L-cysteine transporter-PCR to detect hydrogen sulfide-producing Campylobacter fetus
- Published
- Accepted
- Received
- Academic Editor
- Vasco Azevedo
- Subject Areas
- Agricultural Science, Bioinformatics, Molecular Biology
- Keywords
- PCR, Sulfide production, Molecular differentiation, Bovine genital campylobacteriosis
- Copyright
- © 2019 Farace et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. L-cysteine transporter-PCR to detect hydrogen sulfide-producing Campylobacter fetus. PeerJ 7:e7820 https://doi.org/10.7717/peerj.7820
Abstract
Phenotypic differences between Campylobacter fetus fetus and C. fetus venerealis subspecies allow the differential diagnosis of bovine genital campylobacteriosis. The hydrogen sulfide production, for example, is a trait exclusive to C. fetus fetus and C. fetus venerealis biovar intermedius. This gas that can be biochemically tested can be produced from L-cysteine (L-Cys). Herein, we report a novel multiplex-PCR to differentiate C. fetus based on the evaluation of a deletion of an ATP-binding cassette-type L-Cys transporter that could be involved in hydrogen sulfide production, as previously described. A wet lab approach combined with an in silico whole genome data analysis showed complete agreement between this L-Cys transporter-PCR and the hydrogen sulfide production biochemical test. This multiplex-PCR may complement the tests currently employed for the differential diagnosis of C. fetus.
Introduction
Campylobacter fetus is best known as a major veterinary pathogen that has a detrimental effect on reproductive efficiency of herds. However, in humans, this bacterium can also cause intestinal illness and, occasionally, severe systemic infections and thus the products from cattle and sheep are suspected as sources of transmission (Wagenaar et al., 2014). The classification of C. fetus subspecies relies on clinical features, host specificity, and phenotypic traits. Despite technical limitations and variable success, hydrogen sulfide (H2S) production as well as tolerance to glycine and NaCl, selenite reduction and resistance to antibiotics are the available biochemical tests currently employed as differential diagnosis of C. fetus (OIE, 2018; Schulze et al., 2006).
Members of C. fetus have different tropism, as evidenced in veterinary practice and in the diagnosis. The subspecies C. fetus venerealis (Cfv) is restricted to the bovine reproductive tract, and is associated to the venereal disease bovine genital campylobacteriosis (BGC), whereas C. fetus fetus (Cff) is mainly intestinal and is usually related to sporadic abortion. To date, the bovine products are subjected to strict regulations by the World Organization for Animal Health (OIE) and must be tested for the presence of C. fetus subsp. venerealis before international trading (OIE, 2018). Therefore, its differentiation at the subspecies level is critical. The isolation of the bacteria can confirm BGC and subsequently biochemical tests can determine the particular different isolates. Among the biochemical tests, glycine resistance and hydrogen sulfide(H2S) production are two of the best biochemical performing tests. For example, Cff strains show 1% glycine resistance and produce H2S in L-cysteine (L-Cys) enriched media. By contrast, Cfv strains fail to grow in 1% glycine-containing media and to produce H2S (Véron & Chatelain, 1973). Hence, these traits allow their discrimination. A glycine-tolerant variant of Cfv (C. fetus venerealis biovar intermedius, Cfvi) are frequently isolated in some countries such as USA, UK, South Africa and Argentina, which complicates their accurate identification (Schmidt, Venter & Picard, 2010; Van Bergen et al., 2005; Iraola et al., 2013). A third-host associated subspecies, C. fetus subsp. testudinum, completes the list of subspecies of C. fetus. This subspecies has been isolated from reptiles and humans (Fitzgerald et al., 2014) and therefore would not be relevant for animal production.
In a previous wide genome association study, Van der Graaf-van Bloois et al. (2016a) described a recent diversification of mammalian C. fetus and implicated a genetic factor associated to H2S production. They described a deletion in an ATP-binding cassette-type L-Cys transporter in Cfv strains. The operon structure of this L-Cys transporter has five coding sequences and three of them code for different molecular components of the transporter: the ATP-binding protein, the permease, and the substrate-binding protein (locus tags CFF8240_RS03845, CFF8240_RS03850 and CFF8240_RS03855 in C. fetus 82-40 genome, respectively). This L-Cys importer could be part of the Class 3 ABC-transporters (Licht & Schneider, 2011) and in Cfv the permease and the extracellular binding domain coding genes are deleted. This deletion may impair the transporter assembly, affecting the up-take of L-Cys. This therefore could explain the impaired production of H2S from this amino acid in Cfv strains. On these bases, we aimed to develop a simple molecular technique for detecting the L-Cys transporter-deletion polymorphism with the main purpose of identifying H2S-producing C. fetus strains.
Materials and Methods
Campylobacter fetus isolates and bacterial culture
All the C. fetus isolates (n = 36) were obtained from bovine clinical samples at the Bacteriology Unit (EEA-INTA Balcarce, Argentina). Thirty of these clinical isolates were randomly-selected for this study. In addition, the strains Cfv 97/608, Cfv 98/25 and Cfvi 99/541 were also selected because of the availability of their whole genome sequences (Van der Graaf-van Bloois et al., 2014; Iraola et al., 2013) and three additional isolates were selected to perform whole genome sequencing (see below).
All the C. fetus isolates were grown on 7% blood-Skirrow selective agar plates (Oxoid, Hampshire, UK) with 1.25 IU/ml polymyxin B sulfate, 5 μg/ml trimethoprim, 10 μg/ml vancomycin and 50 μg/ml cycloheximide (Sigma-Aldrich, St. Louis, MO, USA). The plates were incubated under microaerophilic conditions (5% O2, 10% CO2 and 85% N2) for 72 h at 37 °C. C. hyointestinalis NCTC11562 and the field isolate C. sputorum 08/209 were grown under the same conditions. C. coli NCTC11353 and C. jejuni NCTC11392 were cultured on Blood-Columbia agar plates (Oxoid) under microaerophilic condition for 24 h at 42 °C.
Biochemical tests
The classification of the subspecies was performed following standard protocols (OIE, 2018): sodium selenite reduction, 3.5% sodium chloride resistance, 1% glycine tolerance and H2S production in 0.02% L-Cys enriched medium. We also tested 1.3%, 1.5% and 1.9% glycine tolerance. The isolates were identified as Cff if they reduced sodium selenite, produced H2S and showed sodium chloride tolerance and at least 1% glycine resistance.
DNA isolation
A rapid protocol (freeze-thaw cycles) was applied to obtain the DNA template as follows. A loopful of each culture was collected and resuspended in 250 µl of sterile deionized water. Two cycles of freeze and boiling (−80/95 °C) were performed and the cellular debris were discarded after a centrifugation step. Two µl of the supernatant was used as PCR-template. High quality genomic DNA was obtained using mini spin columns (NucleoSpin Tissue; Macherey-Nagel GmbH & Co., Duren, Germany). DNA quality was tested using the Qubit 4 fluorometer (Invitrogen, Carlsbad, CA, USA; Thermo Scientific, Waltham, MA, USA) and further used for sequencing purpose.
L-Cys transporter-PCR
One forward and two reverse primers (Fwd 5′-gtccatttacttatcacgataacagtgg-3′, Rev1 5′-gatattaggctaagaggaatggtgtattg-3′ and Rev2 5′-ctcccgtatctacatgaaagctaatatc-3′) were designed for a multiplex-PCR format using open source Unipro UGENE 1.31 (Okonechnikov et al., 2012) (Fig. 1A). The amplification mix consisted of 1 × GoTaq green Reaction buffer (1.5 mM MgCl2), 0.25 mM of each dNTP, 0.1 μM of each primer, and 1.25 U Taq polymerase (Promega Corp., Madison, WI, USA), nuclease-free water to reach a final volume of 25 µl and Campylobacter DNA template.
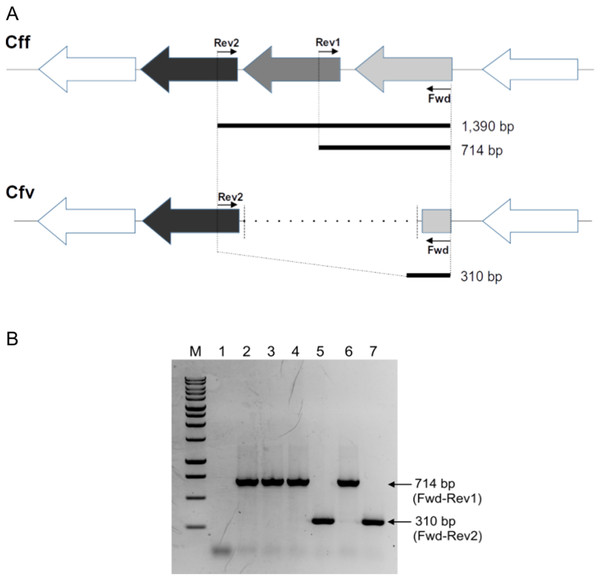
Figure 1: Differential L-Cys Transporter-PCR.
(A) Schematic representation of the organization of the genes encoding the L-Cys transporter in C. fetus. Primer targeting regions and expected PCR-products are shown. The gray arrow represents the permease protein YckJ coding gene (locus tag CFF8240_RS03850 in Cff 82-40 genome) which is deleted in Cfv strains. The light gray arrow represents the extracellular-binding protein YckK coding gene (locus tag CFF8240_RS03855), which is partially deleted in Cfv. ATP-binding protein coding gene (locus tag CFF8240_RS03845) which is another component of the transporter is conserved in both subspecies and is represented by black arrow. (B) Representative agarose gel electrophoresis. Lane 1, negative control (water); lane 2, Cff 08/421; lane 3, Cff 96/136; lane 4, Cfvi 06/341; lane 5, Cfv 97/608; lane 6, Cfvi 03/596 and lane 7, Cfv 95/258. Under the set conditions, the product of 1,390 bp is absent. M: molecular weight marker, 1 kb DNA ladder (Promega).The touch-down amplification program consisted of an initial step at 94 °C for 3 min, 10 cycles at 94 °C for 1 min, followed by annealing temperatures starting at 55 °C for 1 min and decreasing 1 °C per cycle from 55 to 45 °C. Then, an extension step was performed at 72 °C for 1 min, followed by 30 cycles with an annealing at 51 °C, and a final termination step at 72 °C for 8 min.
Under these conditions, the absence of the expected product of 1,390 bp makes the interpretation of the PCR results easy. A product of 714-bp is indicative of Cff and Cfvi strains (which have a complete version of the operon and are H2S-producing strains), whereas a 310 bp product refers to Cfv strains (which contain a partly deleted operon and are non- H2S-producing strains). All the products were resolved in 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining. The PCR-products were submitted to the UGB unit-INTA to confirm their identity through Sanger sequencing.
In silico-PCR: whole genome sequencing and genomic data analysis
We selected three isolates from bovine abortions (Cff 13/344, Cff 08/421 and Cfvi 06/341) of the most productive agricultural areas of Argentina. Paired-end Nextera XT libraries were constructed and sequenced in a MiSeq sequencer (2 × 250 pb, Illumina). A quality trimming step was applied to raw reads using Trimmomatic (Bolger, Lohse & Usadel, 2014). De novo assembly was done using SPAdes v3.11.1 (Bankevich et al., 2012). Contigs were oriented using Mauve (Darling et al., 2004; Rissman et al., 2009) and the genome of C. fetus venerealis 97-608 as a reference (NZ_CP008810.1). The genomes were annotated using PROKKA (Seemann, 2014) and RASTtk (Brettin et al., 2015). The assembly summary statistics is shown in Table S1.
In total, whole-genome sequence data of 214 C. fetus strains (Cff, n = 152; Cfv, n = 42; Cfvi, n = 19 and one strain not identified at the subspecies level, Cf = 1) from 19 countries and different hosts (bovine, n = 117; human, n = 78; ovine, n = 15; monkey, n = 1 and unknown, n = 3) were screened to search for the target sequences of the primers designed for the L-Cys transporter-PCR protocol. These data included the three genomes obtained in this study (Cff 13/344, Cff 08/421 and Cfvi 06/341) and 37 publicly available genomes from GenBank. Additionally, reads from C. fetus strains (n = 174) deposited in ENA database (https://www.ebi.ac.uk/ena/) were also assembled, as mentioned above, and subsequently analyzed as follows. The Primer map software (http://www.bioinformatics.org/sms2/primer_map.html) was used for global searching of Fwd, Rev1 and Rev2 primer sequences. Primer Map output is a textual map showing the annealing positions of PCR primers. Afterwards, several conditions were evaluated, including annealing of both primers of each pair and their orientation. The position of each target annealing site was employed to estimate the amplicon size. The program, by default, does not allow mismatches. Cases where the annealing was confirmed for a single primer were classified as not detected or unknown.
Statistics
The agreement between the H2S production biochemical test and the L-Cys transporter-PCR was tested with Cohen´s Kappa statistic.
Results
L-Cys transporter-PCR: wet-lab assay
The multiplexed PCR-based approach herein designed produced a differential band pattern between the C. fetus isolates with distinct H2S-biochemical test results (Fig. 1B). This protocol was named L-Cys transporter-PCR. We tested 36 biochemically typed isolates with this L-Cys transporter-PCR, followed by electrophoresis of the products in agarose gel to reveal the size of the amplicons. A single amplification product was obtained in all the tested strains. The retrieved band from Cff and Cfv biovar intermedius (Cfvi) strains was of 714 bp. This result coincided with a complete version of the L-Cys transporter operon and this pattern was named “CFF/CFVI.” Amplifications from Cfv strains generated a smaller product of 310 bp, equivalent to a partially deleted operon, and this profile was named “CFV” (Fig. 1B). This L-Cys transporter-PCR allowed a differential testing that avoided a negative result in presence of C. fetus DNA. Indeed, a negative result, sometimes could be indicative of both the absence of the specific target and the presence of inhibitors in the sample. As expected, no product was obtained from DNA of Campylobacter spp. other than C. fetus (C. hyointestinalis, C. coli, C. jejuni and C. sputorum) (Table 1). This result confirmed the specificity of this L-Cys transporter-PCR test for C. fetus.
| Strain | Origin | Biochemical test | Phenotypic ID |
L-cys transporter-PCR pattern | |
|---|---|---|---|---|---|
| 1% Glycine resistance | H2S production | ||||
| Cff 96-136 | Bahía Blanca, BA | + | + | Cff | CFF/CFVI |
| Cff 08-421 | Gral. López, SF | + | + | Cff | CFF/CFVI |
| Cff 14-284 | Pila, BA | + | + | Cff | CFF/CFVI |
| Cff 04-240 | Olavarría, BA | + | + | Cff | CFF/CFVI |
| Cff 13-344 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff 11-572 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff 89-222 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff 90-189 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff CI N3 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff 01-165 | Santa Rosa, LP | + | + | Cff | CFF/CFVI |
| Cff 01-64 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff 05-622 | Cnel. Dorrego, BA | + | + | Cff | CFF/CFVI |
| Cff 11-262 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff 11-295 | Saladillo, BA | + | + | Cff | CFF/CFVI |
| Cff 11-360 | Necochea, BA | + | + | Cff | CFF/CFVI |
| Cff 11-685 | Balcarce, BA | + | + | Cff | CFF/CFVI |
| Cff 11-408 | Necochea, BA | + | + | Cff | CFF/CFVI |
| Cff btu5 | BA | + | + | Cff | CFF/CFVI |
| Cff btu6 | BA | + | + | Cff | CFF/CFVI |
| Cff btu7 | BA | + | + | Cff | CFF/CFVI |
| Cff 18-09 | BA | + | + | Cff | CFF/CFVI |
| Cff 18-100 | BA | + | + | Cff | CFF/CFVI |
| Cfv 97-608 | Hucal, LP | − | − | Cfv | CFV |
| Cfv 95-258 | Mar Chiquita, BA | − | − | Cfv | CFV |
| Cfv 08-382 | Gral. Belgrano, BA | − | − | Cfv | CFV |
| Cfv 05-355 | Balcarce, BA | − | − | Cfv | CFV |
| Cfv 98-25 | Gral. Pueyrredón, BA | − | − | Cfv | CFV |
| Cfv 19-01 | BA | − | − | Cfv | CFV |
| Cfvi 06-341 | Pehuajó BA | − | + | Cfvi | CFF/CFVI |
| Cfvi 03-596 | Pehuajó, BA | − | + | Cfvi | CFF/CFVI |
| Cfvi 02-146 | BA | − | + | Cfvi | CFF/CFVI |
| Cfvi 98-472 | Azul, BA | − | + | Cfvi | CFF/CFVI |
| Cfvi 99-541 | Balcarce, BA | − | + | Cfvi | CFF/CFVI |
| Cfvi 07-379 | Mar Chiquita, BA | − | + | Cfvi | CFF/CFVI |
| Cfvi 00-305 | BA | − | + | Cfvi | CFF/CFVI |
| Cfvi 03-596 | Pehuajó, BA | − | + | Cfvi | CFF/CFVI |
| C. sputorum 08-209 | Balcarce, BA | ND | ND | ND | – |
| C. coli NCTC11353 | National Collection of Type Cultures, England | ND | ND | ND | – |
| C. hyointestinalis NCTC11562 | National Collection of Type Cultures, England | ND | ND | ND | – |
| C. jejuni NCTC11392 | National Collection of Type Cultures, England | ND | ND | ND | – |
Notes:
“CFF/CFVI pattern” means that all the components of the L-Cys transporter are present and therefore, a product of 714 bp is obtained. “CFV pattern” means that the L-Cys transporter is deleted and a product of 310 bp is obtained. “−” means that the amplification product was absent.
BA, Buenos Aires province; LP, La Pampa province; SF, Santa Fe province; ND, Not determined.
The results from the L-Cys transporter-PCR analysis displayed a perfect correlation with the H2S production test (κ = 1). The analysis of concordance between tests is shown in Table S2.
We also addressed an in silico analysis of genomic sequences from mammalian C. fetus to further support this conclusion.
L-Cys transporter-PCR : in silico screening
To study the performance of the L-Cys transporter-PCR in a more diverse panel of strains, we applied an in-silico PCR-strategy by performing searches of the primer targeting sequences in whole genomes of 214 C. fetus strains (three of which were obtained in this study by Next-Generation Sequencing technology). For this purpose, we employed the online Primer map application. The same products as the obtained by the wet lab-PCR were considered among all the predicted PCR products and the same patterns were determined according to the product size. This approach confirmed the primer annealing sites, and consequently, also allowed us to define the type of L-Cys transporter operon in 213 out of 214 C. fetus strains (Table 2). The target annealing sites were highly conserved because of the lack of nucleotide mismatches in these strains. The in silico-PCR was able to predict the annealing sites for Fwd-Rev2 primers in the genome of Cfv Azul-94 but the target sites were located into different contigs. The product size was difficult to estimate and consequently this strain had inconclusive results (Table 2).
| ID | Strain | Host | Source | Country | Accession number | H2S production (Reference) | PCR L-Cys transporter pattern |
|
|---|---|---|---|---|---|---|---|---|
| Cff | 04/554 | Bovine | Foetus | AR | CP008808–CP008809 | + | (Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cff | 08/421 | Bovine | Foetus | AR | SOOT00000000 | + | (This study) | CFF/CFVI |
| Cff | 110800-21-2 | Bovine | Bull | NL | LSZN00000000 | + | (Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cff | 13/344 | Bovine | Foetus | AR | SOYX00000000 | + | (This study) | CFF/CFVI |
| Cff | 82/40 | Human | Blood | US | CP000487 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cff | Cff 98/v445 | Bovine | Bull | UK | LMBH00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cff | ATCC 27374 | Ovine | Foetus (brain) | Unk. | MKEI00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cff | BT 10/98 | Ovine | Unknown | UK | LRAL00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cff | NCTC10842 | Unknown | Unknown | Unk. | LS483431 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cff | B0042 | Bovine | Feces | UK | ERR419595 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0047 | Bovine | Feces | UK | ERR419600 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0066 | Bovine | Feces | UK | ERR419653 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0097 | Bovine | Feces | UK | ERR419653 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0129 | Bovine | Feces | UK | ERR419637 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0130 | Bovine | Feces | UK | ERR419638 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0131 | Bovine | Feces | UK | ERR419639 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0151 | Bovine | Feces | UK | ERR419648 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0152 | Bovine | Feces | UK | ERR419649 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0167 | Bovine | Feces | UK | ERR460866 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | B0168 | Bovine | Feces | UK | ERR460867 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | S0693A | Bovine | Feces | UK | ERR419284 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cff | S0478D | Bovine | Feces | UK | ERR419653 | + | (Van der Graaf-van Bloois et al., 2016a) | CFF/CFVI |
| Cfvi | 01/165 | Bovine | Mucus | AR | CP014568–CP014570 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cfv | 84/112 | Bovine | Genital secretion | US | HG004426–HG004427 | − | (Van Bergen et al., 2005) | CFV |
| Cfv | 97/608 | Bovine | Placenta | AR | CP008810–CP008812 | − | (Van Bergen et al., 2005) | CFV |
| Cfv | ADRI 1362 | Bovine | Unknown | AR | LREX00000000 | + | (Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cfv | ADRI513 | Unknown | Unknown | AU | LRFA00000000 | + | (Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cfv | B10 | Bovine | Unknown | US | LRET00000000 | − | (Van der Graaf-van Bloois et al., 2014) | CFV |
| Cfv | CCUG 33872 | Bovine | Abortion | CZ | LREU00000000 | −/+ | (Willoughby et al., 2005; Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cfv | CCUG 33900 | Bovine | Abortion | FR | LREV00000000 | − | (Van der Graaf-van Bloois et al., 2014) | CFV |
| Cfv | LMG 6570 | Bovine | Unknown | BE | LREW00000000 | − | (Van Bergen et al., 2005) | CFV |
| Cfv | NCTC 10354 | Bovine | Mucus | UK | CM001228 | − | (Van Bergen et al., 2005) | CFV |
| Cfv | WBT 011/09 | Unknown | Unknown | UK | LMBI00000000 | + | (Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cfv | zaf3 | Bovine | Foetus | SA | LREZ00000000 | + | (Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cfv | zaf65 | Bovine | Unknown | SA | LREY00000000 | + | (Van der Graaf-van Bloois et al., 2014) | CFF/CFVI |
| Cfvi | 02/298 | Bovine | Foetus | AR | LRVK00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cfvi | 03/293 | Bovine | Foetus | AR | CP0006999–CP007002 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cfvi | 03/596 | Bovine | Foetus | AR | LRAM00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cfvi | 06/341 | Bovine | Foetus | AR | SOYW00000000 | + | (This study) | CFF/CFVI |
| Cfvi | 92/203 | Bovine | Placenta | AR | LRVL00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cfvi | 97/532 | Bovine | Mucus | AR | LRER00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cfvi/Cfv | 98/25 | Bovine | Foetus | AR | LRES00000000 | +/−/− | (Van Bergen et al., 2005; Van der Graaf-van Bloois et al., 2016a; This study) | CFV |
| Cfvi | 99/541 | Bovine | Prepuce | AR | ASTK00000000 | + | (Van Bergen et al., 2005) | CFF/CFVI |
| Cff | H1-UY | Human | Blood | UY | JYCP00000000 | n.a | CFF/CFVI | |
| Cff | HC1 | Human | Blood | UY | QJTR00000000 | n.a | CFF/CFVI | |
| Cff | HC2 | Human | Cerebrospinal fluid | UY | QJTS00000000 | n.a | CFF/CFVI | |
| Cff | CIT01 | Human | Peripheral blood culture | IR | RBHV00000000 | n.a | CFF/CFVI | |
| Cfv | 642-21 | Bovine | Uterus | AU | AJSG00000000 | n.a | CFF/CFVI | |
| Cfv | 66Y | Bovine | Prepuce | CA | JPQC00000000 | n.a | CFV | |
| Cfv | Azul-94 | Bovine | Abortion | AR | ACLG00000000 | n.a | Unknown | |
| Cfv | B6 | Bovine | Vagina | AU | AJMC00000000 | n.a | CFV | |
| Cfv | TD | Bovine | Prepuce | CA | JPPC00000000 | n.a | CFV | |
| Cf | MMM01 | Human | Sepsis | IN | JRKX00000000 | n.a | CFF/CFVI | |
| Cff | 99/801 | Bovine | Prepuce | AR | ERS739235 | n.a | CFF/CFVI | |
| Cff | 00/398 | Bovine | Foetus | AR | ERS739236 | n.a | CFF/CFVI | |
| Cff | 00/564 | Bovine | Prepuce | AR | ERS739237 | n.a | CFF/CFVI | |
| Cff | 01/320 | Bovine | Foetus | AR | ERS739238 | n.a | CFF/CFVI | |
| Cff | 01/210 | Bovine | Vaginal mucus | AR | ERS739239 | n.a | CFF/CFVI | |
| Cff | 04/875 | Bovine | Foetus | AR | ERS739242 | n.a | CFF/CFVI | |
| Cff | 05/394 | Bovine | Foetus | AR | ERS739243 | n.a | CFF/CFVI | |
| Cff | 05/434 | Bovine | Vaginal mucus | AR | ERS739244 | n.a | CFF/CFVI | |
| Cff | 06/340 | Bovine | Prepuce | AR | ERS739245 | n.a | CFF/CFVI | |
| Cff | 07/485 | Bovine | Vaginal mucus | AR | ERS739248 | n.a | CFF/CFVI | |
| Cff | 08/362 | Bovine | Foetus | AR | ERS739249 | n.a | CFF/CFVI | |
| Cff | 10/247 | Bovine | Prepuce | AR | ERS739250 | n.a | CFF/CFVI | |
| Cff | 10/445 | Bovine | Prepuce | AR | ERS739251 | n.a | CFF/CFVI | |
| Cff | 11/360 | Bovine | Foetus | AR | ERS739252 | n.a | CFF/CFVI | |
| Cff | 11/427 | Bovine | Vaginal mucus | AR | ERS739253 | n.a | CFF/CFVI | |
| Cff | 14/270 | Bovine | Foetus | AR | ERS739254 | n.a | CFF/CFVI | |
| Cff | 15/301 | Bovine | Vaginal mucus | AR | ERS739255 | n.a | CFF/CFVI | |
| Cfvi | 02/146 | Bovine | Foetus | AR | ERS739240 | n.a | CFF/CFVI | |
| Cfvi | 06/195 | Bovine | Foetus | AR | ERS739246 | n.a | CFF/CFVI | |
| Cfvi | 07/379 | Bovine | Foetus | AR | ERS739247 | n.a | CFF/CFVI | |
| Cff | 2006/367h | Human | Cerebrospinal fluid | FR | ERS672242 | n.a | CFF/CFVI | |
| Cff | 2006/479h | Human | Feces | FR | ERS672243 | n.a | CFF/CFVI | |
| Cff | 2006/588h | Human | Cerebrospinal fluid | FR | ERS672244 | n.a | CFF/CFVI | |
| Cff | 2006/621h | Human | Blood | FR | ERS672245 | n.a | CFF/CFVI | |
| Cff | 2006/649h | Human | Feces | FR | ERS672246 | n.a | CFF/CFVI | |
| Cff | 2008/170h | Human | Feces | FR | ERS672247 | n.a | CFF/CFVI | |
| Cff | 2008/568h | Human | Joint fluid | FR | ERS672248 | n.a | CFF/CFVI | |
| Cff | 2008/604h | Human | Feces | FR | ERS672249 | n.a | CFF/CFVI | |
| Cff | 2008/691h | Human | Cerebrospinal fluid | FR | ERS672250 | n.a | CFF/CFVI | |
| Cff | 2008/755h | Human | Blood | FR | ERS672251 | n.a | CFF/CFVI | |
| Cff | 2008/898h | Human | Blood | FR | ERS672252 | n.a | CFF/CFVI | |
| Cff | 2010/41h | Human | Feces | FR | ERS672253 | n.a | CFF/CFVI | |
| Cff | 2010/524h | Human | Kidney | FR | ERS672254 | n.a | CFF/CFVI | |
| Cff | 2010/1094h | Human | Blood | FR | ERS672255 | n.a | CFF/CFVI | |
| Cff | 2010/1119h | Human | Feces | FR | ERS672256 | n.a | CFF/CFVI | |
| Cff | 2010/1180h | Human | Blood | FR | ERS672257 | n.a | CFF/CFVI | |
| Cff | 2012/60h | Human | Feces | FR | ERS672258 | n.a | CFF/CFVI | |
| Cff | 2012/185h | Human | Blood | FR | ERS672259 | n.a | CFF/CFVI | |
| Cff | 2012/286h | Human | Blood | FR | ERS672260 | n.a | CFF/CFVI | |
| Cff | 2012/331h | Human | Blood | FR | ERS672261 | n.a | CFF/CFVI | |
| Cff | 2012/879h | Human | Feces | FR | ERS672263 | n.a | CFF/CFVI | |
| Cff | 2012/1045h | Human | Joint fluid | FR | ERS672264 | n.a | CFF/CFVI | |
| Cff | 2014/52h | Human | Cerebrospinal fluid | FR | ERS672265 | n.a | CFF/CFVI | |
| Cff | 2014/602h | Human | Blood | FR | ERS672266 | n.a | CFF/CFVI | |
| Cff | 2014/790h | Human | Blood | FR | ERS672267 | n.a | CFF/CFVI | |
| Cff | 2014/947h | Human | Blood | FR | ERS672269 | n.a | CFF/CFVI | |
| Cff | 2014/1097h | Human | Feces | FR | ERS672270 | n.a | CFF/CFVI | |
| Cff | 2007/123h | Human | Cerebrospinal fluid | FR | ERS672271 | n.a | CFF/CFVI | |
| Cff | 2009/56h | Human | Cerebrospinal fluid | FR | ERS672272 | n.a | CFF/CFVI | |
| Cff | CF156 | Human | Blood | TR | ERS672273 | n.a | CFF/CFVI | |
| Cfvi | 21-C0091-10-14_2 | Bovine | Prepuce | UK | ERS672276 | n.a | CFF/CFVI | |
| Cff | GTC _08732 | Human | Cerebrospinal fluid | JP | ERS672218 | n.a | CFF/CFVI | |
| Cff | GTC _11236 | Human | Feces | JP | ERS672220 | n.a | CFF/CFVI | |
| Cff | 96-48 | Human | Feces | JP | ERS672224 | n.a | CFF/CFVI | |
| Cff | 01-187 | Human | Blood | JP | ERS672226 | n.a | CFF/CFVI | |
| Cff | 2004/103h | Human | Cerebrospinal fluid | FR | ERS672233 | n.a | CFF/CFVI | |
| Cff | 2004/199h | Human | Cerebrospinal fluid | FR | ERS672234 | n.a | CFF/CFVI | |
| Cff | 2004/359h | Human | Blood | FR | ERS672235 | n.a | CFF/CFVI | |
| Cff | 2004/362h | Human | Placenta | FR | ERS672236 | n.a | CFF/CFVI | |
| Cff | 2004/526h | Human | Feces | FR | ERS672237 | n.a | CFF/CFVI | |
| Cff | 2004/598h | Human | Blood | FR | ERS672238 | n.a | CFF/CFVI | |
| Cff | 2004/605h | Human | Feces | FR | ERS672239 | n.a | CFF/CFVI | |
| Cff | 2004/637h | Human | Joint fluid | FR | ERS672240 | n.a | CFF/CFVI | |
| Cff | 2006/222h | Human | Blood | FR | ERS672241 | n.a | CFF/CFVI | |
| Cff | ID111063 | Human | Blood | CA | ERS739225 | n.a | CFF/CFVI | |
| Cff | ID117228 | Human | Blood | CA | ERS739226 | n.a | CFF/CFVI | |
| Cff | ID129038 | Human | Blood | CA | ERS739227 | n.a | CFF/CFVI | |
| Cff | ID131159 | Human | Feces | CA | ERS739228 | n.a | CFF/CFVI | |
| Cff | ID134381 | Human | Feces | CA | ERS739229 | n.a | CFF/CFVI | |
| Cff | ID136207 | Human | Blood | CA | ERS739230 | n.a | CFF/CFVI | |
| Cff | ID136551 | Human | Blood | CA | ERS739231 | n.a | CFF/CFVI | |
| Cff | ID136656 | Human | Blood | CA | ERS739232 | n.a | CFF/CFVI | |
| Cff | ID136706 | Human | Blood | CA | ERS739233 | n.a | CFF/CFVI | |
| Cff | ID132939 | Human | Blood | CA | ERS739234 | n.a | CFF/CFVI | |
| Cff | 2975 | Human | Blood | TW | ERS739256 | n.a | CFF/CFVI | |
| Cff | 923 | Human | Blood | TW | ERS739257 | n.a | CFF/CFVI | |
| Cff | 7035 | Human | Blood | TW | ERS739258 | n.a | CFF/CFVI | |
| Cff | My5726 | Human | Blood | TW | ERS739259 | n.a | CFF/CFVI | |
| Cff | 1592 | Human | Blood | TW | ERS739260 | n.a | CFF/CFVI | |
| Cff | 1830 | Human | Blood | TW | ERS739261 | n.a | CFF/CFVI | |
| Cff | 8468 | Human | Blood | TW | ERS739262 | n.a | CFF/CFVI | |
| Cff | 0003304-2 | Human | Blood | TW | ERS739263 | n.a | CFF/CFVI | |
| Cff | 2115 | Human | Blood | TW | ERS739264 | n.a | CFF/CFVI | |
| Cff | 2819 | Human | Blood | TW | ERS739265 | n.a | CFF/CFVI | |
| Cff | 5871 | Human | Blood | TW | ERS739266 | n.a | CFF/CFVI | |
| Cff | 1666 | Human | Blood | TW | ERS739267 | n.a | CFF/CFVI | |
| Cff | 9502 | Human | Blood | TW | ERS739270 | n.a | CFF/CFVI | |
| Cfv | 800 | Human | Blood | TW | ERS739271 | n.a | CFF/CFVI | |
| Cff | 8031708 | Human | Blood | TW | ERS739272 | n.a | CFF/CFVI | |
| Cff | 8025552 | Human | Blood | TW | ERS739273 | n.a | CFF/CFVI | |
| Cff | 3069482 | Human | Blood | TW | ERS739274 | n.a | CFF/CFVI | |
| Cfv | C1 | Bovine | Prepuce | SP | ERS739275 | n.a | CFV | |
| Cfv | C2 | Bovine | Prepuce | SP | ERS739276 | n.a | CFV | |
| Cff | C3 | Bovine | Prepuce | SP | ERS739277 | n.a | CFF/CFVI | |
| Cff | C4 | Bovine | Prepuce | SP | ERS739278 | n.a | CFF/CFVI | |
| Cff | C5 | Bovine | Prepuce | SP | ERS739279 | n.a | CFF/CFVI | |
| Cfv | C6 | Bovine | Prepuce | SP | ERS739280 | n.a | CFF/CFVI | |
| Cff | C7 | Bovine | Prepuce | SP | ERS739281 | n.a | CFV | |
| Cff | C8 | Bovine | Prepuce | SP | ERS739282 | n.a | CFF/CFVI | |
| Cff | C11 | Bovine | Prepuce | SP | ERS739285 | n.a | CFF/CFVI | |
| Cfvi | C12 | Bovine | Prepuce | SP | ERS739286 | n.a | CFF/CFVI | |
| Cff | C13 | Bovine | Prepuce | SP | ERS739287 | n.a | CFF/CFVI | |
| Cff | C14 | Bovine | Prepuce | SP | ERS739288 | n.a | CFF/CFVI | |
| Cff | C15 | Bovine | Prepuce | SP | ERS739289 | n.a | CFF/CFVI | |
| Cff | C16 | Bovine | Prepuce | SP | ERS739290 | n.a | CFF/CFVI | |
| Cff | C17 | Bovine | Prepuce | SP | ERS739291 | n.a | CFF/CFVI | |
| Cfv | C19 | Bovine | Prepuce | SP | ERS739293 | n.a | CFV | |
| Cff | C20 | Bovine | Prepuce | SP | ERS739294 | n.a | CFF/CFVI | |
| Cff | C21 | Bovine | Prepuce | SP | ERS739295 | n.a | CFF/CFVI | |
| Cfv | C22 | Bovine | Prepuce | SP | ERS739296 | n.a | CFV | |
| Cfv | C23 | Bovine | Prepuce | SP | ERS739297 | n.a | CFV | |
| Cfv | C24 | Bovine | Prepuce | SP | ERS739298 | n.a | CFV | |
| Cfv | C25 | Bovine | Prepuce | SP | ERS739299 | n.a | CFV | |
| Cfvi | C26 | Bovine | Prepuce | SP | ERS739300 | n.a | CFF/CFVI | |
| Cfv | C27 | Bovine | Prepuce | SP | ERS739301 | n.a | CFV | |
| Cfvi | C28 | Bovine | Prepuce | SP | ERS739302 | n.a | CFF/CFVI | |
| Cff | C29 | Bovine | Prepuce | SP | ERS739303 | n.a | CFF/CFVI | |
| Cfv | C30 | Bovine | Prepuce | SP | ERS739304 | n.a | CFV | |
| Cfvi | C31 | Bovine | Prepuce | SP | ERS739305 | n.a | CFF/CFVI | |
| Cfvi | C32 | Bovine | Prepuce | SP | ERS739306 | n.a | CFF/CFVI | |
| Cfvi | C33 | Bovine | Prepuce | SP | ERS739307 | n.a | CFF/CFVI | |
| Cfv | C34 | Bovine | Prepuce | SP | ERS739308 | n.a | CFF/CFVI | |
| Cfv | BS 201/02 | Bovine | Prepuce | GE | ERS686632 | n.a | CFV | |
| Cfv | BS 76/04 | Bovine | Foetus | GE | ERS686633 | n.a | CFV | |
| Cfv | BS 38/06 | Bovine | Prepuce | GE | ERS686634 | n.a | CFV | |
| Cfv | 07BS020 | Bovine | Prepuce | GE | ERS686635 | n.a | CFV | |
| Cfv | 08CS0024 | Bovine | Prepuce | GE | ERS686636 | n.a | CFF/CFVI | |
| Cfv | 09CS0030 | Bovine | Prepuce | GE | ERS686637 | n.a | CFV | |
| Cfv | 11CS0190 | Bovine | Prepuce | GE | ERS686638 | n.a | CFV | |
| Cfv | 11CS0191 | Bovine | Prepuce | GE | ERS686639 | n.a | CFV | |
| Cfv | 13CS0183 | Bovine | Prepuce | GE | ERS686640 | n.a | CFV | |
| Cfv | 14CS0001 | Bovine | Prepuce | GE | ERS686641 | n.a | CFV | |
| Cff | BS 456/99 | Ovine | Foetus | GE | ERS686642 | n.a | CFF/CFVI | |
| Cff | BS 458/99 | Bovine | Foetus | GE | ERS686643 | n.a | CFF/CFVI | |
| Cff | BS 03/04 | Bovine | Foetus | GE | ERS686644 | n.a | CFF/CFVI | |
| Cff | BS 91/05 | Bovine | Prepuce | GE | ERS686645 | n.a | CFF/CFVI | |
| Cff | 08CS0027 | Bovine | Prepuce | GE | ERS686646 | n.a | CFF/CFVI | |
| Cff | 11CS0098 | Ovine | Placenta | GE | ERS686648 | n.a | CFF/CFVI | |
| Cff | 12CS0302 | Bovine | Prepuce | GE | ERS686649 | n.a | CFF/CFVI | |
| Cff | 13CS0001 | Bovine | Prepuce | GE | ERS686650 | n.a | CFF/CFVI | |
| Cff | 13CS0373 | Monkey | Feces | GE | ERS686651 | n.a | CFF/CFVI | |
| Cff | 001A-0374 | Human | Blood | CA | ERS686652 | n.a | CFF/CFVI | |
| Cff | 001A-0648 | Human | Blood | CA | ERS686653 | n.a | CFF/CFVI | |
| Cff | LR133 | Ovine | Foetus | NZ | ERS846544 | n.a | CFF/CFVI | |
| Cff | 1 | Bovine | Prepuce | UK | ERS846553 | n.a | CFF/CFVI | |
| Cff | 2 | Bovine | Prepuce | UK | ERS846554 | n.a | CFF/CFVI | |
| Cff | 3 | Ovine | Placenta | UK | ERS846555 | n.a | CFF/CFVI | |
| Cff | 4 | Ovine | Placenta | UK | ERS846556 | n.a | CFF/CFVI | |
| Cff | 5 | Ovine | Placenta | UK | ERS846557 | n.a | CFF/CFVI | |
| Cff | 6 | Bovine | Prepuce | UK | ERS846558 | n.a | CFF/CFVI | |
| Cff | 7 | Ovine | Foetus | UK | ERS846559 | n.a | CFF/CFVI | |
| Cff | 8 | Ovine | Foetus | UK | ERS846560 | n.a | CFF/CFVI | |
| Cff | 9 | Ovine | Placenta | UK | ERS846561 | n.a | CFF/CFVI | |
| Cff | 12 | Ovine | Placenta | UK | ERS846562 | n.a | CFF/CFVI | |
| Cff | 13 | Bovine | Prepuce | UK | ERS846563 | n.a | CFF/CFVI | |
| Cff | 14 | Ovine | Placenta | UK | ERS846564 | n.a | CFF/CFVI | |
| Cff | 15 | Ovine | Placenta | UK | ERS846565 | n.a | CFF/CFVI | |
| Cff | 17 | Ovine | Foetus | UK | ERS846566 | n.a | CFF/CFVI | |
| Cfv | JCM_2528 | Bovine | Vaginal mucus | UK | ERS846567 | n.a | CFF/CFVI | |
| Cfv | 161/97 | Bovine | Prepuce | BR | ERS846568 | n.a | CFF/CFVI | |
| Cfv | 515/98 | Bovine | Prepuce | BR | ERS846569 | n.a | CFF/CFVI |
Notes:
“CFF/CFVI pattern” means that a complete L-Cys transporter is present. Hybridization of the primer pair Fwd-Rev1-template should occur and a product of 714 bp is predicted. “CFV pattern” means that the L-Cys transporter is partially deleted. Hybridization of the primer pair Fwd-Rev2-template should occur and a product of 310 bp is predicted.
Country code: US, United States; AR, Argentina; UK, United Kingdom; CZ, Czech Republic; FR, France; AU, Australia; CA, Canada; SA, South Africa; NL, The Netherlands; UY, Uruguay; BE, Belgium; IR, Ireland; IN, India; TR, Turkey; JP, Japan; TW, Taiwan; SP, Spain; GE, Germany; BR, Brazil. N.A: Not available.
The hydrogen sulfide production data were available for 43/214 of the studied strains, three from this study and forty from publicly available results (Van Bergen et al., 2005; Van der Graaf-van Bloois et al., 2014, 2016a; Willoughby et al., 2005). However, two of the evaluated strains have shown discrepant results according to the literature and were excluded from this analysis. Interestingly, all of the H2S-producing strains displayed a CFF/CFVI pattern, whereas the non- H2S-producing strains, with unequivocally results according to the biochemical test, presented a CFV pattern (k = 1) (Table 2). The analysis of concordance between in silico-PCR and H2S production is shown in Table S3.
Despite this concordance with the H2S-production test, 14 out of 43 strains that were identified as Cfv in the database did not match with the criteria of the deleted L-Cys transporter for this subspecies. Instead, these strains displayed a CFF/CFVI pattern (Table 2). This situation is also reflected by the overall analysis where the in silico study was able to assign the expected result in 92% (197/213) of the strains (one strain with inconclusive subspecies identification was excluded from the analysis). This partial discrepancy could be attributed to the different methods employed to determine subspecies and this information is not available for most of the strains used in this analysis.
As a proof of concept, we assessed six local field isolates (Cff 08-421, Cff 13-344, Cfv 97-608, Cfv 98-25, Cfvi INTA 99/541 and Cfvi 06-341) through the wet-lab and in silico-PCR approaches. The strains Cfv 97-608, Cfv 98-25 and Cfvi INTA 99/541 were selected from the C. fetus collection because of their genomic sequence availability (Van der Graaf-van Bloois et al., 2016a; Iraola et al., 2013). The L-Cys-transporter-PCR results perfectly matched the in silico-PCR predictions (Tables 1 and 2).
Altogether, this study showed a strong concordance between the results of the L-Cys transporter-PCR and the H2S-production test for C. fetus analysis. Furthermore, it highlights the lack of consensus in the classification of these bacteria between the different laboratories around the world.
Discussion
To date, phenotypic tests are among the most valuable methods to identify and differentiate microorganisms. However, these tests are usually time-consuming, because they are growth-rate dependent, and the whole process depends on the objectivity and skills of the operator. Furthermore, a proper standardization, which is essential to obtain reliable and reproducible results, is often absent. Despite all this, the replacement of these phenotypic tests by molecular techniques is not always an alternative to date. The antimicrobial resistance constitutes a good example of complementary testing, and this particular phenotypic trait can be tested by bacteriological methods and at molecular level in several pathogens (Fluit, Visser & Schmitz, 2001).
Over the last years, researchers have proposed many genotypic tests to facilitate C. fetus differentiation. For example, different studies have employed molecular techniques such as PCR based on different target genes to differentiate Cfv from Cff (Hum et al., 1997; Van Bergen et al., 2005; Abril et al., 2007). However, to date there is no clear consensus on the best method to assess C. fetus subspecies. The main problems rise from the limited number of tested strains, the failure to identify Cfvi strains and the low concordance with other techniques such as AFLP and, mainly, biochemical tests (Willoughby et al., 2005; Schulze et al., 2006; Schmidt, Venter & Picard, 2010).
A genome-wide association study has proposed the association between candidate gene loci coding for the L-Cys transporter and the H2S production, which together to glycine resistance is one of the phenotypic traits available for assessing C. fetus subspecies to date. According to this, H2S-producing C. fetus strains, commonly classified as Cff and Cfvi, have a complete L-Cys transporter operon, whereas the non-producing H2S C. fetus strains, classically classified as Cfv, have a deleted L-Cys transporter. It is important to mention that C. fetus subsp. testudinum (Cft), the last subspecies proposed of C. fetus, has a complete version of the operon. This is the case of the strain Cft 03/427 (whose genome is the representative of the species) which has been concordantly described as an H2S-producing strain elsewhere (Van der Graaf-van Bloois et al., 2016a). To date, this subspecies has not been described in cattle, and for this reason it was excluded of this study.
In this work, we have designed a multiplex-PCR protocol to provide a molecular tool to contribute to C. fetus characterization and differentiation. This L-Cys transporter-PCR showed an excellent correlation with the H2S production test according to both wet lab and in silico approaches. As other molecular techniques, this PCR failed to differentiate Cff from Cfvi strains. This will limit its use in countries where this biovar is prevalent. However, until more discriminative techniques are developed, its usefulness could be further enhanced by combining this technique with other complementary test, such as the glycine resistance assay.
In addition to practical implications of this tool in the laboratory, this study also contributes to the existing debate around C. fetus subspecies classification.
In this study, we tested C. fetus strains isolated and typed at the Bacteriology Unit of INTA-Balcarce (Argentina), which has a long history in culturing this bacterium and in performing its biochemical based classification. In this way, the wet-lab approach showed a perfect correlation not only with the H2S production test, but also with the C. fetus subspecies. Indeed, a CFF-CFVI pattern, which is indicative of L-Cys complete transporter, was associated with H2S-producing strains typically classified as Cff or Cfvi. By contrast, a CFV pattern, which is indicative of a deleted transporter, was exclusively associated with H2S-non-producing strains, which are typically classified as Cfv.
On the other hand, when we performed the in silico study, we analyzed genomic data from strains classified elsewhere by both molecular based approaches and/or biochemical tests and, as mentioned above, both techniques frequently displayed discordant results. In this way, we have obtained a perfect correlation with the H2S production test, but not with the reported subspecies of the strains.
This discordancy is well reflected by the strain 98-25. Researchers from the Bacteriology Unit of INTA-Balcarce isolated this strain in 1998 from aborted foetus, and originally typed it as Cfv because of its glycine sensitivity and its inability to produce H2S. This strain was included in this study and the PCR-L-Cys result was concordant with the phenotype of this strain. Later studies have also tested this strain and successfully sequenced its genome. Indeed, Van Bergen et al. (2005) typed it as glycine sensitive and H2S positive (typical traits of Cfvi strains). Later, Van der Graaf-van Bloois et al. (2014) reported it as non-H2S-producing strain. However, in this latter work, it has been called Cfvi 98-25 regardless the biochemical traits reported. In our study, the in silico sequence data analysis revealed the polymorphism of the L-Cys transporter (CFV pattern) typical of the non- H2S- producing strains.
Therefore, the same strain could display different biochemical traits when assayed in different labs -or time- and this is the classical bottle-neck of phenotypic tests.
We initially had other discrepancies with some isolates. Remarkably, 14 out of 43 Cfv genomic sequences tested in silico showed a complete version of the L-Cys transporter (CFF/CFVI pattern). Hence, at first glance, the hypothesis that all Cfv isolates do have a deleted L-Cys transporter appeared as not valid, according to the in silico analysis. However, when we searched the biochemical tests reported for some of these strains, we concluded that the in silico results presented here were concordant with the H2S production test. This discrepancy with the subspecies assigned could be due to the classification method of the strains that is frequently based on molecular techniques regardless the biochemical test results and moreover; the chosen method is not always specified (Iraola et al., 2017). Altogether, the in silico analysis also supports the hypothesis that states the occurrence of a deletion in the transporter operon in non-H2S producing strains, which are classified as Cfv according to biochemical methods.
As was mentioned earlier, it is important to highlight that the strains from databases are not typed by the same methodology and this fact is not always taken into account. Consequently, this could be problematic as our study showed. The most widespread molecular-based method is the multiplex-PCR described by Hum et al. (1997). This PCR targets the parA gene to identify Cfv strains. This transfer-associated gene is harbored in a pathogenicity island which encodes a Type 4 Secretion System (T4SS). Although the presence of a T4SS has been previously associated to Cfv strains (Gorkiewicz et al., 2010), it has also been demonstrated later that some Cff strains can harbor the T4SS and their related genes (Van der Graaf-van Bloois et al., 2016b). Furthermore, distinct phylogenetic analyses of C. fetus suggest that the current classification in subspecies must be redefined. A phylogenomic study based on the core genome have shown that the strains are divided in two clusters. While all the Cfv and Cfvi strains were grouped in one genome cluster, the Cff strains were equally distributed in both clusters (Van der Graaf-van Bloois et al., 2014). Additionally, a phylogenetic reconstruction based on the divergence acquired by recombination have also shown that Cfv and Cff strains share the same clade, which differs clearly from the clade of Cft strains of reptile origin (Gilbert et al., 2018). This emphasizes a real need to go further toward current C. fetus classification and identification, which will have a significant impact on the diagnostic practice. As mentioned above, although this issue has been addressed in the literature and genomic studies have broadened and strengthened our knowledge of this bacterium (Van der Graaf-van Bloois et al., 2014, 2016a; Iraola et al., 2017), a concerted action toward C. fetus subspecies classification and differentiation has been neglected. There are no molecular markers associated to tropism or virulence of Cfv that could help with a differential diagnosis of the BGC (Gilbert et al., 2018).
Consequently, in light of these evidences, more research is essential to determine, as a first step, whether the differential diagnosis should be promoted and, if so, to improve or replace definitively the tests currently available. Another point to consider is that, veterinary diagnostic laboratories from developing countries are often refractory to replace those methods that have proven be useful, even if they are not the most suitable ones. Because of that, the adoption of genetic or even genomic-based methods has been delayed. One possible reason is the cost related to each method. The second main reason is the time lapse it takes for scientific knowledge to reach end users. Interdisciplinary research combining genomics, biochemistry, epidemiology and the provision of updated information and training to end users could shed light on this matter in the near future.
Conclusions
Biochemical tests including tolerance to glycine and H2S production are currently recommended by the OIE (2018) for C. fetus subspecies differentiation and are still employed in laboratories around the world. Thus, a molecular tool linked to a phenotypic trait is a valuable tool that could be more accurate and less time consuming than the available phenotypic tests. Mutagenesis and functional studies are essential to associate definitely this putative L-Cys transporter with the H2S production. Meanwhile, this study shows that this transporter constitutes a good marker that is useful for detecting H2S-producing C. fetus. Future actions will be addressed to test the L-Cys-PCR in clinical samples to propose it not only as a typing method, but as a detection technique and, as a second phase of validation, to transfer this technology to other labs to test the reproducibility of the results.
Finally, this work provides a molecular tool linked to H2S production in C. fetus and supports the findings of the pioneering study of Van der Graaf-van Bloois et al. (2016a).
Supplemental Information
Whole Genome Sequencing of three local C. fetus Strains: Overall Assembly Statistics.
Analysis of agreement between the H2S production test and L-Cys-PCR.
Contingency table and calculation of the Cohen’s Kappa coefficient. Data set from Table 1 comprising 36 strains was included. The analysis showed a perfect agreement (ĸ = 1) ( https://idostatistics.com/cohen-kappa-free-calculator/ ).
Analysis of agreement between H2S production test and in silico L-Cys-PCR.
Contingency table and calculation of the Cohen’s Kappa Coefficient. Data set from Table 2 comprising 41 strains with reported-H2S production test was included in the analysis. The analysis showed a perfect agreement (ĸ = 1) (https://idostatistics.com/cohen-kappa-free-calculator/).