Undocumented translocations spawn taxonomic inflation in Sri Lankan fire rasboras (Actinopterygii, Cyprinidae)
- Published
- Accepted
- Received
- Academic Editor
- Marcio Pie
- Subject Areas
- Biodiversity, Conservation Biology, Taxonomy, Zoology, Freshwater Biology
- Keywords
- mtDNA, Rasboroides, Allometry, Biodiversity hotspot, Integrative taxonomy, Systematics, Translocations, Species boundaries, Freshwater fish, Genetic distance
- Copyright
- © 2018 Sudasinghe et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2018. Undocumented translocations spawn taxonomic inflation in Sri Lankan fire rasboras (Actinopterygii, Cyprinidae) PeerJ 6:e6084 https://doi.org/10.7717/peerj.6084
Abstract
A recent (2013) taxonomic review of the freshwater-fish genus Rasboroides, which is endemic to Sri Lanka, showed it to comprise four species: R. vaterifloris, R. nigromarginatus, R. pallidus and R. rohani. Here, using an integrative-taxonomic analysis of morphometry, meristics and mitochondrial DNA sequences of cytochrome b (cytb) and cytochrome oxidase subunit 1 (coi), we show that R. nigromarginatus is a synonym of R. vaterifloris, and that R. rohani is a synonym of R. pallidus. The creation and recognition of unnecessary taxa (‘taxonomic inflation’) was in this case a result of selective sampling confounded by a disregard of allometry. The population referred to R. rohani in the Walawe river basin represents an undocumented trans-basin translocation of R. pallidus, and a translocation into the Mahaweli river of R. vaterifloris, documented to have occurred ca 1980, in fact involves R. pallidus. A shared haplotype suggests the latter introduction was likely made from the Bentara river basin and not from the Kelani, as claimed. To stabilize the taxonomy of these fishes, the two valid species, R. vaterifloris and R. pallidus, are diagnosed and redescribed, and their distributions delineated. We draw attention to the wasteful diversion of conservation resources to populations resulting from undocumented translocations and to taxa resulting from taxonomic inflation. We argue against translocations except where mandated by a conservation emergency, and even then, only when supported by accurate documentation.
Introduction
With a standard length that rarely exceeds 35 mm, Rasboroides and Horadandia are the only two genera of miniaturized cyprinid fishes to occur in Sri Lanka (Britz & Conway, 2009). Horadandia are distributed across the open lowland floodplains of Sri Lanka and south-western India, whereas Rasboroides are endemic to streams draining the rain forests of the island’s south-western quarter (Pethiyagoda, 1991). Known as Fire Rasboras in the ornamental-fish trade, the gentle disposition, attractive coloration and diminutive size of these fishes have made them a favourite among aquarists. The genus Rasboroides was considered to comprise of only a single species, R. vaterifloris (Deraniyagala, 1930), until Meinken (1957), based on aquarium specimens exported from Sri Lanka, described a second species, R. nigromarginata. Shortly thereafter, Deraniyagala (1958) treated R. vaterifloris as consisting—in effect—of five infraspecific taxa: R. v. vaterifloris, R. v. nigromarginatus, R. v. pallidus, R. v. ruber, and R. v. rubioculis.
Recently, Batuwita, De Silva & Edirisinghe (2013) in a taxonomic review of the genus Rasboroides, recognized three of these taxa (R. vaterifloris, R. nigromarginatus and R. pallidus) as valid at the rank of species, and relegated the remaining two to the synonymy of R. pallidus. While these three nominal species are distributed across the Kelani to the Nilwala river basins (Fig. 1D), Batuwita, De Silva & Edirisinghe (2013) described also a new species, R. rohani, from a localized population in the Walawe basin. This population too, was sampled by us in the course of fieldwork associated with a taxonomic assessment of the cypriniform fishes of Sri Lanka.
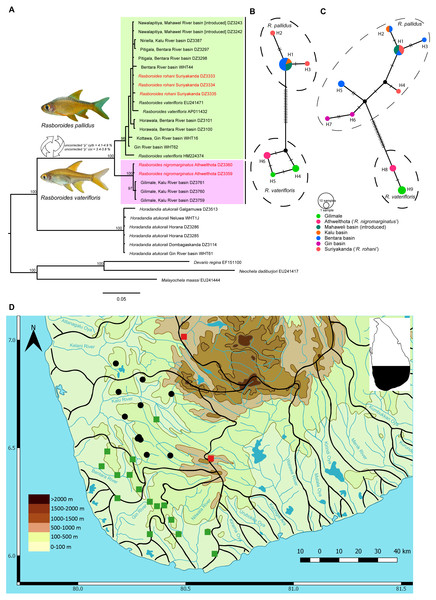
Figure 1: A Phylogram, haplotype networks for two genes and distribution map for species of Rasboroides in Sri Lanka.
(A), Phylogram based on Bayesian inference for the cytb dataset for species of Rasboroides in Sri Lanka. Numbers above represent the Bayesian Posterior Probabilities. The scale bar represents number of changes per site; (B–C) TCS Haplotype network for species of Rasboroides in Sri Lanka based on the analysis of B, 669 bp fragment of the coi, and C, 609 bp fragment of the cytb genes. The sizes of the circles are proportional to the number of individuals sharing a given haplotype. The number of mutational steps are indicated by hatch marks. The black circles are hypothetical nodes; D, Distribution of R. vaterifloris (circles) and R. pallidus (squares) in Sri Lanka. The translocated populations of R. pallidus (red), and natural wild populations of R. pallidus (green).Several factors combine to make R. rohani unusual in comparison with the other species of Rasboroides. First, the genus Rasboroides had not previously been recorded from the Walawe basin (Deraniyagala, 1958; Senanayake, 1980; Pethiyagoda, 1991). Further, the only known population of R. rohani occurs at an elevation of ca 980 m asl, substantially higher than all other natural populations of Rasboroides, which occur in the approximate elevation range 30–230 m asl.
While these factors give rise to suspicion that R. rohani might in fact represent an undocumented introduction, Batuwita, De Silva & Edirisinghe (2013) nevertheless unambiguously distinguished this species from the three other species of Rasboroides they recognized, by an apparently robust suite of morphological characters. The recognition of a new species, however, results from testing the hypothesis that its phenotype and/or genotype, as represented by the examined sample of its population, differs sufficiently from those of congeneric species as to support its novelty according to one or more species concepts (Gaston & Mound, 1993; Winston, 1999). Here we test this hypothesis for R. rohani based on topotypic material collected by us, employing both morphological and molecular analyses, and show that R. rohani in fact represents an undocumented translocation of R. pallidus.
In their review, Batuwita, De Silva & Edirisinghe (2013) omitted to examine another population of Rasboroides in the vicinity of Ginigathena in the Mahaweli basin, from which the genus was absent until an introduction was made by F. R. Senanayake and P. B. Moyle in 1981 (Wikramanayake, 1990; Moyle, in Pethiyagoda, 1991:36). Although of uncertain provenance, this population is now well established and occurs across a range of some 5 km2. Here we show that this too, is the result of a translocation of R. pallidus. We further show also that R. nigromarginatus, considered to be a valid species by Batuwita, De Silva & Edirisinghe (2013), is in fact a synonym of R. vaterifloris. Finally, given the taxonomic problems manifestly resulting from the work of Batuwita, De Silva & Edirisinghe (2013), we diagnose and redescribe the two species of Rasboroides shown by our analyses to be valid: R. vaterifloris and R. pallidus.
Materials & Methods
Ethics statement
Permission to conduct field work was obtained from the Department of Wildlife Conservation (permit no. WL/3/2/59/14) and Forest Department (permit no. R&E/RES/NFSRCM/14-16-4) of Sri Lanka. Methods of specimen collection, euthanisation (using MS-222 Tricaine methanesulfonate), tissue sampling and fixation were approved by the ethical committee of the Postgraduate Institute of Science, University of Peradeniya, at its 27th meeting held on 4 August 2017.
Metrics & meristics
Methods for measurements and counts follow Sudasinghe et al. (2018), except that the lateral-line scale count is given as the number of pored lateral-line scales + the scales between the last pored scale and the base of the hypural plate + the scales on the caudal-fin base. Scales in transverse line were counted diagonally to include the scales between the dorsal-fin origin and the lateral line row, +1, plus the scales between the lateral-line row and the origin of the anal fin. All measurements and counts were taken on the left side of specimens whenever possible. Head length and body measurements are represented as proportions of standard length; and subunits of the head as proportions of head length. Values in parentheses after a count represent the frequency of that count. The names Rasboroides nigromarginatus and R. rohani as used in this text refer to the populations to which these names were applied by Batuwita, De Silva & Edirisinghe (2013).
Material
Specimens referred to in the text are deposited in the collections of the Natural History Museum, London (BMNH); the Zoological Museum Hamburg (ZMH); the Department of Molecular Biology and Biotechnology, University of Peradeniya (DZ); and the Wildlife Heritage Trust of Sri Lanka (WHT), now at the National Museum (NH) of Sri Lanka.
Morphometric analysis
Sex was determined by the presence of tubercles on the anterior margin of the pectoral fin in males. Log-transformed measurements were checked for normality using a Shapiro–Wilk test. Following the results of the Shapiro–Wilk test, independent sample t-tests were carried out to test whether the morphological measurements were independent of sexual dimorphism in each putative species. In order to remove the effect of size-allometry, measurements were standardized by using the equation where Ms is the standardized measurement, Mo is the measured character length, Ls is the overall (arithmetic) mean standard length for all individuals from all populations of all putative species, Lo is the standard length of each specimen, and b is estimated for each character from the observed data by using the allometric-growth equation M = aLb, where b is the gradient of regression of logMo on logLo (Elliott, Haskard & Koslow, 1995). The size-corrected data were used to conduct a Principal Component Analysis (PCA) of a covariance matrix. All statistical analyses were carried out in the software PAST (Hammer, Harper & Ryan, 2001).
Molecular analysis
DNA was extracted from ethanol-preserved fin clips or muscle tissues using a DNeasy Blood & tissue kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s protocols. Details of the specimens used in the molecular analysis are given in Table 1. The partial mitochondrial cytochrome b (cytb) and cytochrome oxidase subunit 1 (coi) were amplified. PCR was carried out in 25 µl reactions, using 2 µl of template DNA, 12.5 µl of mastermix MangoMix™ (Bioline, London, UK), 0.5 µl (for cytb), 0.3 µl (for coi) of each primer and 9.5 µl (for cytb), and 9.9 µl (for coi) of deionized water. The primer pair CB-J-10933 (5′ TATG TTCT ACCA TGAG GACA AATA TC 3′: Simon et al., 1994) and BSF4 (5′ CTTC TACT GGTT GTCC TCCG ATTCA 3′); FishF1 (5′TCAA CCAA CCAC AAAG ACAT TGGC AC3′) and FishR1 (5′TAGA CTTC TGGG TGGC CAA AGAA TCA3′: Ward et al., 2005) were used to amplify ∼615 bp and ∼670 bp of the cytb, and coi genes respectively. The thermal profile for cytb followed an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 40 s, annealing at 45 °C for 50 s, extension at 72 °C for 60 s and a final extension of 72 °C for 5 min; for coi, an initial denaturation at 95 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 0.5 min, annealing at 50 °C for 0.5 min, extension at 72 °C for 1 min and a final extension of 72 °C for 10 min. PCR products were visualized by an electrophoresis on 1.2% agarose gel and then purified using a PCR purification kit (Qiagen) and sequenced in both directions.
| Species | Voucher | Location | Source | cytb | coi |
|---|---|---|---|---|---|
| Rasboroides vaterifloris | DZ3759 | Sri Lanka: Kalu River basin, Gilimale | This study | MH780781 | MH780767 |
| Rasboroides vaterifloris | DZ3760 | Sri Lanka: Kalu River basin, Gilimale | This study | MH780780 | MH780766 |
| Rasboroides vaterifloris | DZ3761 | Sri Lanka: Kalu River basin, Gilimale | This study | MH780779 | MH780765 |
| Rasboroides nigromarginatus | DZ3359 | Sri Lanka: Kalu River basin, Athwelthota | This study | MH780783 | MH780769 |
| Rasboroides nigromarginatus | DZ3360 | Sri Lanka: Kalu River basin, Athwelthota | This study | MH780782 | MH780768 |
| Rasboroides pallidus | DZ3100 | Sri Lanka: Bentara River basin, Horawala | This study | MH780790 | MH780778 |
| Rasboroides pallidus | DZ3101 | Sri Lanka: Bentara River basin, Horawala | This study | MH780789 | MH780777 |
| Rasboroides pallidus | DZ3297 | Sri Lanka: Bentara River basin, Pitigala | This study | MH780788 | MH780776 |
| Rasboroides pallidus | DZ3298 | Sri Lanka: Bentara River basin, Pitigala | This study | MH780787 | MH780775 |
| Rasboroides pallidus | DZ3387 | Sri Lanka: Kalu River basin, Niriella | This study | MH780786 | MH780772 |
| Rasboroides pallidus | DZ3242 | Sri Lanka: Mahaweli River basin, Nawalapitiya | This study | MH780785 | MH780771 |
| Rasboroides pallidus | DZ3243 | Sri Lanka: Mahaweli River basin, Nawalapitiya | This study | MH780784 | MH780770 |
| Rasboroides pallidus | WHT16 | Sri Lanka: Gin River basin, Kottawa | This study | MH780794 | N/A |
| Rasboroides pallidus | WHT44 | Sri Lanka: Bentara River basin | This study | MH780795 | N/A |
| Rasboroides pallidus | WHT62 | Sri Lanka: Gin River basin | This study | MH780796 | N/A |
| Rasboroides pallidus | CTOL00534 | Not available | Genbank | HM224374 | N/A |
| Rasboroides pallidus | NRM 50310 | Not available | Genbank | EU241471 | N/A |
| Rasboroides pallidus | CBM ZF 11546 | Not available | Genbank | AP011432 | AP011432 |
| Rasboroides rohani | DZ3333 | Sri Lanka: Walawe River basin, Suriyakanda | This study | MH780791 | MH780774 |
| Rasboroides rohani | DZ3334 | Sri Lanka: Walawe River basin, Suriyakanda | This study | MH780792 | N/A |
| Rasboroides rohani | DZ3335 | Sri Lanka: Walawe River basin, Suriyakanda | This study | MH780793 | MH780773 |
| Horadandia atukorali | DZ3114 | Sri Lanka: Kalu River basin, Dombagaskanda | This study | MH780801 | MH780764 |
| Horadandia atukorali | DZ3285 | Sri Lanka: Kalu River basin, Remuna | This study | MH780800 | MH780763 |
| Horadandia atukorali | DZ3286 | Sri Lanka: Kalu River basin, Remuna | This study | MH780799 | MH780762 |
| Horadandia atukorali | WHT1J | Sri Lanka: Gin River basin, Neluwa | This study | MH780798 | MH780761 |
| Horadandia atukorali | DZ3513 | Sri Lanka: Mi Oya River basin, Galgamuwa | This study | MH780797 | MH780760 |
| Horadandia atukorali | WHT61 | Sri Lanka: Gin River basin, | This study | MH780802 | N/A |
| Neochela dadiburjori | NRM 50246 | Not available | Genbank | EU241417 | N/A |
| Neochela dadiburjori | LR1689 | Not available | Genbank | NA | FJ753506 |
| Malayochela maassi | NRM 50167 | Not available | Genbank | EU241444 | MF991139 |
| Devario regina | LR1644 | Not available | Genbank | EF151100 | FJ753489 |
Sequenced data were checked and assembled in ChromasPro v1.34 (Technelysium Pty Ltd) and contig sequences of the two strands were prepared using MEGA v. 7.0 (Kumar, Stecher & Tamura, 2016). Additional available GenBank sequences were incorporated in to the phylogenetic analysis (Table 1). The cytb and coi contig dataset were constructed and aligned separately using ClustalW in MEGA v. 7.0 (Kumar, Stecher & Tamura, 2016), improved manually and translated and checked for premature stop codons or frameshift mutations. The uncorrected pairwise genetic distances for the putative species of Rasboroides for the two partial genes cytb and coi were calculated using MEGA. A barcoding gap analysis was conducted, separately for the cytb and coi gene sequences, using the Automatic Barcode Gap Discovery (ABGD) software of Puillandre et al. (2012) to delimit the putative species of Rasboroides by employing the K2P distance and a transition/ transversion ratio of 2.
Appropriate substitution model for the cytb and coi dataset was chosen by jModelTest 2 (Darriba et al., 2012) under the Bayesian Information Criterion (BIC). Bayesian inference for the two genes cytb (609 bp) and coi (669 bp) were carried out independently in MrBayes v3.2 (Ronquist et al., 2012). Four Metropolis-coupled Markov chain Monte Carlo (MCMCMC) chains were run for 1 million generations in two independent runs (chain temperature 0.1; sample frequency 100). Using Tracer (Rambaut et al., 2014), the first 100,000 generations were determined as burn-in and discarded. The frequency of the remaining clades in trees that were sampled every one hundred generations was used as an estimate of the posterior probabilities (PP) of those clades (Huelsenbeck et al., 2001). A maximum likelihood analysis was carried out for each gene using RAxML 8.0 (Stamatakis, 2014) implementing the GTRGAMMA model (see RAxML manual v8.2.X for justification) through the CIPRES Science Gateway (Miller, Pfeiffer & Schwartz, 2010). Clade support was assessed by rapid ML bootstrap analysis with 1,000 iterations. Devario regina, Neochela dadiburjori and Malayochela maassi were designated as outgroup taxa in all the analyses.
To estimate the divergence times between Rasboroides and Horadandia, and between sister-species pairs of Rasboroides, a Bayesian approach using BEAST v.10.0 (Suchard et al., 2018) was implemented. We used the average cyprinid cytb substitution rate of 0.0082 substitutions per site per million years and a standard deviation of 0.0025 substitutions per site per million years to calibrate the mitochondrial cytb tree under a normal prior (Rüber et al., 2004; Rüber et al., 2007). This substitution rate of cytb had been derived for European cyprinids in reference to two independent and well-dated geological events (Zardoya & Doadrio, 1999). Yule Process and strict clock model were specified as the tree prior and the clock type, respectively and two independent runs of 10 million generations, each sampling the Markov Chain Monte Carlo (MCMC) chain every 1,000 generations was carried out. Tracer was used to confirm convergence between the two runs and within each run and the first 0.1% the generations were discarded as burnin. The two runs were later combined and a maximum clade credibility (MCC) tree was constructed from the posterior sample of trees using TREEANNOTATOR, and visualized using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree).
Haplotype network reconstruction for the cytb and coi genes of putative species of Rasboroides was inferred by TCS network (Clement et al., 2002) in PopArt (Leigh & Bryant, 2015). TCS (Templeton, Crandall & Sing, 1992) has been used widely to infer genealogies of populations with low genetic divergences (Clement et al., 2002). GenBank sequences were not used since they derive from aquarium specimens of uncertain provenance. The nucleotide diversity (π) and neutrality tests Tajima’s D (Tajima, 1989) and Fu and Li’s F (Fu & Li, 1993) test statistics were computed using DNAsp v.6 (Rozas et al., 2017) for the species of Rasboroides in Sri Lanka.
Results
Phylogenetic analysis
The Bayesian and Maximum likelihood (Fig. S1) phylogenetic analyses for cytb and coi recovered similar topologies; hence, the Bayesian phylogram for cytb, which includes more samples, is given (Fig. 1A). The uncorrected pairwise distances obtained for putative species of Rasboroides for cytb and coi are given in Table 2. Rasboroides rohani and R. nigromarginatus are nested within the R. pallidus and R. vaterifloris clades, respectively. The minimum uncorrected pairwise cytb and coi distance between samples identified as R. rohani and R. pallidus is 0.0% for both, and between those identified as R. nigromarginatus and R. vaterifloris, is 0.2%. The GenBank sequences identified as deriving from Rasboroides vaterifloris (EU241471, AP011432, HM224374) are all apparently of R. pallidus. The population of Rasboroides introduced in the vicinity of Ginagathena (Wikramanayake, 1990) in the Mahaweli basin too, is R. pallidus. The intraspecific uncorrected pairwise distances for cytb and coi within R. pallidus (considering R. rohani as conspecific) are 0.0–2.4% (when HM224374 is included, 0.0–1.2% if HM224374 is excluded) and 0.0–0.3%, respectively, while those for R. vaterifloris (considering R. nigromarginatus as conspecific) are 0.0–0.2% and 0.0–0.3%, respectively. Rasboroides pallidus and R. vaterifloris differ genetically by a minimum uncorrected pairwise distance of 4.1% and 3.4% for cytb and coi, respectively. The uncorrected pairwise distances between Rasboroides pallidus and H. atukorali for the cytb and coi genes are 16.3% and 12.2%, respectively, while those between R. vaterifloris and H. atukorali are 17.9% and 13.0%, respectively.
| % | R. vaterifloris | R. nigromarginatus | R. pallidus | R. rohani | ||||
|---|---|---|---|---|---|---|---|---|
| coi | cytb | coi | cytb | coi | cytb | coi | cytb | |
| R. vaterifloris | 0.0–0.2 | 0.0 | ||||||
| R. nigromarginatus | 0.2–0.3 | 0.2 | 0.0 | 0.0 | ||||
| R. pallidus | 3.4–3.5 | 4.3–4.9 (when HM224374 included), if not 4.3–4.7 | 3.4 | 4.1–4.7 | 0.0 | 0.0–2.4 (when HM224374 included), if not 0.0–1.2 | ||
| R. rohani | 3.4–3.8 | 4.5–4.9 | 3.4–3.7 | 4.3–4.7 | 0.0–0.3 | 0.0–2.0 (when HM224374 included), if not 0.0–0.8 | 0.3 | 0.0–0.4 |
The Automatic Barcode Gap Detection (ABGD) algorithm, in which groups are empirically found to correspond with species (Puillandre et al., 2012), did not identify R. nigromarginatus and R. rohani as distinct groups. It did, however, identify <R. pallidus + R. rohani >and <R. vaterifloris + R. nigromarginatus>, as distinct groups, supporting their recognition as valid species.
Divergence Timing
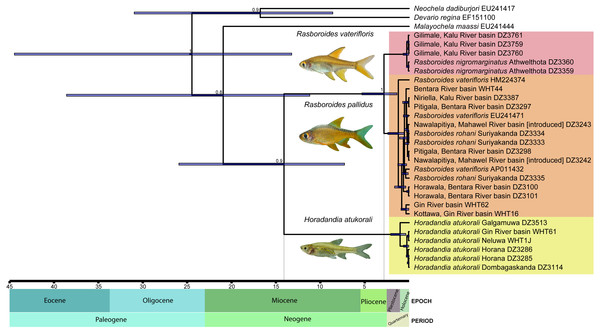
Based on the time-calibrated tree obtained (Fig. 2), the basal split between the common ancestor of Horadandia and Rasboroides occurred ∼14 mya, in the mid-Miocene, while that between R. vaterifloris and R. pallidus occurred ∼2.6 mya, in the late Pliocene (see Fig. 2 for 95% highest posterior densities).
Figure 2: A time calibrated mitochondrial tree of Horadandia arukorali and species of Rasboroides in Sri Lanka.
Bayesian posterior probabilities displayed for each major clade and node heights showing 95% highest posterior densities.Reconstruction of the haplotype network
The TCS networks for the coi (Fig. 1B) and cytb (Fig. 1C) genes formed two clearly-separated haplotype groups for R. pallidus and R. vaterifloris, with minimum 21 and 19 mutational steps, respectively. There was no sharing of haplotypes between the two species. The haplotype network reconstructed for cytb includes samples from all the river basins in which Rasboroides is encountered, other than the Kelani and Nilwala. Within, the R. pallidus haplotype group for cytb, two shared (H1, H2) and five unique haplotypes (H3–H7) are identified. One sample from the population identified as R. rohani formed a unique haplotype (H4), while the other two samples formed a shared haplotype with samples from the Bentara and Mahaweli basins (H1); samples from the Kalu and Bentara basins formed the other shared haplotype (H2). The other two samples from the Bentara (H3, H5) and Gin basins (H6, H7) formed four unique haplotypes. It is possible that the specimens of R. pallidus introduced to the Mahaweli basin near Ginigathena and the Walawe basin at Suriyakanda (R. rohani) descended from a population in the Bentara basin. Within the R. vaterifloris haplotype group for cytb, samples from Gilimale (H9) and Athwelthota (H8, R. nigromarginatus) formed two unique haplotypes. Populations of R. pallidus (13 sequences) included 11 segregating sites and six parsimony-informative sites, while populations of R. vaterifloris (five sequences) each included only a single segregating site and parsimony-informative site. The nucleotide diversity was greater in R. pallidus than in R. vaterifloris (0.00584 vs. 0.00099) and Tajima’s D-test, negative for R. pallidus (−0.76736) and positive for R. vaterifloris (1.22474), was nevertheless insignificant (p > 0.05) in both species. Fu and Li’s F-test too, while negative for R. pallidus (−0.59785) and positive for R. vaterifloris (1.15728), was insignificant (p > 0.02) for both species.
| R. pallidus (considering R. rohani as conspecific), males (n = 22, 22.2–33.6 mm SL) vs females (n = 10, 22.2–29.4 mm SL) | R. pallidus (excluding R. rohani), males (n = 12, 22.2–32.5 mm SL) vs females (n = 8, 22.2–29.4 mm SL) | Topotypic R. vaterifloris, males (n = 3, 21.2–24.2 mm SL) vs females (n = 6, 21.6–25.4 mm SL) | Topotypic R. nigromarginatus, males (n = 4, 24.7–28.9 mm SL) vs females (n = 5, 21.7–26.1 mm SL) | R. vaterifloris (considering R. nigromarginatus as conspecific), males (n = 7, 21.2–28.9 mm SL) vs females (n = 11, 21.6–26.1 mm SL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t | p | Significant (p < 0.05) | t | p | Significant (p < 0.05) | t | p | Significant (p < 0.05) | t | p | Significant (p < 0.05) | t | p | Significant (p < 0.05) | |
| Standard length | 1.580 | 0.124 | no | 0.907 | 0.376 | no | 1.204 | 0.267 | no | 1.443 | 0.192 | no | 0.541 | 0.595 | no |
| Predorsal length | 1.476 | 0.150 | no | 0.863 | 0.399 | no | 0.779 | 0.461 | no | 1.771 | 0.119 | no | 0.939 | 0.361 | no |
| Postdorsal length | 1.791 | 0.083 | no | 1.169 | 0.257 | no | 0.520 | 0.618 | no | 2.200 | 0.063 | no | 1.290 | 0.215 | no |
| Preanal length | 1.251 | 0.220 | no | 0.667 | 0.512 | no | 0.929 | 0.383 | no | 0.773 | 0.464 | no | 0.306 | 0.763 | no |
| Prepelvic length | 1.365 | 0.182 | no | 0.778 | 0.446 | no | 0.870 | 0.412 | no | 1.151 | 0.287 | no | 0.618 | 0.544 | no |
| Caudal peduncle length | 1.722 | 0.095 | no | 1.181 | 0.252 | no | 0.721 | 0.494 | no | 1.462 | 0.186 | no | 0.849 | 0.408 | no |
| Caudal peduncle depth | 2.152 | 0.039 | yes | 1.559 | 0.136 | no | 0.791 | 0.454 | no | 1.721 | 0.128 | no | 0.816 | 0.425 | no |
| Body depth | 2.375 | 0.024 | yes | 1.756 | 0.095 | no | 0.588 | 0.574 | no | 2.211 | 0.062 | no | 1.434 | 0.170 | no |
| Dorsal fin height | 3.001 | 0.005 | yes | 2.506 | 0.022 | yes | 1.047 | 0.329 | no | 3.109 | 0.017 | yes | 3.107 | 0.006 | yes |
| Dorsal fin base length | 2.683 | 0.011 | yes | 2.073 | 0.052 | no | 1.247 | 0.252 | no | 0.517 | 0.620 | no | 1.263 | 0.224 | no |
| Anal fin height | 2.747 | 0.010 | yes | 2.969 | 0.008 | yes | 0.530 | 0.611 | no | 3.996 | 0.005 | yes | 3.091 | 0.007 | yes |
| Anal fin base length | 2.194 | 0.036 | yes | 1.513 | 0.147 | no | 0.106 | 0.918 | no | 3.448 | 0.010 | yes | 2.124 | 0.049 | yes |
| Pelvic fin height | 2.545 | 0.016 | yes | 2.257 | 0.036 | yes | 0.696 | 0.508 | no | 4.131 | 0.004 | yes | 2.804 | 0.012 | yes |
| Pectoral fin height | 3.394 | 0.001 | yes | 3.446 | 0.002 | yes | 0.411 | 0.693 | no | 2.816 | 0.025 | yes | 2.399 | 0.028 | yes |
| Head length | 1.410 | 0.168 | no | 0.942 | 0.358 | no | 1.608 | 0.151 | no | 1.047 | 0.329 | no | 0.217 | 0.830 | no |
| Head depth | 1.396 | 0.172 | no | 0.940 | 0.359 | no | 2.027 | 0.082 | no | 1.902 | 0.098 | no | 0.654 | 0.522 | no |
| Snout length | 0.945 | 0.351 | no | 1.224 | 0.236 | no | 0.732 | 0.487 | no | 2.236 | 0.060 | no | 1.022 | 0.321 | no |
| Eye diameter | 1.286 | 0.208 | no | 0.931 | 0.363 | no | 0.192 | 0.852 | no | 1.302 | 0.234 | no | 0.950 | 0.356 | no |
| Inter orbital width | 1.994 | 0.055 | no | 1.789 | 0.090 | no | 1.099 | 0.307 | no | 2.150 | 0.068 | no | 0.896 | 0.383 | no |
| Inter narial width | 1.528 | 0.136 | no | 1.836 | 0.082 | no | 0.234 | 0.820 | no | 1.306 | 0.232 | no | 0.822 | 0.422 | no |
Within, the R. pallidus haplotype group for coi, the samples identified as R. rohani formed two unique haplotypes (H2, H3) while those from the Bentara, Kalu and Mahaweli basins formed a single shared haplotype (H1). Within the R. vaterifloris haplotype group for coi, samples from Gilimale (H4, H5) and Athwelthota (H6, R. nigromarginatus) formed three unique haplotypes. Populations of R. pallidus (nine sequences) included nine segregating sites but no parsimony-informative sites, while populations of R. vaterifloris (five sequences) included only two segregating sites and two parsimony-informative sites. The nucleotide diversity was similar in both R. pallidus and R. vaterifloris (0.00100 vs. 0.00179) and Tajima’s D-test, negative for R. pallidus (−1.51297) and positive for R. vaterifloris (1.45884), was nevertheless not significant (p > 0.05) in both species. Fu and Li’s F-test was negative for R. pallidus (−1.82046) and positive for R. vaterifloris (1.43161), but insignificant (p > 0.02) for both species.
Statistical analysis
The Shapiro–Wilk test showed all the measurements to be normally distributed (p > 0.05) among the sexes of R. pallidus, R. vaterifloris and the population referred to as R. nigromarginatus by Batuwita, De Silva & Edirisinghe (2013). The results of the independent sample t-tests for each measurement of the species are given in Table 3. Standard length, predorsal length, postdorsal length, preanal length, prepelvic length, caudal peduncle length, head length, head depth, snout length, eye diameter, interorbital width and internarial width were not significantly different (p > 0.05) between the sexes for any of the putative species. When R. rohani and R. nigromarginatus were considered as conspecific with R. pallidus and R. vaterifloris, respectively, dorsal-fin height, anal-fin height, anal-fin base length, pelvic-fin height and pectoral-fin height were significantly different (p < 0.05) between the sexes of each species.
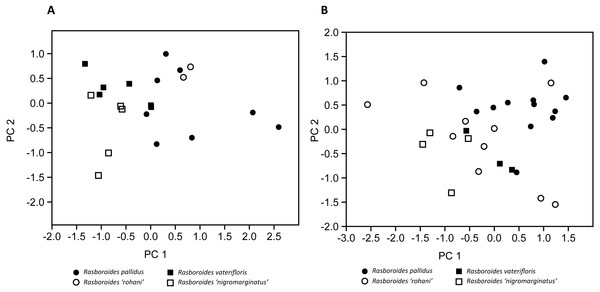
Figure 3: Plot of scores from the principal component analysis of size corrected measurements from putative species of (A) females, and (B) males of Rasboroides in Sri Lanka.
The results of the size corrected PCA are given in Fig. 3 and Table 4: in females, PC1 and PC2 explained 48.30% and 18.35% of the total variance, respectively. The females of R. pallidus and R. rohani are clustered together, while the females of R. vaterifloris and R. nigromarginatus form a separate cluster, with only a slight overlap between the two clusters. In males, PC1 and PC2 explained 33.82% and 19.35% of the total variance, respectively. The males of R. pallidus and R. rohani are clustered together, while the males of R. vaterifloris and R. nigromarginatus form a slightly overlapping cluster. When R. rohani and R. nigromarginatus are considered as conspecific with R. pallidus and R. vaterifloris, respectively, the males form an overlapping cluster, while the females form almost non-overlapping cluster.
Hence, based on the multiple criteria assessed, we consider R. rohani and R. nigromarginatus to be junior synonyms of R. pallidus and R. vaterifloris, respectively.
| Males | Females | |||
|---|---|---|---|---|
| PC 1 | PC 2 | PC 1 | PC 2 | |
| Eigen value | 0.988 | 0.565 | 1.063 | 0.404 |
| Variance explained % | 33.82 | 19.35 | 48.3 | 18.35 |
| Predorsal length | 0.1934 | 0.2198 | 0.3866 | −0.1012 |
| Postdorsal length | 0.0567 | 0.2263 | 0.2073 | −0.0098 |
| Preanal length | 0.2755 | 0.3472 | 0.3444 | −0.4544 |
| Prepelvic length | 0.2928 | 0.2626 | 0.3176 | −0.1383 |
| Caudal peduncle length | −0.0061 | −0.1260 | −0.1504 | 0.0541 |
| Caudal peduncle depth | 0.0836 | 0.1191 | 0.1098 | −0.0500 |
| Body depth | 0.4038 | 0.3988 | 0.4521 | −0.0451 |
| Dorsal fin height | 0.4391 | −0.3579 | 0.1367 | 0.3130 |
| Dorsal fin base length | 0.1058 | −0.0217 | 0.0560 | −0.2556 |
| Anal fin height | 0.5056 | −0.5355 | 0.2670 | 0.6388 |
| Anal fin base length | −0.0001 | 0.0247 | 0.1784 | −0.0824 |
| Pelvic fin height | 0.2618 | −0.154 | 0.1025 | 0.3328 |
| Pectoral fin height | 0.2171 | 0.0893 | 0.3201 | 0.2164 |
| Head length | 0.1237 | 0.0652 | 0.0555 | −0.0255 |
| Head depth | 0.1717 | 0.2032 | 0.2948 | 0.0159 |
| Snout length | 0.0361 | 0.1567 | 0.1236 | −0.0815 |
| Eye diameter | 0.0397 | −0.0051 | −0.0066 | −0.0235 |
| Inter orbital width | 0.0563 | 0.0172 | 0.0462 | −0.1117 |
| Inter narial width | 0.0350 | 0.0594 | 0.0579 | −0.0043 |
Species descriptions
General morphology
The following characters are common to R. pallidus and R. vaterifloris. Head and body laterally compressed. Body depth greatest at dorsal-fin origin. Dorsal profile of head concave behind level of eye; predorsal profile rising gently thereafter to origin of dorsal fin; postdorsal profile slightly concave. Ventral profile slightly convex up to origin of pelvic fin, straight from pelvic-fin origin to anal-fin origin, concave thereafter to base of caudal fin. Snout short, shorter than eye diameter, rounded in dorsal aspect, subtriangular in lateral aspect. Mouth terminal, rictus just passing vertical through anterior margin of eye. Symphysial knob present, minute, rounded, fitting into shallow groove on inner margin of upper jaw with mouth closed. Dorsal fin anterior margin straight, posterior margin slightly concave, its origin located just posterior to vertical through origin of pelvic fin. Tip of longest ray of dorsal fin, when adpressed, reaching beyond vertical through origin of anal fin. Pectoral fin originating ventrolaterally, immediately posterior to opercular membrane, its adpressed tip reaching beyond vertical through origin of pelvic fin. Adpressed tip of pelvic fin reaching vertical through base of 2nd branched anal-fin ray. Adpressed tip of anal fin reaching beyond midpoint of caudal peduncle.
Sexual dimorphism
Male specimens >21 mm SL have a series of conical tubercles along the anterior margin of the pectoral fin, a character absent in females. Male specimens >25 mm SL also have 7–10 bands of minute conical tubercles on the anterior lower jaw, reaching just beyond the rictus; and 1–3 bands of minute conical tubercles along the posterior margin of the preopercle, both characters absent in females. Some male specimens >25 mm SL possess a single row of tubercles beneath the eye, absent in females. Males generally with a greater body depth (31.8–44.4% SL vs. 31.7–35.5, t = 2.375, p = 0.024 in R. pallidus; 29–34.2% SL vs. 27.2–31.9, t = 1.434, p = 0.170 in R. vaterifloris). The male cranium shows a prominent concavity behind the level of the eye, and males have longer pectoral (23.7–30.5% SL vs. 21.5–25.3, t = 3.394, p = 0.001, in R. pallidus; 22.5–24.8% SL vs. 19.9–22.6, t = 2.399, p = 0.028 in R. vaterifloris), pelvic (21.4–31.6% SL vs. 21.1–25.4, t = 2.545, p = 0.016 in R. pallidus; 22.9–27.3% SL vs. 20-23.1, t = 2.804, p = 0.012 in R. vaterifloris), anal (24.5–40.8% SL vs. 22.9–31.5, t = 2.747, p = 0.010 in R. pallidus; 28.1–31.9% SL vs. 20.4–27.6, t = 3.091, p = 0.007 in R. vaterifloris) and dorsal fins (31.4–41.4% SL vs. 30.2–35, t = 3.001, p = 0.005 in R. pallidus; 32.6–36.8% SL vs. 29.1–33.1, t = 3.107, p = 0.006 in R. vaterifloris) than females of the respective species. Further, males of both R. pallidus and R. vaterifloris are more brightly colored than females (Fig. 4).
Figure 4: Live color pattern variation in A–D, R. vaterifloris; E–J, R. pallidus.
(A) topotypes of R. vaterifloris, Kalu basin, Gilimale; (B–D) topotypes of population identified as R. nigromarginatus by Batuwita, De Silva & Edirisinghe (2013), Kalu basin, Athwelthota; (E) Bentara basin, Pitigala; (F) topotypes of population identified as R. rohani by Batuwita, De Silva & Edirisinghe (2013), Walawe basin, Suriyakanda; (G) Bentara basin, Yagirala; (H) Gin basin, Udugama; (I) Bentara basin, Yagirala; (J) Bentara basin, Pitigala. (A, B, D, E, F, G, H) males; (C, I, J) females. Specimens not collected.Rasboroides vaterifloris Deraniyagala, 1930
| Rasbora vaterifloris Deraniyagala, 1930: 129 |
| Rasbora nigromarginata Meinken, 1957: 65–68 |
| Rasbora vaterifloris var. nigromarginatus Deraniyagala, 1958: 137 |
| Rasboroides nigromarginatus (Meinken, 1957): Batuwita, De Silva & Edirisinghe, 2013 |
Material examined (all from Sri Lanka). DZ3768, topotypes of R. vaterifloris, 9, 21.2–25.4 mm SL, Kalu basin, Gilimale, 6°45′44.9″N 80°25′34.4″E; DZ3970, topotypes of ‘R. nigromarginatus’, 9, 21.7–28.9 mm SL, Kalu basin, Athwelthota, 6°32′35.7″N 80°16′38.0″E. Material not included in morphometric analysis: BMNH 1930.10.8.1, Rasbora vaterifloris, syntype, 25.7 mm SL, Kalu basin, near Illukvattai [Illukwatta] ferry on the Ratnapura to Gilimale road; WHT578, Kalu basin, Athwelthota.
Diagnosis. Males of Rasboroides vaterifloris can be distinguished from males of R. pallidus by having the unbranched rays of dorsal, anal, pectoral and pelvic fins black along their entire length, more distinctly evident in the last unbranched ray of the dorsal fin (vs. the mentioned rays being the same color as other rays; in preserved specimens, interradial membranes of dorsal, anal, pelvic and pectoral fins with distinct, scattered melanophores (vs. absent or vaguely present only around the beginning). Females of R. vaterifloris have a lesser body depth (27.2–31.9% SL vs. 31.7–35.5) than those of R. pallidus.
Figure 5: Depiction of Rasboroides species in preservation.
(A) Syntype of Rasboroides vaterifloris, BMNH 1930.10.8.1, 25.7 mm SL, Kalu basin, Illukwatta; (B) holotype of R. nigromarginatus, ZMH 1207, 35.5 mm SL, ‘Ceylon’ (SriLanka); (C–D) topotypes of R. vaterifloris, Kalu basin, Gilimale: (C) male, DZ3768A, 24.2 mm SL, (D) female, DZ3768D, 24.4 mm SL; (E–F) topotypes of population identified as R. nigromarginatus by Batuwita, De Silva & Edirisinghe (2013), Kalu basin, Athwelthota: (E) male, DZ3970A, 28.9 mm SL, (F) female, DZ3970E, 26.1 mm SL; (G–H) R. pallidus: (G) male, DZ3972B, 31.0 mm SL, Bentara basin, Lewwanduwa, (H) female, DZ3973A, 29.4 mm SL, Bentara basin, Pitigala; (I–J) topotypes of population identified as R. rohani by Batuwita, De Silva & Edirisinghe (2013), Walawe basin, Suriyakanda: (I) male, DZ3971B, 31.9 mm SL, (J) female, DZ3971E, 28.9 mm SL.| R. vaterifloris | R. nigromarginatus | R. vaterifloris (considering R. nigromarginatus as conspecific) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| males (n = 3) | females (n = 6) | males (n = 4) | females (n = 5) | males (n = 7) | females (n = 11) | |||||||||||
| min | max | min | max | min | max | min | max | min | max | mean | s.d | min | max | mean | s.d | |
| Standard length | 21.2 | 24.2 | 21.6 | 25.4 | 24.7 | 28.9 | 21.7 | 26.1 | 21.2 | 28.9 | 21.6 | 26.1 | ||||
| In percent of standard length | ||||||||||||||||
| Predorsal length | 51.9 | 53.8 | 50.9 | 53.4 | 52.7 | 54.0 | 50.2 | 53.5 | 51.9 | 54.0 | 53.0 | 0.8 | 50.2 | 53.5 | 52.0 | 1.0 |
| Postdorsal length | 51.9 | 53.4 | 50.0 | 52.1 | 52.3 | 53.9 | 50.0 | 52.2 | 51.9 | 53.9 | 53.0 | 0.7 | 50.0 | 52.2 | 51.3 | 0.8 |
| Preanal length | 60.4 | 62.4 | 58.4 | 63.2 | 58.7 | 61.6 | 59.5 | 65.1 | 58.7 | 62.4 | 60.9 | 1.2 | 58.4 | 65.1 | 61.4 | 2.0 |
| Prepelvic length | 45.8 | 47.2 | 44.7 | 46.7 | 44.2 | 46.6 | 45.1 | 46.7 | 44.2 | 47.2 | 45.9 | 1.0 | 44.7 | 46.7 | 45.8 | 0.7 |
| Caudal peduncle length | 23.6 | 25.5 | 23.2 | 24.7 | 23.6 | 24.9 | 22.4 | 26.8 | 23.6 | 25.5 | 24.4 | 0.7 | 22.4 | 26.8 | 24.1 | 1.2 |
| Caudal peduncle depth | 12.2 | 12.8 | 11.1 | 13.2 | 11.7 | 13.0 | 11.0 | 12.5 | 11.7 | 13.0 | 12.3 | 0.5 | 11.0 | 13.2 | 12.2 | 0.8 |
| Body depth | 29.0 | 31.0 | 27.2 | 31.3 | 31.5 | 34.2 | 27.2 | 31.9 | 29.0 | 34.2 | 31.6 | 1.7 | 27.2 | 31.9 | 29.7 | 1.6 |
| Dorsal fin height | 33.1 | 36.8 | 31.0 | 33.1 | 32.6 | 36.7 | 29.1 | 31.4 | 32.6 | 36.8 | 34.6 | 1.9 | 29.1 | 33.1 | 31.0 | 1.2 |
| Dorsal fin base length | 12.4 | 14.5 | 11.1 | 13.0 | 11.8 | 12.9 | 11.6 | 13.5 | 11.8 | 14.5 | 12.9 | 0.9 | 11.1 | 13.5 | 12.4 | 0.9 |
| Anal fin height | 28.1 | 28.6 | 24.1 | 27.6 | 28.6 | 31.9 | 20.4 | 27.2 | 28.1 | 31.9 | 29.6 | 1.5 | 20.4 | 27.6 | 25.0 | 2.1 |
| Anal fin base length | 15.0 | 15.8 | 13.4 | 15.9 | 15.3 | 16.6 | 13.5 | 15.2 | 15.0 | 16.6 | 15.7 | 0.6 | 13.4 | 15.9 | 14.5 | 0.8 |
| Pelvic fin height | 23.4 | 27.3 | 21.3 | 23.1 | 22.9 | 25.6 | 20.0 | 23.1 | 22.9 | 27.3 | 24.5 | 1.5 | 20.0 | 23.1 | 22.0 | 1.0 |
| Pectoral fin height | 23.2 | 24.8 | 20.9 | 22.6 | 22.5 | 24.6 | 19.9 | 22.5 | 22.5 | 24.8 | 23.8 | 0.8 | 19.9 | 22.6 | 21.5 | 1.0 |
| Head length | 26.1 | 28.8 | 26.0 | 28.1 | 25.2 | 28.4 | 25.4 | 27.2 | 25.2 | 28.8 | 26.5 | 1.5 | 25.4 | 28.1 | 26.9 | 0.8 |
| Head depth | 18.7 | 19.4 | 18.6 | 21.0 | 19.9 | 21.2 | 19.3 | 20.2 | 18.7 | 21.2 | 19.9 | 0.9 | 18.6 | 21.0 | 19.7 | 0.7 |
| In percent of head length | ||||||||||||||||
| Snout length | 18.1 | 25.4 | 19.5 | 23.4 | 21.5 | 28.6 | 20.0 | 23.6 | 18.1 | 28.6 | 22.8 | 3.6 | 19.5 | 23.6 | 21.3 | 1.4 |
| Eye diameter | 42.9 | 46.0 | 39.0 | 43.3 | 39.2 | 44.5 | 38.4 | 42.9 | 39.2 | 46.0 | 42.6 | 2.4 | 38.4 | 43.3 | 41.2 | 1.6 |
| Inter orbital width | 32.2 | 36.6 | 32.3 | 36.4 | 35.8 | 39.7 | 31.5 | 37.2 | 32.2 | 39.7 | 36.2 | 2.4 | 31.5 | 37.2 | 34.6 | 1.9 |
| Inter narial width | 19.7 | 22.3 | 17.0 | 21.3 | 19.2 | 24.4 | 19.2 | 21.9 | 19.2 | 24.4 | 20.9 | 1.9 | 17.0 | 21.9 | 19.9 | 1.4 |
Description. For general appearance, see Figs. 4A–4D, Figs. 5A–5F; morphometric data are provided in Table 5. Largest female 26.1 mm SL. Largest male 28.9 mm SL.
Dorsal fin with three unbranched and 7 (8) branched rays; first unbranched ray minute, less than length of second; second unbranched ray stiff, less than half length of third. Anal fin with three unbranched and 6 (8) branched rays; first unbranched ray minute, less than length of second; second unbranched ray stiff, less than half length of third. Pelvic fin with one simple and seven (8) branched rays. Origin of pelvic fin slightly anterior to vertical through origin of dorsal fin. Pectoral fin with one simple and 10 (1), 11 (6) or 12 (1) branched rays. Caudal fin forked, with 9+8 (7) branched rays in upper and lower lobe, respectively. Lower caudal-fin lobe slightly longer than upper.
Lateral body scales chaotically arranged. Lateral line incomplete, with 26 (2), 27 (3), 28 (1), 29 (1) or 30 (1) + 1 scales; pored scales 2 (2), 3 (4), 4 (2) or 5 (1). Scales in transverse series 8 (8). Circumpeduncular scales 10 (8). Predorsal scales 12 (4), 13 (1) or 15 (2). Prepelvic scales 15 (2), 16 (2) or 17 (3).
Coloration (in life) variable; at least three color morphs present. The male populations of R. vaterifloris from Gilimale, Parakaduwa, Madakada (Kalu basin) and Labugama (Kelani basin) are generally dull colored, with silvery-grey-brown body coloration, becoming lighter ventrally (Fig. 4A). Dorsal fin and lower lobe of caudal fin yellowish. Upper lobe of caudal fin, anal, pectoral, and pelvic fins dull colored. Male R. vaterifloris from Athwelthota, Runakanda, Pahiyangala, Kiriella (Kalu basin) laterally and dorsally bright golden-orange, becoming lighter ventrally (Fig. 4B). Dorsal, anal, pectoral, and pelvic fins golden orange. Lower lobe of caudal fin more golden orange than the upper lobe. A few individuals of R. vaterifloris from Athwelthota, Runakanda, Pahiyangala, Kiriella (Kalu basin) are greyish blue, becoming lighter ventrally (Fig. 4D). Females less brightly colored in all three morphs (Fig. 4C). Unbranched rays of dorsal, anal, pectoral and pelvic fins in males of all three color morphs black along their entire length, more distinctly in last unbranched dorsal-fin ray; these rays the same color as the remainder of the fin in females. Interradial membranes of dorsal, anal, pelvic and pectoral fins with scattered melanophores in males, absent in females. Upper half of sclera generally golden orange. Melanophores densely arranged on sclera above and below iris, giving appearance of a black bar on eye. Opercle tinged with red.
In preservative, overall body color brown, becoming lighter ventrally. Interradial membranes of dorsal, anal, pelvic and pectoral fins with scattered melanophores in males, uniform in females. Caudal fin hyaline. Last unbranched dorsal-fin ray of males blackish along its entire length.
Rasboroides pallidus Deraniyagala, 1958
| Rasbora vaterifloris pallida Deraniyagala, 1958: 136. |
| Rasbora vaterifloris ruber Deraniyagala, 1958: 136. |
| Rasbora vaterifloris rubioculis Deraniyagala, 1958: 136. |
| Rasboroides rohani Batuwita, De Silva & Edirisinghe, 2013 |
Material examined (all from Sri Lanka). DZ3972, 5, 24.3–32.5 mm SL, Bentara basin, Walallawita, 6°24′53.3″N 80°06′35.0″E; DZ3973, 8, 22.8–27.4 mm SL, Bentara basin, Pitigala, 6°22′29.5″N 80°14′22.6″E; DZ3904, 7, 22.2–26.2 mm SL, Gin basin, Ma dola, 6°13′46.3″N 80°21′59.9″E; DZ3971, topotypes of ‘R. rohani’, 12, 25.9–33.6 mm SL, Walawe basin, Suriyakanda, 6°27′02.3″N 80°37′00.6″E. Material not included in morphometric data: 2013.22.01 NH, ‘Rasboroides rohani’, holotype?, 32.0 mm SL, Walawe basin, Suriyakanda; 2013.24.01 NH-2013.24.11 NH, ‘R. rohani’, paratypes, 11, 23.2–29.3 mm SL, Walawe basin, Suriyakanda; NH uncatalogued, ‘R. rohani’, holotype?, 32.2 mm SL, Walawe basin, Suriyakanda; NH uncatalogued, 2, 28.9–29.3 mm SL, ‘R. rohani’, paratypes?, Walawe basin, Suriyakanda; WHT30019, Mahaweli basin, Ginigathena; WHT30667, Gin basin, Udugama ela; WHT127, Bentara basin, Mahakalupahana, Horawala; WHT30049, Bentara basin, Bambarawana; WHT30607, Gin basin, Kottawa.
Diagnosis. The males of Rasboroides pallidus can be distinguished from the males of R. vaterifloris by having the unbranched rays of dorsal, anal, pectoral and pelvic fins the same color as other branched rays (vs. black along their entire length); in preserved specimens, interradial membranes of dorsal, anal, pelvic and pectoral fins without distinct scattered melanophores throughout or with only minute, vague melanophores only around the beginning (vs. melanophores distinctly present). The females of R. pallidus have greater body depth (31.7–35.5% SL vs. 27.2–31.9) than females of R. vaterifloris.
Description. For general appearance, see Figs. 4E–4J, Figs. 5G–5J; morphometric data are provided in Table 6. Largest female 29.4 mm SL. Largest male 33.6 mm SL.
| R. pallidus | R. rohani | R. pallidus (considering R. rohani as conspecific) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| males (n = 12) | females (n = 8) | males (n = 10) | females (n = 2) | males (n = 22) | females (n = 10) | |||||||||||
| min | max | min | max | min | max | min | max | min | max | mean | s.d | min | max | mean | s.d | |
| Standard length | 22.2 | 32.5 | 22.2 | 29.4 | 25.6 | 33.6 | 28.8 | 28.9 | 22.2 | 33.6 | 22.2 | 29.4 | ||||
| In percent of standard length | ||||||||||||||||
| Predorsal length | 52.8 | 55.7 | 52.3 | 58.2 | 50.9 | 54.9 | 52.6 | 52.8 | 50.9 | 55.7 | 53.8 | 1.4 | 52.3 | 58.2 | 54.2 | 2.0 |
| Postdorsal length | 50.5 | 56.7 | 51.4 | 54.0 | 52.3 | 56.0 | 52.8 | 54.0 | 50.5 | 56.7 | 53.9 | 1.8 | 51.4 | 54.0 | 52.9 | 0.9 |
| Preanal length | 58.6 | 66.2 | 61.0 | 66.7 | 59.4 | 64.3 | 61.9 | 62.7 | 58.6 | 66.2 | 62.3 | 2.0 | 61.0 | 66.7 | 63.1 | 1.8 |
| Prepelvic length | 45.1 | 50.2 | 46.3 | 50.0 | 45.4 | 52.1 | 47.5 | 47.6 | 45.1 | 52.1 | 47.6 | 1.6 | 46.3 | 50.0 | 47.9 | 1.3 |
| Caudal peduncle length | 21.4 | 25.4 | 21.8 | 23.3 | 21.1 | 27.8 | 23.6 | 24.4 | 21.1 | 27.8 | 23.5 | 1.8 | 21.8 | 24.4 | 22.9 | 0.8 |
| Caudal peduncle depth | 11.8 | 14.8 | 11.1 | 14.0 | 11.8 | 13.9 | 11.8 | 13.2 | 11.8 | 14.8 | 13.1 | 0.7 | 11.1 | 14.0 | 12.6 | 1.0 |
| Body depth | 32.9 | 44.4 | 31.7 | 34.7 | 31.8 | 40.3 | 35.0 | 35.5 | 31.8 | 44.4 | 36.6 | 3.2 | 31.7 | 35.5 | 33.4 | 1.3 |
| Dorsal fin height | 32.2 | 37.9 | 30.2 | 35.0 | 31.4 | 41.4 | 32.3 | 33.6 | 31.4 | 41.4 | 36.1 | 2.4 | 30.2 | 35.0 | 32.7 | 1.6 |
| Dorsal fin base length | 12.0 | 14.2 | 10.6 | 14.2 | 11.5 | 15.0 | 11.9 | 12.9 | 11.5 | 15.0 | 13.4 | 0.9 | 10.6 | 14.2 | 12.4 | 1.2 |
| Anal fin height | 28.2 | 34.2 | 22.9 | 28.7 | 24.5 | 40.8 | 28.9 | 31.5 | 24.5 | 40.8 | 31.5 | 3.7 | 22.9 | 31.5 | 27.3 | 2.3 |
| Anal fin base length | 14.6 | 18.2 | 13.2 | 16.7 | 13.9 | 18.5 | 15.6 | 16.4 | 13.9 | 18.5 | 16.5 | 1.2 | 13.2 | 16.7 | 15.4 | 1.2 |
| Pelvic fin height | 22.8 | 27.4 | 21.1 | 24.3 | 21.4 | 31.6 | 23.2 | 25.4 | 21.4 | 31.6 | 25.7 | 2.3 | 21.1 | 25.4 | 23.1 | 1.3 |
| Pectoral fin height | 24.4 | 28.6 | 21.5 | 25.3 | 23.7 | 30.5 | 24.7 | 25.0 | 23.7 | 30.5 | 27.1 | 2.0 | 21.5 | 25.3 | 23.8 | 1.3 |
| Head length | 24.4 | 28.2 | 25.5 | 27.8 | 24.7 | 28.6 | 27.4 | 27.5 | 24.4 | 28.6 | 26.9 | 1.2 | 25.5 | 27.8 | 26.8 | 0.9 |
| Head depth | 20.6 | 24.0 | 20.7 | 23.6 | 19.6 | 23.7 | 21.5 | 22.3 | 19.6 | 24.0 | 22.0 | 1.2 | 20.7 | 23.6 | 21.9 | 1.0 |
| In percent of head length | ||||||||||||||||
| Snout length | 20.7 | 31.2 | 21.0 | 32.9 | 21.6 | 25.4 | 25.4 | 29.2 | 20.7 | 31.2 | 25.3 | 3.0 | 21.0 | 32.9 | 25.6 | 3.5 |
| Eye diameter | 36.1 | 44.5 | 38.5 | 43.1 | 36.6 | 44.5 | 38.0 | 39.3 | 36.1 | 44.5 | 39.9 | 2.7 | 38.0 | 43.1 | 40.7 | 1.6 |
| Inter orbital width | 34.3 | 40.0 | 29.2 | 39.5 | 33.0 | 36.8 | 33.0 | 36.8 | 33.0 | 40.0 | 36.0 | 1.8 | 29.2 | 39.5 | 35.1 | 3.0 |
| Inter narial width | 20.0 | 26.7 | 17.8 | 23.1 | 17.5 | 22.8 | 22.8 | 22.8 | 17.5 | 26.7 | 21.5 | 1.9 | 17.8 | 23.1 | 21.0 | 2.0 |
Dorsal fin with three unbranched and 7 (8) branched rays; first unbranched ray minute, less than length of second; second unbranched ray stiff, less than half length of third. Anal fin with three unbranched and (11) branched rays; first unbranched ray minute, less than length of second; second unbranched ray stiff, less than half length of third. Pelvic fin with one simple and seven (11) branched rays. Origin of pelvic fin slightly anterior to vertical through origin of dorsal fin. Pectoral fin with one simple and 10 (6) or 11 (5) branched rays. Caudal fin forked, with 9+8 (11) branched rays in upper and lower lobe, respectively. Lower caudal-fin lobe slightly longer than upper.
Lateral body scales chaotically arranged. Lateral line incomplete, with 24 (1), 25 (4), 26 (3), 27 (1), 28 (3), 29 (1) or 30 (1) + 1 scales; pored scales 2 (2), 3 (3), 4 (5), 5 (3) or 6 (1). Scales in transverse series 7 (3), 8 (5) or 8 (6). Circumpeduncular scales 8 (7) or 10 (7). Predorsal scales 10 (1), 11 (3), 12 (7), or 13 (2). Prepelvic scales 12 (1), 14 (2), 15 (6), 16 (1), 17 (1), 18 (1) or 19 (1).
Coloration (in life), variable, at least four color morphs present: orange, blue-grey, yellow and dull red (Figs. 4E–4J). Out of these, most common is the orange morph. However, in most of localities, 2–3 color morphs of R. pallidus generally occur together. Females less brightly colored in all morphs. Upper half of sclera generally golden orange. Melanophores densely arranged on sclera above and below iris, giving the appearance of a black bar on eye. Opercle tinged with red. Lower lobe of caudal fin more brightly colored than upper lobe.
In preservative, overall body color brown, becoming lighter ventrally. All fins hyaline.
Habitat, distribution and natural history
Based on the present data, R. vaterifloris is restricted primarily to the Kalu basin, where it is recorded in several scattered localities in the elevation range of 31–169 m asl (Fig. 1D). However, we also recorded a small population of R. vaterifloris close to Labugama, which lies within the Kelani basin.
Rasboroides pallidus is more widely distributed than R. vaterifloris, being recorded from several localities within and between the Kalu, Bentara, Gin and Nilwala basins, in the elevation range 17–231 m asl. We have not encountered the two species in syntopy, though R. pallidus and R. vaterifloris occur in close proximity in some localities around Badureliya, near Mathugama, within the Kalu basin. The translocated population of R. pallidus in the Mahaweli basin occurs at an elevation of about 600 m asl and appears to have established itself successfully in the vicinity of Ginigathhena. The population described by Batuwita, De Silva & Edirisinghe (2013) as ‘R. rohani’ from Suriyakanda in the Walawe basin is evidently an undocumented translocation of R. pallidus (see ‘Discussion’). Both R. vaterifloris and R. pallidus usually occur in clear, shaded, slow-flowing streams and rivers. They are found often in large groups of more than 100 individuals, usually occupying the upper half of the water column, close to the margins.
Discussion
Translocations
As explained by P.B. Moyle in Pethiyagoda (1991:36) and Wikramanayake (1990), Senanayake and Moyle conducted an experimental study to establish refugial populations of several freshwater-fish species they considered to be threatened, by translocating them to a waterway near Ginigathena, in the upper regions of the Mahaweli basin. The fishes they targeted were all lowland-rainforest species—Pethia reval (as Barbus cumingii), P. nigrofasciata (as B. nigrofasciatus), Puntius titteya (as B. titteya) and R. pallidus (as Rasbora vaterifloris). Although the translocated fishes had been caught from the wild, their provenance was not in every case documented. However, the approximate locality and river basin of origin for three species is mentioned in Wikramanayake (1990), based on which the founder population of the translocated Rasboroides was stated to have been from Parakaduwa in the Kelani basin (Wikramanayake, 1990, table 2). Parakaduwa, however, drains to the Kalu basin, and Deraniyagala (1958) and Batuwita, De Silva & Edirisinghe (2013) both recorded Rasboroides vaterifloris from this locality. However, our molecular and morphological analysis shows the introduced Ginigathena population to be R. pallidus, casting doubt on the origin declared by Wikramanayake (1990). Based on the concordance of our haplotype network for coi and cytb, it seems plausible that the Ginigathena population of R. pallidus was founded by individuals drawn from the Bentara basin, in which it remains relatively common. In the almost four decades since these introductions were made, R. pallidus, Pethia nigrofasciata, Pethia reval and Puntius titteya, all of which were until then absent from the Mahaweli basin, have established substantial populations around Ginigathena. To this extent, the goal of establishing a refugial population of these species appears to have been met.
We show here that the Suriyakanda population of Rasboroides that Batuwita, De Silva & Edirisinghe (2013) named ‘R. rohani’ is in fact R. pallidus. As a result of selecting a series of R. pallidus and ‘R. rohani’ of different size ranges for either sex (see below), these authors evidently inferred morphological differences that should correctly have been attributed to allometry. What is more, the type material of ‘R. rohani’ declared by Batuwita, De Silva & Edirisinghe (2013) to be lodged in National Museum of Sri Lanka is discrepant with the specimens present in NH. The holotype specimen, according to Batuwita, De Silva & Edirisinghe (2013), measures 34.0 mm SL and was collected by S. Udugampala and R. Krishantha on 3 June 2013. The specimen catalogued as being the holotype of R. rohani (2013.22.01 NH), however, measures 32.0 mm SL and has been collected by S. Batuwita and S. Udugampola on 2 June 2012. There is a further uncatalogued specimen of ‘R. rohani’ labeled ‘holotype’, which measures 32.2 mm SL and has been collected by S. Udagampola and R. Krishantha on 1 July 2013. Similarly, the 11 paratypes of ‘R. rohani’ catalogued as 2013.24.01 NH-2013.24.11 NH measure 23.2–29.3 mm SL and have the same collector and date of collection as 2013.22.01 NH (the specimen presently registered as the holotype). According to Batuwita, De Silva & Edirisinghe (2013), WHT 9712 included 11 paratypes which measure 28.2–32.0 mm SL and were collected by R. Krishantha on 9 Feb 2012. The NH collection has in addition, two uncatalogued specimens of ‘R. rohani’ labeled ‘paratypes’, which measure 28.9–29.3 mm SL and were collected by S Udagampola and R Krishantha on 1 July 2013. Given the differences in the sizes, dates and collectors between those declared in Batuwita, De Silva & Edirisinghe (2013) and those actually registered at NH, it is impossible to identify with certainty the name-bearing type of R. rohani. Based on the topotypic material examined herein, and our molecular analysis, however, there is no doubt that the population Batuwita, De Silva & Edirisinghe (2013) described as ‘R. rohani’ derives from a translocated individuals of R. pallidus, the genus Rasboroides having been absent from the Walawe basin prior to this introduction.
In June 2018, a conservationist disclosed to two of us (HS and RP) on condition of anonymity that he had ca 2003 translocated, apparently in good faith, several specimens each of Rasboroides (evidently R. pallidus), Devario pathirana, Puntius titteya, Pethia nigrofasciata and Malpulutta kretseri to the stream at Suriyakanda that is the type locality of both ‘R. rohani’ and Schistura madhavai (Sudasinghe, 2017). Except for Pethia nigrofasciata, these species were previously not known from the Walawe basin (Deraniyagala, 1930; Senanayake, 1980; Pethiyagoda, 1991; present data). Our sampling of this stream suggests that M. kretseri and D. pathirana failed to establish there. Puntius titteya and Pethia nigrofasciata, however, still persist at this location, together with R. pallidus.
While translocation is recognized as a valid intervention in the conservation of freshwater fishes threatened in their natural habitat (Galloway et al., 2016), the translocation of freshwater organisms, especially if poorly planned or involving the transfer of species between basins, risks confronting the host community with threats including competition for resources, predation, and the unintentional transfer of pathogens, parasites and non-target species (Minckley, 1995; Arthington et al., 2004). The case of ‘R. rohani’ illustrates also the wasted time and effort, by Batuwita, De Silva & Edirisinghe (2013) and ourselves, first in describing a translocated species as new, and then in working to determine its provenance and, as it happens, ‘sink’ it. Had we not done so, ‘R. rohani’, by being a species with a highly restricted range, would almost certainly have been assessed as Critically Endangered and attracted substantial conservation attention, to the cost of more deserving threatened species. Indeed, Batuwita, De Silva & Edirisinghe (2013) advocated much the same, writing: “we hope that the present work will, by drawing attention to the previously unsuspected diversity within this genus and clarifying its taxonomy, lead to a fresh assessment of these fishes and actions to assure their conservation”.
Rasboroides pallidus and ‘R. rohani’
Deraniyagala (1958) described three “races” of Rasboroides in addition to the forma typica, R. v. vaterifloris, namely, R. v. pallidus, R. v. ruber and R. v. rubioculis. Batuwita, De Silva & Edirisinghe (2013) considered these names to be simultaneous synonyms and, as first reviser, gave R. pallidus precedence over R. rubioculis and R. ruber. No type material is known for R. pallidus, R. ruber and R. rubioculis (Pethiyagoda, 1991: 336).
The population of R. pallidus at Suriyakanda, which Batuwita, De Silva & Edirisinghe (2013) described as ‘R. rohani’, is 980 m above sea level. This is substantially higher than the upper elevation limit of all naturally-occurring populations of Rasboroides, around 230 m asl. Deraniyagala (1958) suggested that Rasboroides may have had a wider distribution within the island during the Pleistocene. Had the species described as ‘R. rohani’ been an isolated population persisting in a rainforest refugium, it would be expected to have a substantial genetic divergence from its sister species. For example, the R. vaterifloris from the Kalu basin differs genetically from the R. pallidus occurring in Kalu, Bentara and Gin basins by over 3% for both coi and cytb genes. However, ‘R. rohani’ and R. pallidus are genetically indistinguishable.
Consideration must be given also to the grounds on which Batuwita, De Silva & Edirisinghe (2013) misdirected themselves in considering the Suriyakanda population of R. pallidus to be a distinct species. Perhaps most significantly, the size range of the specimens of R. pallidus and ‘R. rohani’ examined by these authors contains no overlap: males, 21.5–24.6 mm SL in R. pallidus, vs. 25.3–35.5 mm SL in ‘R. rohani’; females, 20.2–20.7 mm SL in R. pallidus, vs. 23.0–30.8 mm SL in ‘R. rohani’. In our dataset, the females of R. pallidus are relatively smaller when compared with the females of ‘R. rohani’ (7 ex., 22.2–26.6 mm SL and 1 ex. 29.4 mm SL in R. pallidus, vs. 28.8–28.9 mm in ‘R. rohani’) and show differences in caudal-peduncle length (21.8–23.3% SL vs. 23.6–24.4, respectively), body depth (31.7–34.7% SL vs. 35.0–35.5, respectively) and anal-fin height (22.9–28.7% SL vs. 28.9–31.5, respectively). In the case of males of R. pallidus and ‘R. rohani’ examined by us, however, the series are of similar size (22.2–32.5 mm SL vs. 25.6–33.6, respectively) and relatively homogenous morphologically. We infer from this that the differences observed by Batuwita, De Silva & Edirisinghe (2013) between R. pallidus and ‘R. rohani’ were the result of allometry. Further, these authors distinguished ‘R. rohani’ from R. pallidus by the former having more scales in transverse line on body (8 vs. 6–7) and more lateral-line scales (25–28 vs. 20–24). However, both these counts overlap in the material examined by us and the material available at NH.
Batuwita, De Silva & Edirisinghe (2013) noted also that ‘R. rohani’ attained a greater size (35.5 and 30.8 mm SL in males and females, respectively) than R. pallidus (24.6 and 20.7 mm SL, respectively). Indeed, the material examined by us too, reflects this condition, with a maximum size of 33.6 and 28.9 mm SL, respectively, for males and females of ‘R. rohani’, and 32.5 and 29.4 mm SL, respectively, for R. pallidus. We are unable to explain this phenomenon except by way of noting that the population of R. pallidus in the Yagirala Forest Reserve (Bentara basin) too, reaches ∼35mm SL. Unusually, at both these locations, R. pallidus is numerically the dominant cyprinid. We conjecture that its large size in such environments may be correlated with the paucity of larger and more numerous cofamilial competitors, possibly a case in which reduced interspecific competition enhances niche breadth (Robinson, Wilson & Margosian, 2000; Bolnick et al., 2010; Wootton, 2012).
Rasboroides vaterifloris and ‘R. nigromarginatus’
The type locality of R. vaterifloris given by Deraniyagala (1930) is “near Illukvattai ferry on the Ratnapura to Gilimale road”. Consistent with this, Batuwita, De Silva & Edirisinghe (2013) identified the Gilimale population of Rasboroides as R. vaterifloris. Rasboroides nigromarginatus was described by Meinken (1957) based on aquarium specimens, and the exact origin of his material was unknown: “Heimat: Ceylon, genauer Fundort nicht zu ermitteln” (trans. “Home: Ceylon, exact location cannot be determined”). Batuwita, De Silva & Edirisinghe (2013), however, identified the population at Athwelthota in the Kalu basin as ‘R. nigromarginatus’ based primarily on coloration: “The coloration in life of the population we identify as R. nigromarginatus too, is exactly the same as that described by Meinken, including the characteristic blackened first ray of the dorsal, pectoral and pelvic fins, and the orange upper half of the sclera (Fig. 5A). We are confident therefore that our conception of R. nigromarginatus is the same as Meinken’s, with Atweltota being the likely type locality of this species”.
In his description of R. vaterifloris, however, Deraniyagala (1930) was careful to describe the color pattern as: “Dorsal fin with a black anterior edge, rest a bright orange as is the lower lobe of caudal...”. This description appears to have been overlooked by Batuwita, De Silva & Edirisinghe (2013), who described the male coloration of R. vaterifloris as “upper body golden brown, lightening on side to silvery, scattered with melanophores; belly silver. Dorsal, anal, pectoral and pelvic fins hyaline with scattered melanophores.” Further, they illustrated only a female (their fig. 3A). Deraniyagala (1958) further mentioned that the specimens of R. vaterifloris from Parakaduwa (from where Batuwita, De Silva & Edirisinghe (2013) examined material) as being “large with plenty of red on the upper half of the orbit [sic] upon the anal fin and lower lobe of the caudal”. At present, however, the R. vaterifloris at Gilimale are mostly dull-colored when compared with the population at Athwelthota. Similarly, dull-colored morphs of R. vaterifloris were also observed at Labugama (Kelani basin) and Madakada, Ingiriya (Kalu basin). Though the individuals of R. vaterifloris in these populations are less vividly colored, the last unbranched dorsal-fin ray of males shows the distinct black margin described in R. vaterifloris by Deraniyagala (1930) and in ‘R. nigromarginatus’ by Meinken (1957). This character is therefore of no value in distinguishing these nominal taxa.
It appears that Meinken (1957) was misled into describing R. nigromarginatus as a new species because the specimens he considered to be R. vaterifloris were in fact R. pallidus. This caused him to regard the specimens that were in fact R. vaterifloris as belonging to a new species. He stated: “Das zum Vergleich mitgeschickte junge Mannchen von R. vaterifioris ist im ganzen viel hiiher gebaut…der obere Teil des Auges ohne den orangeroten Glanz” (“the young male of the R. vaterifloris, which was sent for comparison, is much higher built…the upper part of the eye is without the orange-red gloss”). Meinken’s illustration of R. vaterifloris too, shows a fish with a higher profile, resembling R. pallidus.
In addition to neglecting to note Meinken’s confusion, Batuwita, De Silva & Edirisinghe (2013), as in the case of R. pallidus and ‘R. rohani’, were apparently misled by the different size-ranges of the series of R. vaterifloris and ‘R. nigromarginatus’ they examined: males, 23.9–28.4 mm SL in R. vaterifloris, vs. 26.2–30.2 mm SL in ‘R. nigromarginatus’; and females, 22.1–25.5 mm SL in R. vaterifloris vs. 26.5–27.3 mm SL in ‘R. nigromarginatus’. In our series too, the males of R. vaterifloris from the type locality (Gilimale) are smaller than those of ‘R. nigromarginatus’ sensu Batuwita, De Silva & Edirisinghe (2013) from Athweltota (21.2–24.2 mm SL, vs. 24.7–28.9). The proportional body depths in these series do not overlap: 29.0–31.0% SL in topotypical R. vaterifloris vs. 31.5–34.2 in ‘R. nigromarginatus’), as do not also the anal-fin height (28.1–28.6% SL in R. vaterifloris vs. 28.6–31.9 in ‘R. nigromarginatus’) and head depth (18.7–19.4% SL in R. vaterifloris vs. 19.9–21.2 in ‘R. nigromarginatus’). Females of R. vaterifloris and ‘R. nigromarginatus’ in our series were, however, similar in size (21.6–25.4 mm SL vs. 21.7–26.1, respectively) and did not differ in any of the mentioned proportions. It appears likely, therefore, that the proportional differences between R. vaterifloris and ‘R. nigromarginatus’ observed by Batuwita, De Silva & Edirisinghe (2013) were a coupling of selective sampling and allometric growth, not indicative of a morphometric signal.
Further, the minimum uncorrected pairwise distances for the partial genes cytb and coi between topotypical R. vaterifloris and ‘R. nigromarginatus’ are only 0.2%, in addition to which there appears to be no geographic barrier separating the two populations. We are confident, therefore, in referring ‘R. nigromarginatus’ to the synonymy of R. vaterifloris. The GenBank sequences earlier identified as R. vaterifloris (HM224374, EU241471, AP011432) were all shown to be R. pallidus when our newly generated sequences were included in the molecular analysis. Such species misidentifications are common in GenBank (Conte-Grand et al., 2017) and care should be taken when interpreting GenBank records with no specific specimen data.
Conclusion
As a freshwater-fish genus endemic to Sri Lanka and restricted largely to streams draining the island’s dwindling rainforest estate, Rasboroides attracts considerable conservation attention. The National Red List (MOE, 2012) treats ‘R. nigromarginatus’ as Critically Endangered and R. vaterifloris as Endangered. The synonymy of these two nominal species demonstrated here allows their ranges to be combined, widening their extent of occurrence and area of occupancy and hence potentially lowering the threat-status of R. vaterifloris. Although ‘R. rohani’ has not as yet been assessed for conservation purposes, its restriction to a small population at a single locality would almost certainly have caused it to be ranked as Critically Endangered. Given that we show here that it represents only an undocumented translocation of R. pallidus, its population is now only of marginal conservation concern. Indeed, of the two valid species of Rasboroides, R. pallidus enjoys the wider range and hence warrants less conservation concern, especially given its successful translocation to two river basins (Mahaweli and Walawe) in which it did not previously occur.
In describing ‘R. rohani’ as a new species, Batuwita, De Silva & Edirisinghe (2013) were misled by apparently collecting only the largest specimens for their sample while neglecting to account for allometric growth. It is additionally regrettable that the type series of ‘R. rohani’ designated by these authors cannot be identified in the collection of the National Museum of Sri Lanka, in which it was stated to be deposited.
Both translocations referred to in this paper were made by well-meaning citizens but without the safeguards that should apply in such cases. Perhaps most egregiously, no records were published of the rationale for translocation or the precise identity and origin of the source population. We urge that any future attempts to introduce species to novel habitats be guided by IUCN/SSC (2013) and that the intentional release or introduction of species without legal sanction be prohibited in Sri Lanka.
Supplemental Information
Raw data of Rasboroides
Raw data of Rasboroides used in the morphological analyses along with sampling localities and voucher numbers.
coi alignment
coi alignment of Rasboroides used in the phylogenetic analysis
Maximum Likelihood phyolograms
Maximum Likelihood phylograms based on cytb and coi datasets for species of Rasboroides in Sri Lanka
cytb alignment of Rasboroides
cytb alignment of Rasboroides used in the phylogenetic analysis