Jatropha curcas ortholog of tomato MADS-box gene 6 (JcTM6) promoter exhibits floral-specific activity in Arabidopsis thaliana
- Published
- Accepted
- Received
- Academic Editor
- Genlou Sun
- Subject Areas
- Biotechnology, Molecular Biology, Plant Science, Forestry
- Keywords
- TM6, Promoter, Flower, Arabidopsis, Physic nut
- Copyright
- © 2020 Wang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits using, remixing, and building upon the work non-commercially, as long as it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Jatropha curcas ortholog of tomato MADS-box gene 6 (JcTM6) promoter exhibits floral-specific activity in Arabidopsis thaliana. PeerJ 8:e9827 https://doi.org/10.7717/peerj.9827
Abstract
Background
Jatropha curcas L., a perennial oilseed plant, is considered as a promising feedstock for biodiesel production. Genetic modification of flowering characteristics is critical for Jatropha breeding. However, analysis of floral-specific promoters in Jatropha is limited.
Methods
In this study, we isolated the Jatropha ortholog of TM6 (JcTM6) gene from Jatropha flower cDNA library and detected the expression pattern of JcTM6 gene by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). We isolated a 1.8-kb fragment from the 5’ region of the JcTM6 gene and evaluated its spatiotemporal expression pattern in Arabidopsis using the β-glucuronidase (GUS) reporter gene and Arabidopsis ATP/ADP isopentenyltransferase 4 (AtIPT4) gene, respectively.
Results
JcTM6 was identified as a flower-specific gene in Jatropha. As expected, JcTM6 promoter was only active in transgenic Arabidopsis flowers with the strongest activity in stamens. Moreover, JcTM6:AtIPT4 transgenic Arabidopsis showed a phenotype of large flowers without any alterations in other organs. Furthermore, deletion of the region from –1,717 to –876 bp resulted in the disappearance of promoter activity in stamens but an increase in promoter activity in young leaves, sepals, and petals. Deletion analysis suggests that the –1,717- to –876-bp promoter fragment contains regulatory elements that confer promoter activity in stamens and inhibit activity in young leaves, sepals, and petals.
Introduction
Promoter plays a significant role in gene expression regulation. Three types of promoters are currently employed in plant genetic engineering: constitutive, tissue-specific, and inducible promoters (Muthusamy et al., 2017; Potenza, Aleman & Sengupta-Gopalan, 2004). Tissue-specific promoters drive transgene expression in a specific spatiotemporal pattern, which is effective in the modification of agronomic traits of crop plants. For example, the rice (Oryza sativa L.) gene OsGA2ox1 encodes a gibberellin (GA) catabolic enzyme, GA 2-oxidase (Lester et al., 1999; Martin, Proebsting & Hedden, 1999; Thomas, Phillips & Hedden, 1999). When the expression of OsGA2ox1 was driven by the constitutive Actin promoter, transgenic rice plants failed to set grains. To prevent sterility, the promoter of a GA biosynthesis gene, OsGA3ox2, which encodes GA 3-oxidase and is specifically active in shoots, was used to control the expression of OsGA2ox1. As expected, transgenic rice exhibited a semi-dwarf phenotype with normal yield (Sakamoto et al., 2003). GA 20-oxidase is a GA biosynthetic enzyme in plants (Coles et al., 1999). In poplar (Populus spp.), overexpression of the Pinus densiflora GA 20-oxidase gene (PdGA20ox) under the control of the constitutive 35S promoter increased GA levels, thereby accelerating stem growth and plant biomass; however, transgenic poplar plants showed poor leaf development and root growth. When the PdGA20ox gene was driven by a xylem-specific promoter DX15 from poplar, the undesirable phenotypes were reduced (Jeon et al., 2016).
Physic nut (Jatropha curcas L.) is an oilseed plant belonging to the Euphorbiaceae family. The seed oil of Jatropha is a promising feedstock for biodiesel production (Kumar & Sharma, 2008). However, low seed yield, which is mainly caused by low female: male ratio, is a long-standing problem in Jatropha (Raju & Ezradanam, 2002; Rao et al., 2008). Jatropha is a monoecious plant species with male and female flowers on the same inflorescence, and the average ratio of female to male flowers is 1:13−1:29 (Raju & Ezradanam, 2002; Tewari et al., 2007). There are 100−300 flowers in each inflorescence of Jatropha, which only produce approximately 10 fruits (Kumar & Sharma, 2008; Pan & Xu, 2011). Hence, genetic modification of flowering characteristics is critical for Jatropha breeding. Floral-specific promoters play crucial roles in this modification because they can drive efficient expression of functional genes in flowers without affecting the vegetative growth of plants. In pea (Pisum sativum), the PsEND1 promoter exhibits anther-specific activity. Expression of the ribonuclease gene barnase (Gardner, Felsheim & Smith, 2009) in Arabidopsis and Brassica napus under the control of the PsEND1 promoter causes anther ablation at an early developmental stage, leading to male sterility (Roque et al., 2007). Arabidopsis APETALA3 (AP3) promoter was identified as a floral-specific promoter in petunia (Petunia x hybrida). Expression of the Agrobacterium tumefaciens isopentenyltransferase (ipt) gene under the control of the AtAP3 promoter in petunia increased the flower size, without affecting vegetative development (Verdonk et al., 2008). However, analysis of promoters, especially floral-specific promoters, in Jatropha is limited. Although the Jatropha APETALA1 (JcAP1) promoter was recently identified as a reproductive tissue-specific promoter showing high activity in inflorescence buds and seeds (Tao et al., 2016), it is not sufficient to address transgene expression analysis in Jatropha.
In this study, we isolated the promoter of the Jatropha ortholog of TOMATO MADS-BOX GENE 6 (JcTM6), a floral-specific gene. The activity of JcTM6 promoter was evaluated in Arabidopsis using the β-glucuronidase (GUS) reporter gene. The results of GUS staining showed that the JcTM6 promoter was active only in flowers, with the highest activity in stamens. By using this promoter directed a cytokinin biosynthesis gene, Arabidopsis ATP/ADP isopentenyltransferase 4 (AtIPT4) gene (Li et al., 2010), only flower phenotype was changed in transgenic Arabidopsis. Furthermore, deletion analysis showed that an approximately 0.85-kb fragment of the JcTM6 promoter (–1717 to –876 bp) is critical for maintaining its floral-specific expression pattern.
Materials & Methods
Plant materials
Plants of Jatropha curcas and Arabidopsis thaliana ecotype Columbia (Col-0) were used in this study. Jatropha plants were cultivated in Xishuangbanna, Yunnan Province, China, as described previously (Pan & Xu, 2011). Arabidopsis plants were grown in an environmentally controlled room at 22 °C under 16-h light/8-h dark photoperiod.
JcTM6 expression analysis
The JcTM6 gene (GenBank accession no. MN820724) was identified in the Jatropha flower cDNA library (Chen et al., 2014). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to examine the expression level of JcTM6 in different organs of Jatropha (roots, stems, young leaves, mature leaves, inflorescence buds, female flowers, male flowers, pericarps and seeds at 42 days after pollination (DAP), male sepals and petals, stamens, female sepals and petals, and pistils) and Arabidopsis (leaves and flowers). Total RNA from each organ was isolated using the silica particle extraction method (Ding et al., 2008). Then, qRT-PCR was performed as previously described in Tao et al. (2015). The JcGAPDH and AtActin were used as an internal control for data normalization. Primers used for qRT-PCR are listed in Table 1. The results of qRT-PCR were obtained from three biological replicates and three technical replicates.
| Name | Sequence (5′ to 3′) | Feature |
|---|---|---|
| GSP1 | CTCTTGGAATAAGTAACCTGTCTGTTGG | JcTM6 gene-specific primer for genome walking |
| GSP2 | CAAAACCCACTACTACAAAACCGAAGA | JcTM6 gene-specific primer for genome walking |
| XT95 | GCTGCTAAGGCTGTTGGGAA | JcGAPDH gene primer for qRT-PCR |
| XT96 | GACATAGCCCAATATTCCCTTCAG | JcGAPDH gene primer for qRT-PCR |
| XK712 | TATCTCTTCGGTTTTGTAGTAGTGGG | JcTM6 gene primer for qRT-PCR |
| XK713 | TCTCTTGGAATAAGTAACCTGTCTGT | JcTM6 gene primer for qRT-PCR |
| XT405 | TGCTCTAGAAATAGCTATAAAATCAATT | For cloning the full-length promoter and construction of JcTM6:GUS |
| XT408 | CGCGGATCCTTTTCCTTTCTTCTTGATA | For cloning the full-length promoter and construction of JcTM6:GUS |
| XD548 | GCTCTAGACGCTTACAGAATTTGCGA | For construction of D:GUS |
| XB994 | CAATCTTTCCACGACCCATTTTTCCTT | JcTM6 gene-specific primer for 5′-RACE |
| XK718 | TGTGCCAATCTACGAGGGTTT | AtActin gene primer for qRT-PCR |
| XK719 | TTTCCCGCTCTGCTGTTGT | AtActin gene primer for qRT-PCR |
| XK984 | TCGCTGAGTTCCACCGCTCTAAG | AtIPT4 gene primer for qRT-PCR |
| XK985 | AGGGTCCCATTTATCCATGTCATTG | AtIPT4 gene primer for qRT-PCR |
| XE815 | CCTTGTCAATGGCAAGAAGAGGCAA | AHK2 gene primer for qRT-PCR (Nishimura et al., 2004) |
| XE816 | CACCTTCTGCAACTCGTCTGTT | AHK2 gene primer for qRT-PCR |
| XE819 | TCAGAGAACATCTTGCCTCGT | ARR5 gene primer for qRT-PCR |
| XE820 | AGCTGCGAGTAGATATCATTAGCTT | ARR5 gene primer for qRT-PCR |
Cloning of the upstream region of JcTM6
The 5′region of JcTM6 was isolated from Jatropha genomic DNA by genome walking (Siebert et al., 1995) according to the Genome Walker™ Kit Universal User Manual (Clontech). Then, the full-length JcTM6 promoter was amplified using the primers, XT405 and XT408. The PCR product was cloned into the pGEM-T Easy vector. Putative cis-acting elements in the JcTM6 promoter were analyzed using the PLACE database (Higo et al., 1999). The transcriptional start site of JcTM6 was identified as previously described in Tao et al. (2016). Primers employed for genome walking and 5′-RACE are listed in Table 1.
Construction of JcTM6 promoter-GUS fusion and Arabidopsis transformation
To generate the JcTM6:GUS plasmid, Xba I and Bam HI were used to digested pBI101 (Jefferson, Kavanagh & Bevan, 1987), and the pGEM -T Easy vector containing the JcTM6 promoter, respectively. The resulting fragments were ligated using the T4 DNA Ligase (Promega) to generate the JcTM6:GUS fusion construct. Then, the JcTM6:GUS plasmid was introduced into Agrobacterium tumefaciens EHA105 by electroporation (GenePulser Xcell; Bio-Rad), and the transformed A. tumefaciens cells were used to transform Arabidopsis plants by the floral dip method (Clough & Bent, 1998).
Histochemical GUS staining assay
To perform GUS staining, various tissues of transgenic Arabidopsis were submerged in the GUS assay buffer (50 mM sodium phosphate [pH 7.0], 0.5 mM K3Fe (CN)6, 0.5 mM K4Fe (CN)6⋅ 3H2O, 0.5% Triton X-100, and 1 mM X-Gluc) and vacuum-infiltrated for 15 min. Then, tissues were incubated overnight at 37 °C, cleared in 70% ethanol (Jefferson, Kavanagh & Bevan, 1987), and examined under a stereomicroscope (Leica M80). The results of GUS staining were obtained from five biological replicates and three technical replicates.
Results
JcTM6 expression in Jatropha
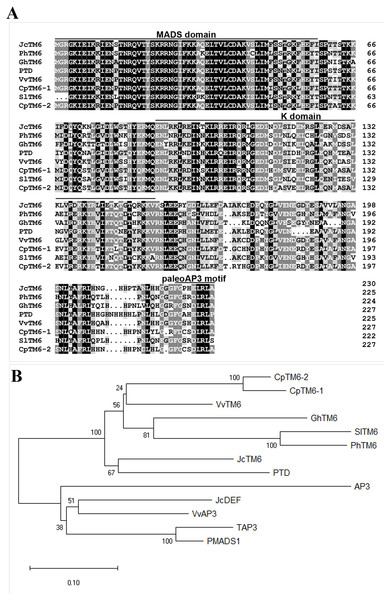
We identified the JcTM6 cDNA (GenBank accession no. MN820724) from our Jatropha flower cDNA library constructed previously (Chen et al., 2014). JcTM6 encodes a 230-amino acid protein, which shows high similarity to TM6 homologs from other plant species (Fig. 1A). Phylogenetic analyses showed that JcTM6, which contains the paleoAP3 motif, belongs to the TM6 group, rather than the euAP3 group (Fig. 1B).
Figure 1: A comparison of JcTM6 and its homologs.
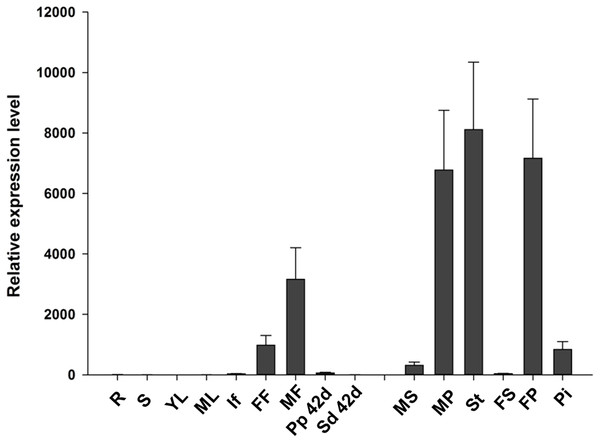
(A) The alignment of the deduced amino acid sequences of JcTM6 with that of Vitis vinifera VvTM6 (accession number DQ979341), Carica papaya CpTM6-1 (accession number ABQ51321), and CpTM6-2 (accession number ABQ51322), Populus trichocarpa PTD (accession number AAC13695) , Gossypium hirsutum GhTM6 (accession number ADX60056), Petunia x hybrida PhTM6 (accession number AF230704) and Solanum lycopersicum SlTM6 (accession number CAA43171) . Identically and partially conserved amino acid sequences are shown in black and gray, respectively. The conserved regions, MADS domain and K domain and paleoAP3 C-terminal motif in JcTM6 are underlined. (B) A phylogenetic analysis of JcTM6 and other homologs. Jatropha curcas JcDEF (accession number XP_012071964), Solanum lycopersicum TAP3 ( accession number ABG73412), Vitis vinifera VvAP3 (accession number NP_001267960), Arabidopsis thaliana AP3 (accession number BAA04665), Petunia hybrida PMADS1 (accession number Q07472). The tree was constructed using MEGA 7.0 software and the neighbor-joining (N–J) method. The N-J unrooted dendrogram was generated from an alignment of the deduced amino acids with the ClustalW program. One thousand replicates were used for the Bootstrap test. The scale bar indicates the average number of substitutions per site.To analyze the expression pattern of JcTM6 in Jatropha, qRT-PCR was performed using total RNA extracted from various tissues including roots, stems, leaves, inflorescences, female and male flowers, and pericarps and seeds at 42 DAP. The JcTM6 gene was predominantly expressed in female and male flowers (Fig. 2), indicating that JcTM6 is a flower-specific gene. Furthermore, JcTM6 showed high expression in the stamens of male flowers and petals of male and female flowers but low expression in sepals and pistils (Fig. 2). Thus, the expression pattern of JcTM6 in floral organs is consistent with that of class B genes (Weigel & Meyerowitz, 1994).
Isolation and sequence analysis of JcTM6 promoter
A 1.8-kb fragment of the JcTM6 promoter (Fig. 3A, –1717 to +103 bp; GenBank accession no. MN044579) was isolated from Jatropha genomic DNA by genome walking (Siebert et al., 1995). The transcription start site of JcTM6 was located 103 nt upstream of the translation start codon (Fig. 3A). Analysis of the JcTM6 promoter using the PLACE database (Higo et al., 1999) revealed various putative cis-elements in the 1.8-kb JcTM6 promoter fragment (Fig. 3A) including two CArG boxes, which act as binding sites for MADS-box transcription factors (Irish & Yamamoto, 1995), some pollen-specific elements, including five GTGANTG10 motifs (GTGA) and eight POLLEN1LELAT52 motifs (AGAAA) (Muschietti et al., 1994; Rogers et al., 2001), and a Q element (TGACCT), which shows enhancer-like activity for the pollen-specific expression of maize (Zea mays L.) ZM13 gene (Hamilton, Schwarz & Mascarenhas, 1998).
Figure 2: Expression pattern of JcTM6 in Jatropha.
Samples from adult plants: roots (R), stems (S), young leaves (YL), mature leaves (ML), inflorescence buds (If), female flowers (FF), male flowers (MF), pericarps at 42 days after pollination (DAP) (Pp 42d), seeds at 42 DAP (Sd 42d), male sepals (MS), male petals (MP), stamens (St), female sepals (FS), female petals (FP), and pistils (Pi). qRT-PCR results were obtained from three biological replicates. The errors denote the SD. The values were normalized to the expression of JcGAPDH (Zhang et al., 2013). The relative expression level of young leaves was set as the standard value of 1.Figure 3: JcTM6 promoter sequence and promoter-reporter gene construct.
(A) The nucleotide sequence of the JcTM6 promoter. The transcription start site (+1) is in red. The start codon ATG is in bold and boxed. Putative regulatory elements on both strands are shown in bold and underlined. (B) A schematic of the T-DNA regions of the JcTM6:GUS binary vector used for transformation.Activity of the JcTM6 promoter in Arabidopsis
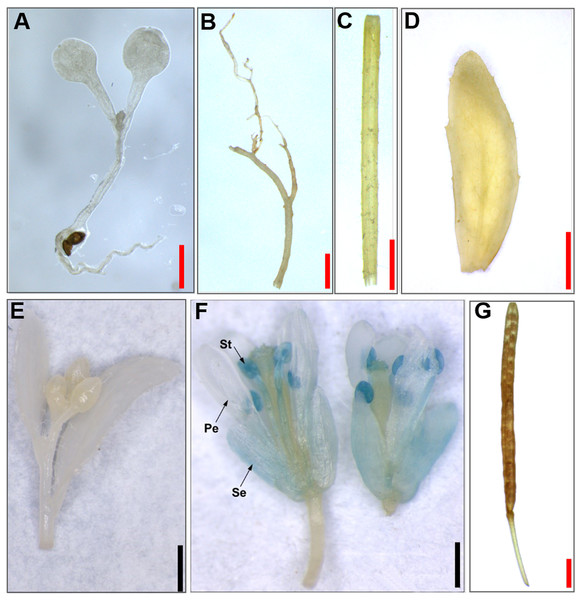
To detect the activity of JcTM6 promoter, a JcTM6 promoter-GUS fusion construct (Fig. 3B) was expressed in Arabidopsis, and GUS staining was monitored in homozygous T3 plants (Fig. 4). No GUS staining was observed in 10-day-old Arabidopsis seedlings (Fig. 4A). Among the five tissues of adult plants examined (including roots, stems, leaves, flowers, and green siliques), GUS staining was detected only in flowers (Figs. 4B–4G). Among all floral organs, GUS staining intensity was the strongest in stamens, followed by sepals and petals, with faint staining in carpels (Fig. S1). Based on the results of GUS staining, we conclude that the JcTM6 promoter functions as a flower-specific promoter in Arabidopsis.
Figure 4: Histochemical GUS staining of transgenic Arabidopsis harboring the JcTM6:GUS fusion.
(A) Ten-day-old seedlings, (B) roots, (C) stems, (D) leaves, (E) inflorescence buds, (F) open flowers, (G) green siliques. Pe, petals; Se, sepals; St, stamens. Red bars = one mm, black bars = two mm.Deletion analysis of the JcTM6 promoter
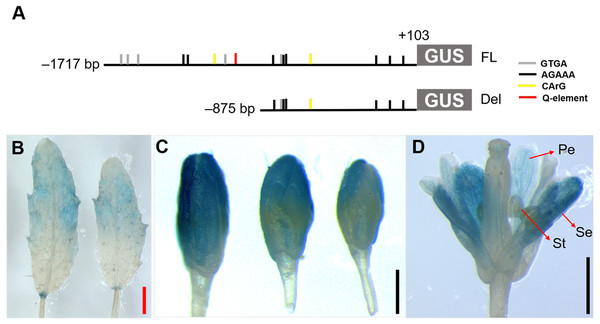
To analyze the region essential for flower-specific activity of the JcTM6 promoter, we carried out a deletion analysis. A deletion variant of the JcTM6 promoter lacking the region from –1,717 to –876 bp was fused to the GUS gene and transformed into Arabidopsis (Fig. 5A). Compared with the full-length JcTM6 promoter, the deletion was not only active in flowers but also in young leaves (Fig. 5B). Moreover, the deletion showed no promoter activity in stamens but increased activity in sepals and petals (Fig. 5C and 5D). These results indicate that the region from −1,717 to −876 bp is critical for JcTM6 promoter activity in stamens and inhibition of promoter activity in young leaves, sepals, and petals.
Figure 5: Histochemical GUS staining of transgenic Arabidopsis harboring the JcTM6 deletion.
(A) Schematic representation of JcTM6 promoter deletion. FL, full length JcTM6 promoter, Del, deletion. GTGA: GTGANTG10 motif (gray vertical bars), AGAAA: POLLEN1LELAT52 motif (black vertical bars), CArG box: CWWWWWWWWG (yellow vertical bars), Q-element: TGACCT (red vertical bar). (B) young leaves, (C) flower buds, (D) flowers. Pe, petals; Se, sepals; St, stamens. Red bar = one mm, black bars = 0.5 mm.JcTM6:AtIPT4 transgenic Arabidopsis produced large flowers
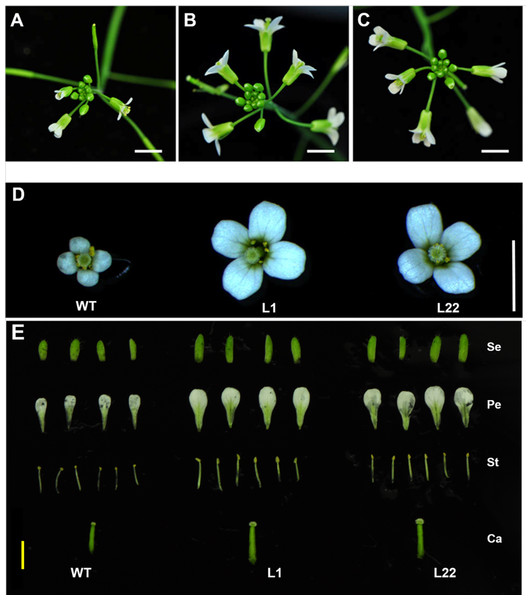
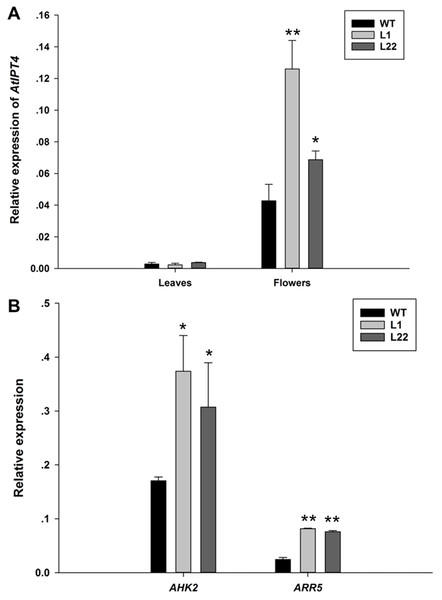
To further verify the floral specificity of JcTM6 promoter, a cytokinin biosynthetic gene (AtIPT4) was expressed under the control of JcTM6 promoter in Arabidopsis. JcTM6:AtIPT4 vector was constructed and was transformed into Arabidopsis plants. A total of 25 independent JcTM6:AtIPT4 lines were obtained. As expected, all transgenic lines showed no vegetative difference from the wild type and most of them produced larger flowers (Fig. 6). Furthermore, the development of siliques was also unaffected. To verify the morphological alteration in flowers that is caused by the transgene, we examined the expression levels of AtIPT4 and the cytokinin signaling genes Arabidopsis histidine kinase 2 (AHK2) (Nishimura et al., 2004) and Arabidopsis response regulator 5 (ARR5) (D’Agostino, Deruère & Kieber, 2000) in wild type and JcTM6:AtIPT4 transgenic plants. The expression level of AtIPT4 in flowers of transgenic lines is significantly higher than that in wild type, whereas the AtIPT4 expression in the leaves of transgenic plants was not different from that in leaves of wile-type plants (Fig. 7A). As expected, higher expression levels of AHK2 and ARR5 were detected in the flowers of transgenic lines (Fig. 7B). These results indicate that the morphological alteration in flowers of JcTM6:AtIPT4 transgenic plants is caused by the flower-specific expression of the transgene driven by the JcTM6 promoter. JcTM6 promoter is indeed a flower-specific promoter.
Figure 6: Flower size is increased in transgenic JcTM6:AtIPT4 Arabidopsis.
Inflorescences of wild-type (A) and transgenic L1 (B) and L22 (C) lines. Flowers of wild-type and transgenic L1 and L22 lines (D). Dissected flowers of WT and transgenic L1 and L22 lines (E). Se, sepals; Pe, petals; St, stamens; Ca, carpels; WT, wild-type. White bars = three mm, yellow bar = two mm.Figure 7: The expression analysis of AtIPT4, AHK2 and ARR5 in JcTM6:AtIPT4 transgenic Arabidopsis.
(A) The expression levels of AtIPT4 in the leaves and flowers of wild type (WT) plants and transgenic lines (L1 and L22). (B) The expression levels of AHK2 and ARR5 in the flowers of wild type (WT) plants and transgenic lines (L1 and L22). The values represent the means ± standard deviation (n = 3). Student’s t-test was used to determine significant differences. ∗p ≤ 0.05, ∗∗p ≤ 0.01.Discussion
TM6 is a member of the MADS-box gene family, which belongs to the paleoAP3 lineage (Pnueli et al., 1991; Rijpkema et al., 2006; Wu et al., 2011). In tomato (Solanum lycopersicum) and petunia, TM6 functions as a class B gene that plays an essential role in stamen development, although it is mainly expressed in whorls 3 and 4, similar to that of a class C gene (Martino et al., 2006; Rijpkema et al., 2006). In trioecious papaya (Carica papaya) plants, which produce male, female, and hermaphrodite flowers, two TM6 genes were isolated previously (CpTM6-1 and CpTM6-2). Both genes are predominantly expressed in the petals of all sex types and stamens of hermaphrodite and male flowers, although CpTM6-2 is also expressed in leaves (Ackerman et al., 2008). In this study, we identified JcTM6 as a flower-specific gene in Jatropha, with high expression in female and male flowers (Fig. 2). Similar to CpTM6-1, the JcTM6 gene showed high expression in the petals of female and male flowers and stamens of male flowers. Because JcTM6 showed flower-specific expression, we isolated its upstream region from Jatropha genomic DNA and analyzed its activity in Arabidopsis by GUS staining.
In transgenic Arabidopsis, GUS staining showed that the JcTM6 promoter was active only in flowers (Fig. 4), suggesting that the JcTM6 promoter is a flower-specific promoter. AtIPT4 is a cytokinin biosynthesis gene encoding ATP/ADP isopentenyltransferase. The expression of this gene under the control of AP1 promoter results in the alterations in flower number and organs (Li et al., 2010). However, the AtIPT4 driven by JcTM6 promoter only gave rise to the changes in flower organs (Fig. 6), indicating that JcTM6 promoter is active at the late stage of flower development rather than floral meristem. This activity is consistent with the expression pattern of the JcTM6 gene in Jatropha. Recently, Ming et al. (2020) showed that JcTM6 promoter has a high activity in female flowers of Jatropha, suggesting that JcTM6 promoter can drive flower-specific expression of transgenes in different plant species.
When the 842-bp fragment of the JcTM6 promoter (−1,717 to −876 bp) was deleted, the promoter was not only active in flowers but also in young leaves (Fig. 5B). We found that the deleted region contained one of the two CArG box motifs, which are very important for mediating the regulatory effect of MADS-box transcription factors (Dolan & Fields, 1991; Treisman, 1992). In Jatropha, a fragment of the JcAP1 promoter (from −1,313 to −1,057 bp), which contains a CArG box motif, is required for promoter activity in inflorescence buds (Tao et al., 2016). The Arabidopsis AP3 promoter contains three CArG boxes: CArG1 is essential for AP3 promoter activity at all stages of flowering; CArG2 is critical for AP3 expression in petals, and CArG3 represents the binding site of a transcription factor that represses the activity of AP3 promoter during early floral stages (Tilly, Allen & Jack, 1998). Therefore, we propose that the CArG box motif in JcTM6 promoter plays an important role in conferring floral-specific activity in transgenic plants.
Among the floral organs, stamens exhibited the highest activity of JcTM6 promoter (Fig. 4F). This expression pattern could be regulated by pollen-specific elements contained in this promoter, including five GTGA and eight AGAAA motifs. The GTGA motif is critical for the expression of g10 promoter in tobacco pollen because mutation of the GTGA motif reduced g10 promoter activity in pollen (Rogers et al., 2001). The AGAAA motif, which was identified in the tomato late-stage pollen-specific LAT52 promoter, is necessary for promoter activity during pollen maturation (Bate & Twell, 1998). In potato (Solanum tuberosum L.), the GTGA and AGAAA motifs present in the promoter of SBgLR, a pollen-specific gene, are critical for high-level gene expression in pollen (Lang et al., 2008). In the current study, deletion of an 842-bp fragment of the JcTM6 promoter, containing four GTGA and two AGAAA motifs, abolished promoter activity in stamens (Fig. 5D). We assumed that these motifs are essential for the activity of the JcTM6 promoter in stamens. Given the importance of CArG box motifs, it is possible that the GTGA and AGAAA motifs cooperate with the CArG box to regulate JcTM6 promoter activity in stamens. In addition, although the deleted region contained six AGAAA motifs, these motifs do not seem to be required for JcTM6 promoter activity in stamens. Furthermore, the deleted region also contained a 6-bp quantitative element (Q-element), which plays an enhancer-like role (Hamilton, Schwarz & Mascarenhas, 1998). In maize, deletion of the Q-element from the pollen-specific ZM13 promoter reduced the promoter activity by 10-fold (Hamilton et al., 2000). Deletion of the Q-element probably also contributed to the loss of JcTM6 promoter activity in stamens in this study (Fig. 5D). In addition, the deletion variant of the JcTM6 promoter exhibited increased activity in sepals and petals (Fig. 5C and 5D), indicating the presence of potential negative elements in the deleted region, which inhibit promoter activity in sepals and petals. By the deletion analysis of the JcTM6 promoter, we demonstrate the combination of these elements are of great importance to the promoter activity in the flowers, and detailed studies of the functions of these elements will be conducted in the future.
Conclusions
Floral-specific promoters play crucial roles in genetic modification of flowering characteristics. In this study, a 1.8-kb JcTM6 promoter fragment was isolated from Jatropha and characterized as a flower-specific promoter in transgenic Arabidopsis plants. When the region from –1,717 to –876 bp in the JcTM6 promoter was deleted, the promoter lost its flower-specific activity and gained activity in young leaves. Our results suggest that the JcTM6 promoter could be used to drive flower-specific expression of transgenes in plants.
Supplemental Information
Histochemical GUS staining in different floral organs of transgenic Arabidopsis harboring the JcTM6:GUS fusion
<!–[if !supportLists]–¿(A) <!–[endif]–>sepals, (B) petals, (C) stamens, (D) carpels. Bars = 1 mm.