp for 28S. The ML phylogenetic tree inferred from this data is shown in Fig. 3.
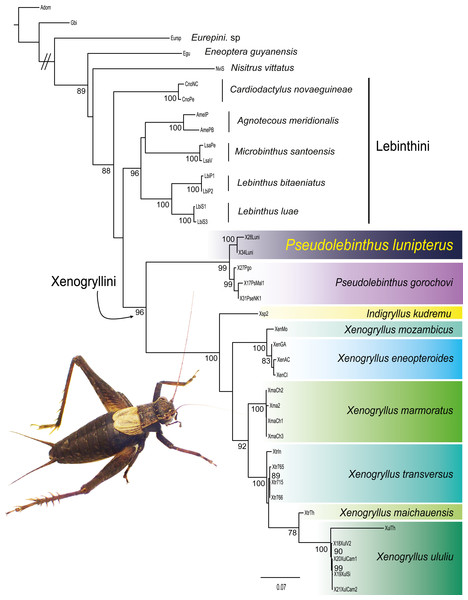
Figure 3: Phylogenetic position of Pseudolebinthus lunipterus sp. nov.
Maximum likelihood tree of Xenogryllini tribe based on the concatenated dataset of eight genetic markers. ML bootstrap (BS) values higher than 75% are indicated for each node on the left. Clades corresponding to species are shaded with a color scale. Represented species: male of Pseudolebinthus lunipterus sp. nov. Photo Tony Robillard.Most nodes of the tree have high bootstrap supports and are consistent with the recent study of Jaiswara, Dong & Robillard (2019). All species represented by two or more terminals are found monophyletic. The subfamily Eneopterinae and the tribe Xenogryllini are recovered as monophyletic with high support values (BS of 100% for Eneopterinae, 99% for Xenogryllini). The relationship of tribes Lebinthini and Xenogryllini are also supported, although the Lebinthini are not found monophyletic. The new species occurs as the sister species of P. gorochovi with high support (BS of 100%). Branches between the two species crown groups are short, suggesting that the new species should be considered as a member of the genus Pseudolebinthus.
Taxonomy
Insecta Linnaeus, 1758
Orthoptera Olivier, 1789
Gryllidae Laicharting, 1781
Eneopterinae Saussure, 1874
Xenogryllini Robillard, 2004
Genus Pseudolebinthus Robillard, 2006
Type species: Pseudolebinthus africanus Robillard, 2006
P. lunipterus sp. nov.
(Figs. 1–11)
Type material
Holotype male, MALAWI. N. Malawi, Mt. Uzumara, 6,500 ft. R.J. Murphy col. 2.i.2001 (MNHN-EO-ENSIF10715). Allotype female, same information as holotype (MNHN-EO-ENSIF10716). Paratypes (9♂, 6♀): MALAWI. Same information as holotype, 1♂, #233RJM (MNHN-EO-ENSIF10717). N. Malawi, Mount Uzumara (MAL1), S10°52′19,3″ E34°07′44,7″, 1,941 m (MAL1), 2-4.x.2018, nuit (night), plante, élevage F0 (collected as juveniles on plant, reared to final moult in captivity), T. Robillard, K. Salazar & R.J. Murphy: 3♂, molecular sample X34, X45, X28 (MNHN-EO-ENSIF10718-10720); 2♂, video of mating behavior (♂#3, ♂#4) (MNHN-EO-ENSIF10721-10722); 3♂ (MNHN-EO-ENSIF10721-10722); 2♀, video of mating behavior (♀#1, ♀#3) (MNHN-EO-ENSIF10723-10724); 4♀ (MNHN-EO-ENSIF10725-10728).
Additional material examined. MALAWI. N. Malawi, Mount Uzumara (MAL1), S10°52′19,3″ E34°07′44,7″, 1,941 m (MAL1), 2019, élevage F1, T. Robillard: 1♂, 1♀, 3 juveniles (MNHN).
Type locality. North Malawi, Mount Uzumara, S10°52′19,3″ E34°07′44,7″, 1,941 m.
Distribution. The species is only known from the type locality in Northern Malawi (Fig. 1C).
Etymology. The species name refers to the whitish wings, rounded in males and crescent-shaped in females, which look like tiny moons on the back of the dark body of these crickets when encountered at night.
Diagnosis
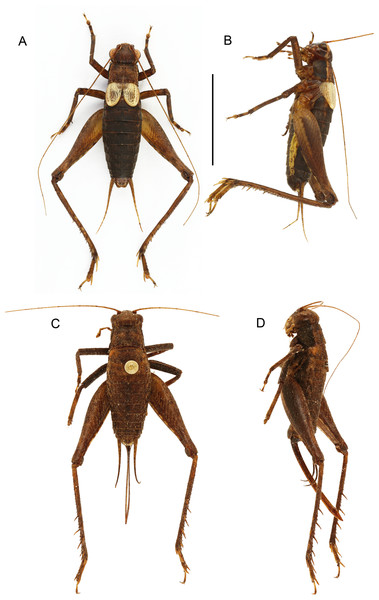
Size small, mostly dark brown with pale wings (Figs. 1, 3 and 4). Among Eneopterinae genera, the new species presents the characteristics of Pseudolebinthus: large lateral eyes (Figs. 5A–5C); brachypterous FWs barely reaching quarter of abdomen length in males (Figs. 4 and 6), shorter in females where it forms pale narrow crescents (Figs. 4 and 5D); male genitalia with long sclerotized lophi, close to that of P. gorochovi (Figs. 8 and 9); female ovipositor little differentiated but less pointed and thicker than in P. gorochovi (Fig. 10A). The new species is characterized by complete absence of tympana (unique feature among eneopterines) (Figs. 7A and 7B), absence of stridulatory apparatus on male FWs (Fig. 6), abdomen ventrally yellow with a wide black stripe (Fig. 5F), thick and short female ovipositor (Fig. 4D), and differences in male genitalia, including shape of pseudepiphallic parameres, shape of sclerite in ectophallic fold and endophallic apodeme with anterior lateral expansions.
Figure 4: Male and female of Pseudolebinthus lunipterus sp. nov.
(A and B) Male and (C and D) female in dorsal (A and C) and lateral (B and D) views. Scale bar = 1 cm. Photos (A and B) Karen Salazar. (C and D) Simon Poulain.Figure 5: Morphology of Pseudolebinthus lunipterus sp. nov.
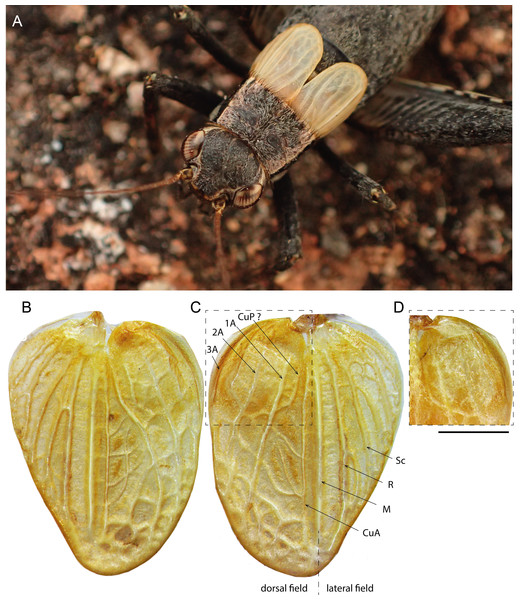
(A–C, E and F) Male and (D) female; (A–C) head in dorsal (A) and facial (C) views; (B) head, pronotum and wings in lateral view; (D) lateral view of wings; (E) metanotal glands; (F) ventral face of tip of abdomen, subgenital plate and cerci. Scale bars = 1 mm. Photos Karen Salazar.Figure 6: Male forewings and venation of Pseudolebinthus lunipterus sp. nov.
(A) Male habitus showing FW coloration in their natural context; (B–D) detailed view of left (B) and right (C) FW in dorsal view; (D) anterior part of right FW in ventral view. Abbreviations: see “Material and Methods”. Scale bar = 1 mm. Photos Tony Robillard.Figure 7: Legs of Pseudolebinthus lunipterus sp. nov.
(A and B) Posterior and anterior views of fore leg showing no trace of tympanum on upper part of tibia (see arrows); (C) lateral external view of hind leg; (D) detail of hind tibia in dorsal view. Scale bar = 5 mm. Photos Karen Salazar.Figure 8: Male genitalia of Pseudolebinthus lunipterus sp. nov.
(A) Dorsal and (B) ventral views; dotted parts represent membranous areas. Abbreviations: see “Material and Methods” section. Scale bar = 1 mm. Drawing Karen Salazar & Tony Robillard.Figure 9: Details views of male genitalia.
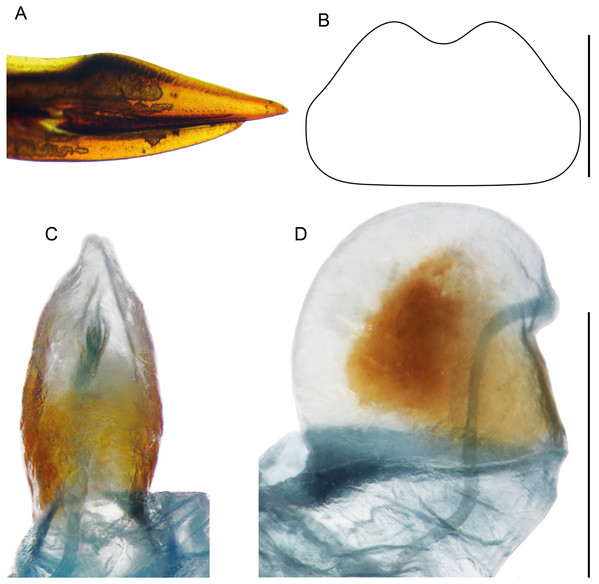
(A) Complete ventral view; (B) lateral view of pseudepiphallic lophi; (C and D) pseudepiphallic lophi and parameres, and ectophallic fold in ventral (C) and posterior views. Scale bar = 0.5 mm. Photos Karen Salazar.Figure 10: Female structures of Pseudolebinthus lunipterus sp. nov.
(A) Apex of ovipositor in lateral view; (B) shape of subgenital plate; (C and D) copulatory papilla in ventral (C) and lateral (D) views. Scale bar = 0.5 mm. Photos and drawing Karen Salazar.Description
In addition to the characters of the genus: body mostly dark brown with pale wings (Figs. 1, 3 and 4). Head rounded dorsally, with five black bands separated by thin yellow lines, including two thin bands posterior to eyes and three wider median bands, sometimes partly faded in median area (Figs. 5A–5C). Fastigium black, setose, with thin yellow margins, its apex with a wide yellow band surrounding median ocellus. Eyes occupying ca. 40% of total head width in dorsal view. Ocelli forming a wide triangle. Scapes short, wider than long, black with a yellow spot on upper inner side; rest of antennae brown. Face triangular (Fig. 5C), almost entirely black except: a median vertical line connected to the yellow band at fastigium apex, two yellow spots above epistomal suture, a yellow spot below each eye; mouthparts black, mottled with yellow. Lateral part of head entirely black (Fig. 5B). Pronotum dark brown dorsally, its lateral margins thinly underlined with yellow (Figs. 5B and 6A); lateral field black (Fig. 5B). TI with no tympana (Figs. 7A and 7B). FWs very short in both sexes (Fig. 4), hind wings absent. Fore and median legs with black femora, dark brown tibiae (Fig. 4); base of tarsomeres yellow, apex darker. FIII external face bicolor: dorsal half black, ventral half yellow; knees black (Fig. 7C); TIII dark brown; tarsomere III yellow basally, apex dark brown. Abdomen rather long and fusiform (Fig. 4), yellow ventrally with a wide black stripe, including subgenital plate (Fig. 5F). Subgenital plate slightly indented apically in both sexes (Figs. 5F and 10B).
Males. FWs short, reaching one quarter of abdomen length (Figs. 4A, 4B and 6); without stridulatory apparatus; veins and cells soft, without pigmentation (whitish in the living, turning yellow when drying). Dorsal field (Figs. 6B and 6C) with two main strong longitudinal veins corresponding to veins 1A and 2A; CuA weak, with two apical expansions delimiting possible cell alignment of apical field; and one longitudinal vein along inner margin (3A?). Area delimited by 1A and CuA slightly widened posteriorly, sometimes with a faint anterior vein possibly corresponding to vein CuP; area with variable transverse veins in posterior region, possibly corresponding to diagonal vein or posterior limit of cells of mirror area in crickets having a stridulatory apparatus. Ventral face of 1A without trace of stridulatory teeth (Fig. 6D). Lateral field narrow (Figs. 6B and 6C), with three main longitudinal veins, including R, Sc and M, the latter separating dorsal and lateral fields; one more ventral vein sometimes near FW base. Hind wings vestigial. Metanotum with glandular structures (Fig. 5E); gland morphology close to that of other Pseudolebinthus, with a bunch of long setae on basal margin, a wide median process on scutum, and basal edge of scutellum raised medially and carrying a bunch of setae orientated anteriorly; posterior part of mesonotum setose and extended posteriorly, covering anterior part of metanotal scutum. Male subgenital plate elongate (Fig. 5F).
Male genitalia (Figs. 8 and 9). Pseudepiphallic sclerite as long as rami, widened laterally near base of rami. Pseudepiphallic lophi thin and parallel, longer than in P. gorochovi, twisted ventro-apically; their apex with a small dorsal expansion. Pseudepiphallic parameres dorsal lobe triangular, longer than ventral lobe; ventral lobe oriented anteriorly forming an apical fold. Ectophallic fold with a strong ventro-lateral sclerotization forming a transversal bridge (Figs. 8B, 9C and 9D) slightly extended posteriorly within pointed membranous apex. Ectophallic apodemes long and parallel. Endophallic sclerite with a small median area, with wide lateral arms; endophallic apodeme including two small dorso-anterior arms at anterior apex of endophallic sclerite and a narrow apical transverse crest extended laterally, underlying arms of endophallic sclerite.
Females (Figs. 4C, 4D, 5D, 10 and 11A). FWs forming very small lateral whitish leaflets (Fig. 5D), not overlapping and not reaching posterior margin of first tergite (Figs. 4C, 4D and 5D). Ovipositor short and thick, shorter than FIII (Figs. 4C and 4D), its apex little differentiated, pointed and not denticulate dorsally (Fig. 10A). Female copulatory papilla flattened laterally, mostly sclerotized, its apex membranous folded ventrally (Figs. 10C and 10D).
Figure 11: Live photos of Pseudolebinthus lunipterus sp. nov.
(A) Female on vegetation; (B) subadult male eating a dead insect on a leaf at night; (C) first instar juvenile. Photos (A and B) Tony Robillard and (C) Karen Salazar.Juveniles. First instars body cylindrical (Fig. 11C), mostly dark brown with a light dorsal stripe; antennae brown with yellow rings. Later instars with similar coloration pattern as adults, usually lighter brown. Last instar with characteristic whitish wing buds, similar to wing coloration in adults (Fig. 11B).
Habitat and life history. Pseudolebinthus lunipterus lives on low vegetation in herbaceous areas near forest hedge or in open areas along trails in forest (Figs. 1A and 1B). Adults and juveniles have been found active at night on top of vegetation, but can also be found lower within vegetation during the day. Remarkably, the species lives in syntopy with P. gorochovi in the type locality, where adults and juveniles of both species are quite abundant. One juvenile specimen of P. lunipterus has been observed eating a dead insect on a low leaf on vegetation (Fig. 11B).
Females maintained in controlled laboratory conditions (20–22 °C, 14–10 day–night cycle) with a single male produced 46–50 offspring (n = 2) during their life; first hatchings started 42–49 days after first mating and occurred on a period of 35–66 days.
Measurements (in mm). See Table 3.
Key to Pseudolebinthus species modified from Jaiswara, Dong & Robillard (2018)
1. TI each with one pair of tympana, male FWs longer than one quarter of abdomen length, with a stridulatory apparatus2
– TI without tympana, male FWs about one quarter of abdomen length, without stridulatory apparatusP. lunipterus sp. nov.
2. Dorsal margin of eye with small ommatidia narrow. Male FW venation: mirror small, hardly distinct from surrounding cells. Male genitalia: pseudepiphallic lophi long and thinP. africanus Robillard, 2006
– Dorsal margin of eye with small ommatidia wider. Male FW venation: mirror larger, wider than long, well differentiated. Male genitalia: pseudepiphallic lophi shorter3
3. Coloration little contrasted; FIII homogeneously brown on external face. Male FW venation: c1 and c2 cells sub-equal, thin. Vein Sc with four projections along its length. Male genitalia: apex of pseudepiphallic lophi bilobateP. whellani Robillard, 2006
– Coloration more contrasted; FIII external face bicolor, dorsal half dark brown, ventral half yellow. Male FW venation: c1 cell wide, c2 twice wider than c1, square and prolonging shape of miror. Vein Sc with two projections along its length. Male genitalia: apex of pseudepiphallic lophi twisted ventrally, not bilobateP. gorochovi Robillard, 2018
| BL | PronL | PronW | FWL | FWW | FIIIL | FIIIW | |
|---|---|---|---|---|---|---|---|
| Holotype male | 13.6 | 1.9 | 3.1 | 2.8 | 1.6 | 9.1 | 2.5 |
| Males (n = 5) | 13.5–14 | 1.8–2.1 | 2.9–3.3 | 2.8–3.5 | 1.6–1.9 | 9.1–11.2 | 2.5–3 |
| (Male mean) | (13.7) | (1.9) | (3.1) | (3.1) | (1.8) | (10.5) | (2.8) |
| Female Allotype | 15.1 | 2.2 | 3.5 | 1.1 | – | 11.8 | 3.2 |
| Females (n = 5) | 15.1–16.9 | 2.2–2.5 | 3.2–3.8 | 0.5–1.4 | – | 11–12.1 | 3.2–3.5 |
| (Female mean) | (15.7) | (2.3) | (3.6) | (0.9) | – | (11.5) | (3.3) |
| TIIIL | TIIIs | TaIIIs | Ols | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Ias | Ibs | Oas | Obs | Ids | Ods | OL | |||
| Holotype male | 9 | 7 | 7 | 10 | 8 | 1 | 3 | 2 | – |
| Males (n = 5) | 9–11 | 6–7 | 7–11 | 8–11 | 8–12 | 0–1 | 3–5 | 2–3 | – |
| (Male mean) | (9.8) | (7) | (9) | (10) | (10) | (1) | (4) | (3) | – |
| Female Allotype | 10.5 | 5 | 8 | 11 | 8 | 0 | 4 | 3 | 10.8 |
| Females (n = 5) | 10.5–12.1 | 5–8 | 7–10 | 9–12 | 8–11 | 0 | 3–5 | 3 | 9.3–10.8 |
| (Female mean) | (11.1) | (6) | (9) | (11) | (9) | (0) | (4) | (3) | (10.1) |
Discussion
Multiple losses of acoustic communication
In this article we described the species P. lunipterus sp. nov., a new eneopterine cricket from Northern Malawi being both mute and deaf. This new species is the first reported case showing complete absence of stridulatory apparatus (no stridulatory file, harp and mirror) in this cricket clade, associated with absence of tympana on both sides of fore tibiae.
Our taxonomic study suggests that P. lunipterus belongs to the tribe Xenogryllini despites all its special morphological features. The phylogenetic analysis shows that it is closely related with at least one other species of the genus Pseudolebinthus (Fig. 3). This phylogenetic position has two interesting consequences:
First, this is the first case of muteness documented in the clade Xenogryllini. Two other cases of loss of acoustic communication were previously reported in eneopterines (Table 4): one occurred in the tribe Nisitrini and concerns the apterous species of the genus Paranisitra, which has diverged from its sister genus Nisitrus ca. 55 Ma, before diversifying in the Philippines after 12.5 Myrs (Vicente et al., 2017; Baroga-Barbecho et al., 2019). The second other mute lineage is the genus Swezwilderia in the tribe Lebinthini; species of Swezwilderia possess long wings, but lack stridulatory structures. This genus has diverged from its sister group ca. 48.4 Ma, and has diversified in Fiji and Samoa after 21.6 Myrs (Vicente et al., 2017). In addition to their phylogenetic independence, these three losses of calling abilities are structurally different, one occurring through the loss of complete wings and the two others consisting of losses of stridulatory structures either on fully formed wings (Swezwilderia) or on reduced wings (P. lunipterus). These different combinations of traits support the hypothesis that these three losses of acoustic communication are convergent in Eneopterinae. The phylogenetic context of each loss may explain these different configurations in relation with two other functions of the wings of insects: flight and protection. In Swezwilderia, the wings might have been retained in association with keeping flying capacities, while short wings might have been necessary in P. lunipterus to protect the metanotal glands that are shared among all the Xenogryllini species (Jaiswara, Dong & Robillard, 2018, 2019; Jaiswara et al., 2019). Interestingly, the short wings in the new species are shorter than that of other Pseudolebinthus species, but remain just long enough to cover the glands beneath (Figs. 5B, 5E and 6). In contrast, Paranisitra is apterous but has also lost the metanotal glands while diverging from Nisitrus.
| Tribe | Taxon | Wings | Flight | Metanotal glands | Stridulatory structures | Inner tympanum | Outer tympanum | Hearing |
|---|---|---|---|---|---|---|---|---|
| Xenogryllini | P. lunipterus | Short | No | Present | Absent | Absent | Absent | Absent |
| Pseudolebinthus (other species) | Short | No | Present | Present | Present | Present | ||
| Xenogryllus | Long | Yes | Present | Present | Present | Present | ||
| Nisitrini | Paranisitra | Absent | No | Absent | Absent | Present | Present | Present |
| Nisitrus | Long | Yes | Present | Present | Present | Present | ||
| Lebinthini | Swezwilderia | Long | Yes | Absent | Absent | Present | Present | Present |
| Lebinthini (other genera) | Long/short | Yes/no | Absent | Present | Present | Present | ||
| Eneopterini | No mute taxon | Long | Yes | Present | Present | Present | Present | Present |
| Eurepini | No mute taxon | Long/short | Yes/no | Variable | Present | Present | Absent | Present |
Note:
Mute taxa discussed in the text are highlighted in gray.
The second interesting observation about the phylogenetic position of P. lunipterus is that this is the only mute species occurring within a genus, while others mute cases concern clear-cut genera which show strong divergence from their sister lineages. Even if genera do not represent evolutionary units, it is interesting to notice that P. lunipterus and P. gorochovi are separated by very short branches in the phylogenetic tree (Fig. 3). A molecular dating analysis of Pseudolebinthus will be necessary with a better taxonomic sampling, but this situation suggests that the loss of acoustic communication in P. lunipterus is likely a recent event, which is recalling the loss of sound production structures occuring convergently and very rapidly within some populations of T. oceanicus as a result to strong selective pressures by a parasitoid fly (Zuk, Rotenberry & Tinghitella, 2006; Pascoal et al., 2014). Analogous selective pressures might be responsible for the loss of sound production in P. lunipterus.
The most unique feature of P. lunipterus at the scale of the subfamily is the deafness of the species. Other cases of deaf crickets have been documented in other clades, but this is the only case known in eneopterines. In many mute lineages of crickets, auditory tympana are retained after the stridulatory mechanism is lost (Otte, 1992). In species that are still able to fly, but in which males have lost the stridulum (such as in species of Swezwilderia), the tympanum is usually retained, which is supposed to be linked with avoidance of bat predation in flight (Otte & Alexander, 1983; Otte, Alexander & Cade, 1987). Species becoming both mute and deaf, such as P. lunipterus, are less common. This combination of traits might be explained by predator avoidance selecting for mute crickets in lineages having ancestrally lost their flying capacities (all Pseudolebinthus). In such cases, maintaining tympana might not be necessary. Interestingly, this hypothesis does not hold with the case of Paranisitra, which lost the wings while retaining hearing capacities (or at least external organs, since the hearing capacities of Paranisitra have never been evaluated).
Generic allocation
The morphological study of the new species shows that it shares all the characteristics of the genus Pseudolebinthus in terms of general morphology, body size and main features of male genitalia. On the other hand, the new species differs by important characters such as FW length, absence of stridulatory file and tympana, and by the shape of female copulatory papilla. Such differences suggest that the new species might have been considered as an easily recognized new genus close to Pseudolebinthus. This hypothesis has been tested using the molecular data and the phylogenetic relationships. Although it is too early to conclude that the new species is nested within Pseudolebinthus (only one previously described species was successfully sequenced here), our results clearly show that the new species is very close to P. gorochovi, and the short stem branches in the phylogeny leading to the new species and P. gorochovi strongly support the hypothesis that the new species should be considered as a particular species of Pseudolebinthus.
Conclusion: Crickets of Malawi
The diversity of crickets in Eastern Africa in general, and Malawi in particular, has been underestimated, understudied and undersampled. This is at least the case for the members of the tribe Xenogryllini which were recently revised (Jaiswara, Dong & Robillard, 2018, 2019; Jaiswara et al., 2019). Despite the large amount of data considered in these systematic studies (several hundreds of specimens studied across the study of the largest natural history museum collections), they gathered very little information about the species of Pseudolebinthus, known by a few specimens each.
A single recent field trip in Malawi allowed us to re-discover two of the previously described species of the genus, which are in fact common species, and it allowed documenting the acoustic features of their calling songs and their ecology (T. Robillard et al., 2020, in prep.). Interestingly, these findings allowed us to discover P. lunipterus, a completely different new species belonging to the Xenogryllini lineage, but with strikingly new morphological features. This finding reveals that more taxa probably remain unrecorded in the whole Eastern African region, as suggested by the large amount of new species and genera recently discovered in this region for other clades of orthopteran insects (Hemp et al., 2018; Hemp & Heller, 2019). More taxonomic surveys with appropriate collecting methods in regions where there is zero record about these crickets, such as other regions of Malawi, but also Zimbabwe, Zambia, Western Mozambique and Northern South Africa, are thus necessary to explore this part of African biodiversity.
Supplemental Information
Sequences submitted to GenBank.
Accession numbers processed by GenBank after sumission. This supplementary material will be deleted when accession numbers will be made available.