Evidence for an oncogenic role of HOXC6 in human non-small cell lung cancer
- Published
- Accepted
- Received
- Academic Editor
- Yegor Vassetzky
- Subject Areas
- Cell Biology, Molecular Biology, Oncology
- Keywords
- HOXC6, Non-small cell lung cancer, Molecular marker, Bioinformatics, Malignancy
- Copyright
- © 2019 Yang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Evidence for an oncogenic role of HOXC6 in human non-small cell lung cancer. PeerJ 7:e6629 https://doi.org/10.7717/peerj.6629
Abstract
Background
Identification of specific biomarkers is important for the diagnosis and treatment of non-small cell lung cancer (NSCLC). HOXC6 is a homeodomain-containing transcription factor that is highly expressed in several human cancers; however, its role in NSCLC remains unknown.
Methods
The expression and protein levels of HOXC6 were assessed in NSCLC tissue samples by Quantitative real-time PCR (qRT-PCR) and immunohistochemistry, respectively. HOXC6 was transfected into the NSCLC cell lines A549 and PC9, and used to investigate its effect on proliferation, migration, and invasion using CFSE, wound healing, and Matrigel invasion assays. Next-generation sequencing was also used to identify downstream targets of HOXC6 and to gain insights into the molecular mechanisms underlying its biological function.
Results
HOXC6 expression was significantly increased in 66.6% (20/30) of NSCLC tumor samples in comparison to normal controls. HOXC6 promoted proliferation, migration, and invasion of NSCLC cells in vitro. RNA-seq analysis demonstrated the upregulation of 310 and 112 genes in A549-HOXC6 and PC9-HOXC6 cells, respectively, and the downregulation of 665 and 385 genes in A549-HOXC6 and PC9-HOXC6 cells, respectively. HOXC6 was also found to regulate the expression of genes such as CEACAM6, SPARC, WNT6, CST1, MMP2, and KRT13, which have documented pro-tumorigenic functions.
Discussion
HOXC6 is highly expressed in NSCLC, and it may enhance lung cancer progression by regulating the expression of pro-tumorigenic genes involved in proliferation, migration, and invasion. Our study highlighted the oncogenic potential of HOXC6, and suggests that it may be a novel biomarker for the diagnosis and treatment of NSCLC.
Introduction
Lung cancer has the highest cancer incidence in the world (Chen et al., 2016b). Non-small cell lung cancer (NSCLC) accounts for 80–85% of the total number of lung cancer cases (Lee et al., 2013; Zienolddiny & Skaug, 2011). In spite of improvements in imaging science, radical surgical resection, and NSCLC auxiliary detection technologies, most patients have advanced disease at diagnosis, and the overall 5-year survival rate is less than 15% (National Lung Screening Trial Research T et al., 2011; Ye & Zhao, 2016). Therefore, more effective molecular makers are needed for the diagnosis and treatment of NSCLC.
HOX genes belong to the homeobox gene superfamily (Cillo et al., 2001). The human HOX gene family is made up of 39 members in four clusters (A–D) located on chromosomes 7, 17, 12, and 2, with each cluster containing 9 to 13 loci (Apiou et al., 1996). Many HOX genes have been found to be closely associated with the progression of cancer (Shah & Sukumar, 2010). HOXA10 can inhibit the invasion of breast cancer cells by enhancing the expression of TP53 (Chu, Selam & Taylor, 2004). HOXB7 can also promote tumor growth by upregulating the expression of angiogenic growth factors (Care et al., 2001). HOXC6 is a transcription factor that regulates cell differentiation during embryonic development (Maroulakou & Spyropoulos, 2003). Aberrant expression of HOXC6 may result in the malignant transformation of normal cells (DeInnocentes et al., 2015; Feng et al., 2009; Moon et al., 2012; Wright et al., 1989; Zhang et al., 2013), and elevated HOXC6 expression has been observed in several types of cancers, including prostate, gastrointestinal, colorectal, and hepatocellular cancers (Chen et al., 2016a; Ji et al., 2016; Sui et al., 2016; Vinarskaja et al., 2011). However, the biological function of HOXC6 has not been well understood. Here, we report that HOXC6 is highly expressed in NSCLC cells, and overexpression of HOXC6 promotes the proliferation, migration, and invasion of NSCLC cells. The phenotypic effects of HOXC6 may be mediated by genes that have been previously reported to be involved in the progression of cancer. Our data also suggest that HOXC6 is a potential molecular marker for the diagnosis and treatment of NSCLC.
Materials & Methods
Cell lines
NSCLC cell lines A549 and PC9 were obtained from the Stem Cell Bank of the Chinese Academy of Sciences. A549 was maintained in RPMI 1640 Medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). PC9 and 293FT cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). All of the cell lines were housed in 37 °C incubators with 5% CO2 saturation.
Human clinical specimens
Clinical specimens were collected from patients at the Affiliated Hospital of Southwest Medical University (Luzhou, China). Tissue samples were surgically retrieved from NSCLC patients after obtaining written consent and with the approval of the ethics committee of the Affiliated Hospital of Southwest Medical University (k2018003-r). Clinical samples were immediately separated into lung tumor tissues (T) and adjacent non-tumor lung tissues (N), flash frozen in liquid nitrogen, and then stored at −80 °C until further analysis.
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded sections were treated with 3% H2O2 for 10 min after deparaffinization in xylene and rehydration in decreasing concentrations of ethanol from 100 to 75%. Antigen retrieval was performed by using heated sodium citrate. Sections were then blocked using 10% normal goat serum to prevent non-specific antibody reactions. The sections were then incubated overnight at 4 °C, with a mouse monoclonal antibody against human HOXC6 (dilution, 1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA) After incubation, the HOXC6 antigen-antibody reaction was performed using an immunoperxodase-based kit (ZSGB Bio, Beijing, China). To quantify the expression of HOXC6, two specialist pathologists scored the stained samples based on the product of the intensity and degree of staining (0–100%). The staining intensity was categorized as follows: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). Only cytoplasmic staining was counted as positive staining.
RNA isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from NSCLC cell lines and human samples using TRIzol (TaKaRa Bio, Otsu, Japan). Then, 0.5 µg total RNA was reverse transcribed into cDNA using a PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa Bio, Otsu, Japan) according to the manufacturer’s instructions. qRT-PCR was performed using SYBR® Premix Ex Taq™ II (TaKaRa Bio, Otsu, Japan) system on an ABI QuantStudio™ 7 Flex Real-Time PCR (Applied Biosystems, USA). For the PCR reaction, specific primers were selected based on PrimerBank HOXC6 sequences. The following primers were synthesized by Invitrogen: human HOXC6 forward primer 5′-ACAGACCTCAATCGCTCAGGA-3′, and reverse primer 5′-AGGGGTAAATCTGGATACTGGC-3′; GAPDH forward primer 5′-ATGCTGGCGCTGAGTACGTC-3′, and reverse primer 5′-GGTCATGAGTCCTTCC- ACGATA-3′. Gene expression levels were normalized to GAPDH using the 2−ΔΔCt method.
Lentivirus packaging
The lentiviral HOXC6- expressing vector pCDH-HOXC6 and empty control vector pCDH-NEO were obtained from Experimental Medicine Center, The Affiliated Hospital of Southwest Medical University. The 293FT cells were used for lentiviral packaging. A total of three vectors (pCDH-HOXC6 or pCDH-NEO, psPAX2 and pMD2G) were co-transfected into 293FT cells using Lipofectamine3000 (Invitrogen, Carlsbad, CA, USA). After 72 h, viral supernatant was collected and stored at −80 °C.
Cell transfection
A549 and PC9 cells were plated into a 6-well plate. Cells were then transfected with the lentiviral plasmids at 80% confluence. After 8 h, the transfection medium was replaced with complete growth medium supplemented with 500 µg/ml neomycin. qRT-PCR and Western blot were performed to evaluate transfection efficiency.
Immunoblotting
Cells were lysed on ice using RIPA buffer (Cell Signaling Technology, USA). Protein concentration was measured by BCA kit (Beyotime, Shanghai, China). Equal amounts of total protein were resolved using 12% SDS-PAGE gel and transferred onto PVDF membranes (Millipore, Burlington, MA, USA). Membranes were incubated with anti-HOXC6 (Santa Cruz, CA, USA, dilution, 1:1,000) and anti-β-actin (Santa Cruz, CA, USA, dilution, 1:1,000) overnight at 4 °C followed by incubation with a secondary antibody. Protein bands were visualized using ECL chemiluminescence (Millipore, Billerica, MA, USA) according to the manufacturer’s recommendation.
Cell proliferation assay
Cell proliferation was performed using Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE) flow cytometry analysis on a BD FACSVerse™ (BD Biosciences, Frankin Lakes, NJ, USA). A final concentration of 5 µM CFSE was added to a single cell suspension of 10 × 106 cells/ml. The dye-cell suspension was then incubated in a 37 °C water bath for 10–15 min and then analyzed by flow cytometry. Cell proliferation was also assessed using the IncuCyte Live Cell Imaging System. Cells were plated in IncuCyte ImageLock 96-well plates (Essen BioScience, USA) at 1 × 104 cells per well. Cell confluency was monitored using the IncuCyte Live Cell Imaging System (Essen Bioscience, Ann Arbor, MI, USA) every 2 h.
Cell migration and invasion assay
Transfected cells were plated in IncuCyte ImageLock 96-well plates (Essen BioScience, Ann Arbor, MI, USA). After cells adhered to the wells, an approximately 600-µm wide scratch was made in the cell layer using a WoundMaker (Essen Bioscience, Ann Arbor, MI, USA). The cells were then washed twice with PBS to remove floating cells. To assess migration, 100 µl of growth medium (with 2% FBS) was added to the cells. For invasion, the plate was coated with 30% Matrigel for 30 min at 37 °C, with an additional 50 µl of normal growth medium overlaid on to the plates. Wound closure was monitored using the IncuCyte Live Cell Imaging System (Essen Bioscience, Ann Arbor, MI, USA) every 2 h. Data analysis was conducted using Incucyte 2016A software.
Construction of Sequence libraries
Total RNA was extracted from NSCLC cells A549 and PC9. Dynabeads Oligo (dT) 25 beads were used for isolating mRNA. RNA libraries were constructed with NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) with 3 independent replicates following the manufacturer’s instructions. Paired-end sequencing (150 bp) was performing using Illumina HiSeq Xten. Raw data was uploaded to the GEO database (GSE121896).
Bioinformatics analysis
We used HISAT2 (version 2.1.0) (Kim, Langmead & Salzberg, 2015) with default parameters to map the RNA-seq data to the GRCh37.p13 genome from GENCODE (Harrow et al., 2012). We aggregated the read counts at the gene level using HTseq (Anders, Pyl & Huber, 2015) and called differentially expressed genes (DEGs) with R package DESeq2 (Love, Huber & Anders, 2014). Genes were considered significantly differentially expressed when the Log2 fold change was >1 or <−1, and the adjusted P value was <0.05. Gene Ontology analysis of the DEGs was performed using the R package ClusterProfiler (Yu et al., 2012) with P < 0.05.
Statistical analysis
All of the statistical analyses were carried out using GraphPad Prism 7 and SPSS 21.0. Numerical data are presented as mean ± standard error, and were calculated using two-tailed Student’s t test and Chi-square test. Two-way analysis of variance (ANOVA) was used to assess the statistical significance of multiple continuous variables. P < 0.05 was considered statistically significant.
Results
High expression of HOXC6 in Human NSCLC tissues
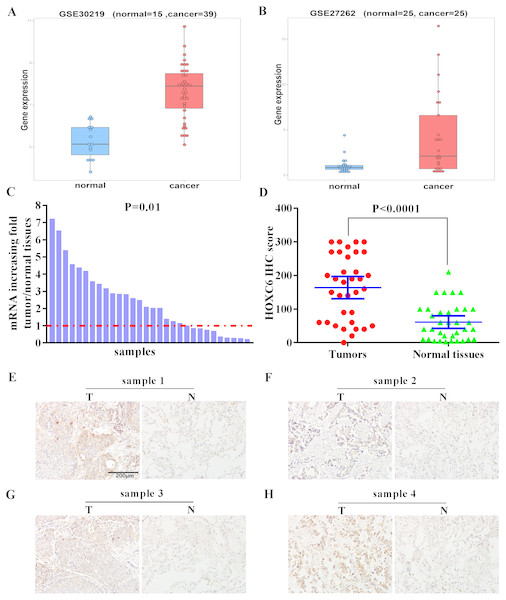
We examined the expression level of HOXC6 in lung cancer using in silico approaches. Analysis of publicly available data revealed that there was a 5-fold and 8.8-fold increase in the expression of HOXC6 in 2 independent lung cancer datasets, GSE30219 and GSE27262, respectively (Figs. 1A and 1B). To validate these findings, we used qRT-PCR to determine HOXC6 expression in tumor samples from NSCLC patients. As shown in Fig. 1C, in 30 pairs of NSCLC tissue samples and adjacent non-tumor tissue samples, 66.6% (20/30) had elevated levels of HOXC6 in comparison to their adjacent normal counterparts (P = 0.01). IHC analysis was consistent with the gene expression data, as HOXC6 protein was elevated in NSCLC tumor samples in comparison to adjacent normal controls (Figs. 1D and 1E). These results indicate that HOXC6 is highly expressed in NSCLC tissues.
Figure 1: Elevated expression of HOXC6 in NSCLC in comparison to adjacent normal tissues.
(A, B) The bioinformatic analysis of publicly available gene expression data sets for HOXC6 expression in lung cancer tissues. Microarray dataset GSE30219 (normal = 15, cancer = 39) and GSE27262 (normal = 25, cancer=25) were download from Gene Expression Omnibus(GEO). HOXC6 expression distribution in (A) and (B), with a fold change of 5 (q. value = 1.4332e–07) and 8.8 (q. value = 0.0039),which were calculated by R package limma ( Ritchie et al., 2015). (C) Relative expression of HOXC6 mRNA in 30 NSCLC and paired adjacent normal tissues as determined by qRT-PCR. The data were assessed using Chi-square test. (D) Expression of HOXC6 protein as determined by IHC. The IHC score of HOXC6 was calculated as the staining intensity (0, 1, 2, or 3) × the staining extent (0–100%). We compared matched samples as in (D) using paired two-tailed Student’s t test. (E) Representative images of IHC staining of HOXC6 in NSCLC tumor (T) and adjacent normal tissues (N), Scale bar = 200 µm.The effect of HOXC6 overexpression on the proliferation of NSCLC cells
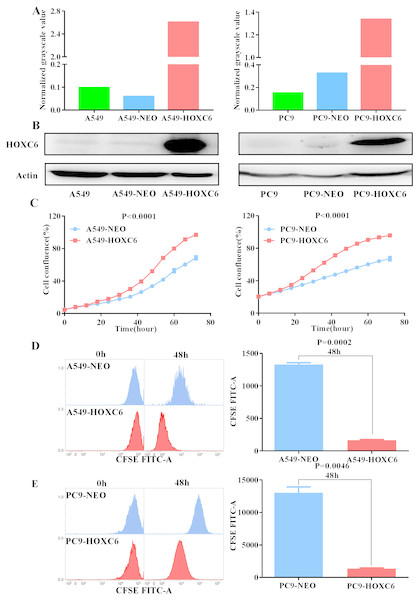
To further explore the biological function of HOXC6, we used lentiviral vectors to establish HOXC6- overexpressing NSCLC cell lines. The resulting cell lines were named A549-HOXC6 and PC9-HOXC6. Control cell lines transfected with the empty vector were named A549-NEO and PC9-NEO. Transfection efficiency was then assessed by qRT-PCR and Western blot (Figs. 2A and 2B). We next examined the effect of HOXC6 on the growth of NSCLC cells, by using an image-based proliferation assay. As shown in Fig. 2C, the proliferation rate was significantly increased in HOXC6- overexpressing A549 and PC9 cells in comparison to control cells. We also utilized the CFSE assay as a secondary proliferation assay to validate this observation. There was an obvious shift in the fluorescence signal in A549-HOXC6 and PC9-HOXC6 cells 48 h after CFSE labeling, indicating that HOXC6 promoted the proliferation of both cell lines (Fig. 2D). These results demonstrate that overexpression of HOXC6 increased the proliferation of NSCLC cells.
Figure 2: Overexpression of HOXC6 promotes NSCLC cell proliferation.
(A, B) Western Blot analysis of HOXC6 in A549 and PC9 HOXC6-expressing or control cells. (C) IncuCyte Live Cell Imaging System analysis of NSCLC cell lines A549 and PC9 transfected with HOXC6 or NEO lentiviral vectors. Cells (1 × 104) were seeded in 96-well plates and monitored at 2-hour intervals, three replicates for each sample. (D) Flow cytometry analysis of CFSE-labeled HOXC6- expressing or control NSCLC cell lines, three replicates for each sample.The promotion of migration and invasion by HOXC6 in NSCLC cells
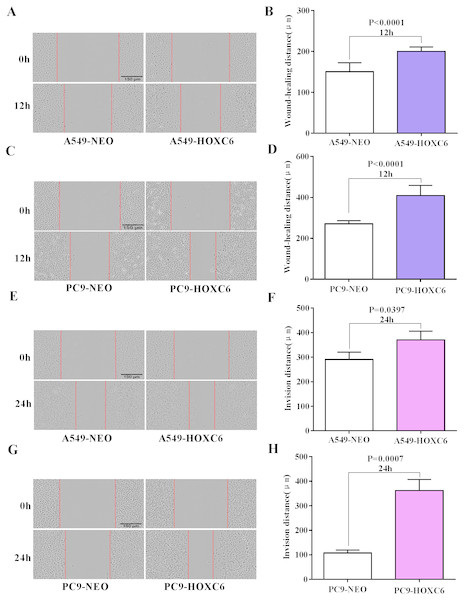
Migration and invasion are recognized as important hallmarks of cancer (Hanahan & Weinberg, 2011). To investigate the effect of HOXC6 on the migration and invasion of NSCLC cell lines, we made use of the wound healing and Matrigel assays. We observed significantly faster motility in A549-HOXC6 and PC9-HOXC6 cells in comparison to A549-NEO and PC9-NEO control cells in the wound healing experiment (Figs. 3A and 3B). Additionally, we observed that HOXC6 increased the invasive capacity of NSCLC cells in Matrigel as shown in Figs. 3C and 3D. These results suggest that HOXC6 may have a pro-tumorigenic role in NSCLC.
Figure 3: Overexpression of HOXC6 promotes migration and invasion of NSCLC cells.
(A, B) Wound healing assay of A549 and PC9 cells expressing HOXC6 or NEO. Scale bar = 150 µm. (C, D) Invasion of HOXC6-expressing or control A549 and PC9 cells was assessed in a wound healing assay using 30% Matrigel. Scale bar = 150 µm.The downstream targets of HOXC6 revealed by gene expression profiling in NSCLC cells
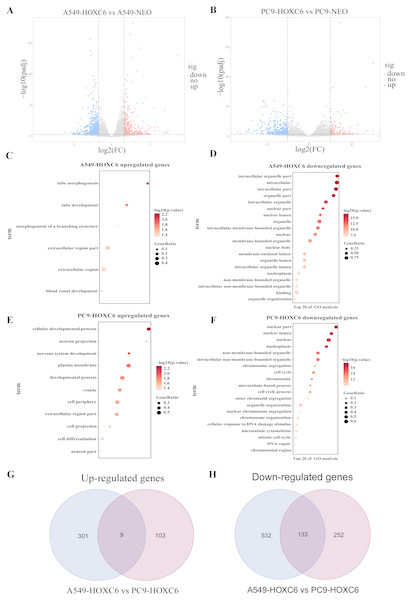
Just like other members of the homeobox superfamily, HOXC6 is a transcription factor that regulates the expression of several downstream target genes. RNA-seq analysis was performed to identify genes modulated by HOXC6 in NSCLC cells. The results (GSE121896) showed that there were 310 genes upregulated and 665 genes downregulated in A549-HOXC6 cells in comparison to A549-NEO control cells. In addition, we found that there were 112 genes upregulated and 385 genes downregulated in PC9-HOXC6 cells in comparison to PC9-NEO control cells (Fig. 4A). Gene Ontology analysis indicated that these genes modulated by HOXC6 may take part in various biological processes, such as morphogenesis and development, which is consistent with the previously reported functions of the homeobox gene superfamily (Fig. 4B). When we compared the genes regulated by HOXC6 in the two cell lines examined in this study, there were 9 common upregulated genes and 133 common downregulated genes (Fig. 4C). However, most of the genes whose expression levels were changed after HOXC6 transfection were different in these two cell lines. This unexpected result suggests that transcriptional regulation by HOXC6 is cell context-dependent.
Figure 4: Analysis of RNA-seq.
(A) The volcano map shows 310 upregulated genes and 665 downregulated genes in A549-HOXC6 cells in comparison to A549-NEO cells, and 112 upregulated genes and 385 downregulated genes in PC9-HOXC6 cells in comparison to PC9-NEO cells. Log2 fold change (Log2FC) >1 or <−1, and P < 0.05 were considered to be statistically significant. (B) Gene ontology (GO) analysis identified biological processes impacted by HOXC6-regulated genes in NSCLC cells. (C) Overlap of upregulated and downregulated genes in A549-HOXC6 and PC9-HOXC6 cells.Pro-tumorigenic genes controlled by HOXC6 in NSCLC cells
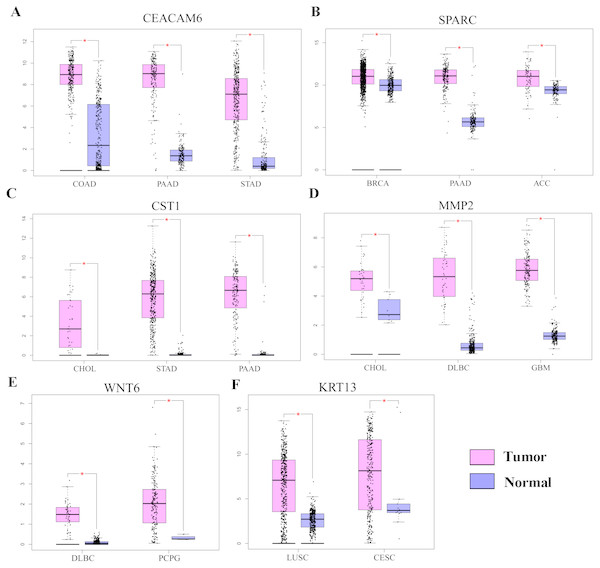
Gene Ontology analysis did not clearly identify the molecular mechanisms underlying the pro-tumorigenic effect of HOXC6. Therefore, we supplemented this analysis by manually screening critical tumor-associated HOXC6-upregulated genes using functional analysis and literature review. Using this approach, we successfully identified several genes closely associated with tumor progression. CEACAM6, SPARC, WNT6, CST1, and MMP2 were identified in A549-HOXC6 cells, while CEACAM6 and KRT13 were identified in PC9-HOXC6 cells. These genes have well documented roles in promoting cancer development (Table 1). We further analyzed the expression of these genes in various cancers through GEPAI (Tang et al., 2017). The results clearly demonstrate that the expression of these genes is associated with the progression of cancers (Fig. 5A), and that they are primarily involved in the regulation of cell proliferation, migration, and invasion. Furthermore, when we tried to analyze the function of the common upregulated genes, i.e., HOXC6, SGK1, S100A4, MALL, CEACAM6, SLCO4A1-AS1, C11orf86, ENSG00000268621, and ENSG00000129270, at least 5 of them, i.e., SGK1, S100A4, MALL, CEACAM6 and SLCO4A1-AS1, have been reported to be implicated in the malignant phenotype of various cancers, while the remaining genes have not been well studied (Egeland et al., 2017; Liang et al., 2017; Rizeq, Zakaria & Ouhtit, 2018; Yu et al., 2018). We also analyzed the common downregulated genes and found that many of these genes such as ANKRD12, KIAA1551, and APC encode proteins with tumor suppressive functions (Bai et al., 2013; Cheng et al., 2017; Lv et al., 2019). These data indicate that HOXC6 could exert its oncogenic function by both activation of oncogenes and downregulation of the expression of tumor suppressive genes.
| Gene symbol | Gene description | Gene functiona | References |
|---|---|---|---|
| CEACAM6 | Tumor marker | A, M, I, P, T | Rizeq, Zakaria & Ouhtit (2018) Zhang et al. (2013) |
| SPARC | Matrix-associated protein | P, M, CC, SF | Chang et al. (2018) Yusuf et al. (2014) |
| WNT6 | A family of highly conserved developmental control genes | P, M, A, CC, T | Yuan et al. (2013) Zheng & Yu (2018) |
| CST1 | The cystatin superfamily | P, M, I | Choi et al. (2009) Dai et al. (2017) |
| MMP2 | The major structural component of basement membranes | M, I | Kalhori & Törnquist (2015) Kuo et al. (2014) |
| KRT13 | Encoded a member of the keratin gene family | M, I, CC, A | Man et al. (2014) Li et al. (2016) |
Notes:
Figure 5: The expression of pro-tumorigenic genes in various cancers.
(A) The expression of CEACAM6, SPARC, WNT6, CST1, MMP2, and KRT13 in various cancers, ∗P < 0.05. Breast invasive carcinoma (BRCA), pancreatic adenocarcinoma (PAAD), adrenocortical carcinoma (ACC), cholangiocarcinoma (CHOL), stomach adenocarcinoma (STAD), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), pheochromocytoma and paraganglioma (PCPG), colon adenocarcinoma (COAD), lung squamous cell carcinoma (LUSC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC).Discussion
Lung cancer is the most frequently occurring malignancy worldwide. Due to the lack of effective measures for early diagnosis and treatment, the 5-year overall survival rate for NSCLC patients is only 16%–18% (Siegel, Miller & Jemal, 2018). The development of NSCLC involves both environmental and genetic changes, and the activation of oncogenes is also an important factor. Therefore, the identification of novel biomarkers is critical for the improvement of clinical outcomes for NSCLC patients.
In recent years, it has been found that the HOX gene is closely associated with the development and prognosis of various cancers. HOXC6 is a member of the HOX family that acts as a transcription factor and participates in the regulation of a number of genes during development. HOXC6 has also been found to be highly expressed in several cancers. In this report, we provide several lines of evidence demonstrating that HOXC6 plays an oncogenic role in human NSCLC. First, HOXC6 expression was shown to be elevated in NSCLC tissues. Second, HOXC6 promoted the proliferation of NSCLC cells. Third, HOXC6 enhanced the migration and invasion of NSCLC cells, which are fundamental hallmarks of cancer. Fourth, HOXC6 can upregulate the expression of genes critical for the development and progression of various cancers. Therefore, our work has established the basis for further investigation of HOXC6 and its oncogenic roles in NSCLC.
To gain insight into the molecular mechanisms underlying the pro-tumorigenic functions of HOXC6, RNA-seq was performed to identify its downstream targets. A number of genes have been found to be modulated by HOXC6, but interestingly, we observed that there was very little overlap in the genes modulated by HOXC6 in two NSCLC cell lines. This result suggests that there is a cell context-dependent mechanism underlying the function of HOXC6. Since most transcription factors need to form complexes to specifically regulate their target genes, the cell context-specificity of HOXC6 may be caused by binding to different cofactors in different cell lines. Despite the fact that different sets of genes are regulated by HOXC6 in various cell lines, transfection of HOXC6 into both cell lines used in this study generated similar phenotypic effects. Consistent with this result, we found that HOXC6 is a master regulator of many genes, which have documented pro-tumorigenic functions. In our studies, we identified several genes, including CEACAM6, SPARC, WNT6, CST1, MMP2, and KRT13, which have been extensively studied and demonstrated to be involved in the regulation of tumor growth, migration and invasion, cell cycle, and apoptosis (Chang et al., 2018; Choi et al., 2009; Dai et al., 2017; Kuo et al., 2014; Rizeq, Zakaria & Ouhtit, 2018; Wang et al., 2018; Yusuf et al., 2014; Zheng & Yu, 2018). In other types of cancers, HOXC6 has also been shown to have the capacity to regulate these cancer-related genes. For example, HOXC6 directly regulates gene expression of Bone morphogenetic protein 7 (BMP7), Fibroblast growth factor receptor 2 (FGF2), and Platelet-derived growth factor receptor (PDGFR) in prostate cancer (McCabe et al., 2008). HOXC6 promotes the migration, invasion, and progression of gastric cancer by upregulating Matrix metalloproteinase 9 (MMP9) (Chen et al., 2016a). These results suggest that HOXC6 can promote tumorigenesis by regulating distinct sets of genes in various cellular contexts.
Conclusion
In conclusion, we demonstrate that HOXC6 was highly expressed in NSCLC tissues and correlated with the malignant phenotype of NSCLC cells. In addition, bioinformatics analyses showed that HOXC6 may enhance lung cancer progression by regulating the expression of pro-tumorigenic genes involved in proliferation, migration, and invasion. Our study highlighted the oncogenic potential of HOXC6 and suggests that it is a candidate molecular marker for the diagnosis and treatment of NSCLC.