Multilocus phylogeny of the parasitic wasps in the tribe Euphorini (Hymenoptera: Braconidae) with revised generic classifications
- Published
- Accepted
- Received
- Academic Editor
- Ilaria Negri
- Subject Areas
- Entomology, Taxonomy
- Keywords
- Leiophron, Mama, Peristenus, Taxonomy, Euphorinae, Phylogenetics, Parasitoid, Euphoriella, Euphorus, Euphoriana
- Copyright
- © 2018 Zhang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2018. Multilocus phylogeny of the parasitic wasps in the tribe Euphorini (Hymenoptera: Braconidae) with revised generic classifications. PeerJ 6:e4783 https://doi.org/10.7717/peerj.4783
Abstract
Background
Parasitic wasps in the family Braconidae are important regulators of insect pests, particularly in forest and agroecosystems. Within Braconidae, wasps in the tribe Euphorini (Euphorinae) attack economically damaging plant bugs (Miridae) that are major pests of field and vegetable crops. However, the evolutionary relationships of this tribe have been historically problematic. Most generic concepts have been based on ambiguous morphological characters which often leads to misidentification, complicating their use in biological control.
Methods
Using a combination of three genes (COI, 28S, and CAD) and 80 taxa collected worldwide, we conducted Bayesian inference using MrBayes, and maximum likelihood analyses using RAxML and IQ-Tree on individual gene trees as well as the concatenated dataset.
Results
The monophyly of the tribe Euphorini and the two genera Peristenus and Leiophron were confirmed using maximum likelihood and Bayesian inference. The subgeneric classifications of Leiophron sensu lato were not supported, and the monotypic genus Mama was also not supported.
Discussion
Euphoriella, Euphoriana, Euphorus, and Mama syn. n, have been synonymized under Leiophron. Mama mariae syn. n was placed as a junior synonym of Leiophron reclinator. The generic concepts of Peristenus and Leiophron were refined to reflect the updated phylogeny. Further we discuss the need for revising Euphorini given the number of undescribed species within the tribe.
Introduction
Braconid wasps in the large and diverse subfamily Euphorinae is divided into 14 tribes and 52 genera (Stigenberg, Boring & Ronquist, 2015). Euphorines attack a variety of host life stages ranging from nymphal/larval hosts to adults of seven different orders of insects: Coleoptera, Hemiptera, Hymenoptera, Neuroptera, Orthoptera, Psocodea, and Lepidoptera (Shaw, 1988; Chen & Van Achterberg, 1997; Stigenberg, Boring & Ronquist, 2015). The tribe Euphorini Foerster contains koinobiont endoparasitoids of Hemiptera and Psocodea, which attack young nymphs (1st or 2nd instar) and feed internally on the hemolymph of their hosts (Loan, 1974b). Mature parasitoid larvae emerge from mature host nymphs or teneral adults, and overwinter as pupae in soil (Loan, 1974b). Several species of Euphorini have been extensively studied for their use in biological control programs because they attack many serious agricultural pests such as Lygus Hahn (Day, 1987; Haye et al., 2005; Haye et al., 2007). Despite the research interest using Euphorini wasps in applied entomology, the classification and identification of these parasitoids remains challenging. Variable morphological characters such as the degree of completion of the occipital carina, and the presence or absence of vein (RS + M)a and cu-a were used as defining characters (Chen & Van Achterberg, 1997; Stigenberg, Boring & Ronquist, 2015), leading to taxonomic uncertainty and misidentification.
The taxonomic history of Euphorini is a long and convoluted one. Euphoriana Gahan, Euphoriella Ashmead, Euphorus Nees, and Peristenus Foerster have all been synonymized under or treated as subgenera of Leiophron in a variety of combinations by different authors (Marshall, 1887; Muesebeck, 1936; Loan, 1974a; Shaw, 1985; Shaw, 1987; Chen & Van Achterberg, 1997; Stigenberg, Boring & Ronquist, 2015). In addition, Aridelus Marshall, Chrysopophthorus Goidanich, Cryptoxilos Viereck, Holdawayella Loan, Mama Belokobylskij, and Wesmaelia Foerster were also included in Euphorini until the most recent comprehensive revision of the entire subfamily Euphorinae by Stigenberg, Boring & Ronquist (2015). Only a few exemplars of Euphorini were included in the study, but the monophyly of the tribe was strongly supported. Currently there are three recognized genera within Euphorini, including Leiophron sensu lato, Peristenus, and the monotypic Mama (Stigenberg, Boring & Ronquist, 2015). Leiophron is further divided into four subgenera, Euphoriana, Euphoriella, Euphorus, and Leiophron sensu stricto (Stigenberg, Boring & Ronquist, 2015).
Here, we extensively sample Euphorini wasps and reconstruct the evolutionary relationships among its members using a multi-locus dataset. We reassess the generic and subgeneric concepts of Euphorini and revise the classification to reflect the phylogeny. The results of this study provide a comprehensive framework for phylogenetic relationships among Euphorini wasps and we provide taxonomic clarity and identification resources to aid future applied research and biological control programs.
Materials & Methods
Sample collection
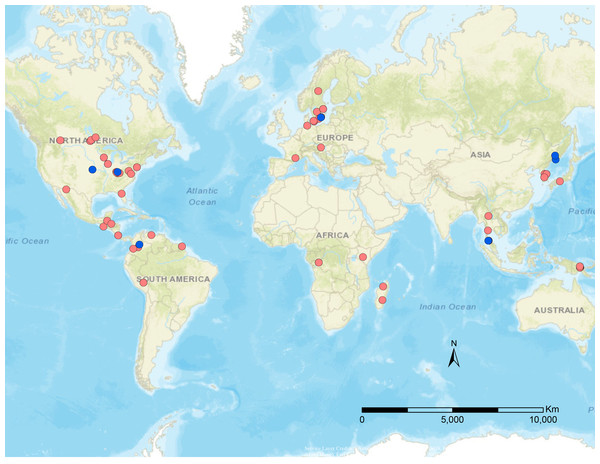
Specimens were borrowed from the following institutions and curators: Hymenoptera Institute Collection, University of Kentucky (HIC, M. Sharkey), French National Museum of Natural History (MNHN, C. Villemant), Swedish Museum of Natural History (NHRS, H. Vårdal), and Zoological Institute of Russian Academy of Sciences (ZIN, S. Belokobylskij). Additional specimens were collected via sweep netting or Malaise trap samples from Canada, USA, and Peru. Specimens were identified using Chen & Van Achterberg (1997), Loan (1974a), Loan (1974b), Stigenberg & Van Achterberg (2016). Outgroups included representatives of the most closely related tribes to Euphorini, based on Stigenberg, Boring & Ronquist (2015): Microctonus Wesmael (Perilitini), Townesilitus Haeselbarth & Loan (Townsilitini), and Chrysopophthorus Goidanich (Helorimorphini). A list of the specimens utilized in this study is provided in Table 1, and detailed locality information in Table S1. A map of the distribution of these specimens is depicted in Fig. 1, which was generated using ArcMap v10.5.1. For ease of interpretation of results, specimen information was added to taxon labels for the phylogenetic analyses, including country and lowest identification. The subgeneric names within Leiophron s.l. are used as specimen names to avoid confusion with Leiophron s.s. (eg. Leiophron (Leiophron) uniformis is listed as Leiophron uniformis, whereas Leiophron (Euphoriana) dispar is listed as Euphoriana dispar).
| Taxon Label | COI | 28S | CAD | Locality | Voucher location |
|---|---|---|---|---|---|
| 07_Yves_Leiophron_PNG | MG926854 | MG913702 | – | Papua New Guinea | MNHN |
| 08_Yves_Leiophron_PNG | MG926855 | MG913703 | – | Papua New Guinea | MNHN |
| 10_Yves_Leiophron_PNG | MG926856 | MG913704 | – | Papua New Guinea | MNHN |
| AB016_Peristenus_KS | KJ591487 | KJ591282 | – | USA | HIC |
| AB020_Peristenus_KY | KJ591488 | KJ591283 | – | USA | HIC |
| AB023_Peristenus_KS | KJ591489 | KJ591284 | – | USA | HIC |
| Euph_001_Euphorus_pallidistigma_SWE | MG926857 | – | MG913762 | Sweden | NHRS |
| Euph_017_Peristenus_JAP | MG926858 | – | MG913763 | Japan | NHRS |
| Euph_020_Peristenus_SWE | MG926859 | MG913705 | MG913764 | Sweden | NHRS |
| Euph_083_Peristenus_HUN | MG926860 | MG913706 | – | Hungary | NHRS |
| Euph_162_Leiophron_apicalis_SWE | MG926861 | – | – | Sweden | NHRS |
| JS01000238_Leiophron_fascipennis_SWE | MG926862 | MG913713 | – | Sweden | NHRS |
| JS01000242_Leiophron_SWE | KJ591452 | KJ591243 | – | Sweden | NHRS |
| JS01000267_Leiophron_FRGU | MG926863 | MG913707 | – | French Guiana | NHRS |
| JS01000499_Mama_mariae_RUS | KJ591460 | KJ591250 | – | Russia | ZIN |
| JS01000515_Euphoriana_dispar_RUS | KJ591458 | MG913708 | – | Russia | ZIN |
| JS01000538_Euphorus_duploclaviventris_SWE | MG926864 | – | – | Sweden | NHRS |
| JS01000539_Euphorus_oblitus_SWE | MG926865 | – | – | Sweden | NHRS |
| JS01000540_Leiophron_deficiens_SWE | MG926866 | MG913709 | – | Sweden | NHRS |
| JS01000542_Leiophron_reclinator_SWE | MG926867 | – | – | Sweden | NHRS |
| JS01000547_ Leiophron _MAD | MG926868 | MG913710 | – | Madagascar | NHRS |
| JS01000552_Peristenus_SWE | MG926869 | MG913711 | MG913765 | Sweden | NHRS |
| JS01000553_Euphorus_basalis_SWE | MG926870 | – | MG913766 | Sweden | NHRS |
| JS01000554_Euphorus_fulvipes_SWE | MG926871 | MG913712 | MG913767 | Sweden | NHRS |
| JS068_Leiophron_COL | KJ591455 | KJ591246 | KJ591362 | Colombia | HIC |
| JS120_Leiophron_THA | KJ591456 | KJ591247 | KJ591363 | Thailand | HIC |
| JS129_Leiophron_THA | KJ591457 | KJ591248 | KJ591364 | Thailand | HIC |
| PNG_5_Leiophron | MG926872 | MG913714 | – | Papua New Guinea | MNHN |
| PNG_6_Leiophron | MG926873 | MG913715 | – | Papua New Guinea | MNHN |
| PNG_7_Leiophron | MG926874 | MG913716 | – | Papua New Guinea | MNHN |
| YMZ038_Peristenus_GER | MG926875 | MG913717 | MG913768 | Germany | UCFC |
| YMZ077_Leiophron_uniformis_MB | – | MG913718 | MG913769 | Canada | UCFC |
| YMZ081_Euphoriella_MB | MG926876 | MG913719 | MG913770 | Canada | UCFC |
| YMZ124_Peristenus_mellipes_MB | KY566090 | MG913720 | MG913771 | Canada | UCFC |
| YMZ132_Leiophron_KY | MG926877 | MG913721 | MG913772 | USA | UCFC |
| YMZ133_Leiophron_KY | MG926878 | MG913722 | MG913773 | USA | UCFC |
| YMZ134_Leiophron_WV | MG926879 | MG913723 | MG913774 | USA | UCFC |
| YMZ136_Leiophron_KY | MG926880 | MG913724 | MG913775 | USA | UCFC |
| YMZ139_Leiophron_uniformis_FRA | MG926881 | MG913725 | – | France | UCFC |
| YMZ141_Leiophron_THA | MG926882 | MG913726 | MG913776 | Thailand | UCFC |
| YMZ142_Peristenus_MAD | MG926883 | MG913727 | – | Madagascar | UCFC |
| YMZ145_Leiophron_COL | MG926884 | MG913728 | MG913777 | Colombia | UCFC |
| YMZ146_Leiophron_COL | MG926885 | MG913729 | – | Colombia | UCFC |
| YMZ148_Euphoriella_GUA | MG926886 | MG913730 | MG913778 | Guatemala | UCFC |
| YMZ211_Peristenus_dayi_MB | KY566098 | MG913731 | MG913779 | Canada | UCFC |
| YMZ335_Peristenus_howardi_AB | KY566100 | MG913732 | MG913780 | Canada | UCFC |
| YMZ341_Peristenus_relictus | KY566106 | MG913733 | MG913781 | USA | UCFC |
| YMZ343_Peristenus_digoneuti | MG926887 | MG913734 | MG913782 | USA | UCFC |
| YMZ345_Leiophron_KY | MG926888 | MG913735 | – | USA | UCFC |
| YMZ346_Peristenus_WI | MG926889 | MG913736 | – | USA | UCFC |
| YMZ349_Peristenus_IL | MG926890 | MG913738 | – | USA | UCFC |
| YMZ351_Leiophron_VA | MG926891 | MG913739 | MG913783 | USA | UCFC |
| YMZ356_Peristenus_WI | MG926892 | MG913740 | – | USA | UCFC |
| YMZ358_Euphoriella_KY | MG926893 | MG913741 | – | USA | UCFC |
| YMZ359_Euphoriella_FL | MG926894 | MG913742 | – | USA | UCFC |
| YMZ361_Leiophron_AZ | MG926895 | MG913743 | – | USA | UCFC |
| YMZ363_Euphoriella_COL | MG926896 | MG913744 | MG913784 | Colombia | UCFC |
| YMZ364_ Leiophron _CR | MG926897 | MG913745 | – | Costa Rica | UCFC |
| YMZ365_Euphoriella_CR | MG926898 | MG913746 | – | Costa Rica | UCFC |
| YMZ366_Euphoriella_GUA | MG926899 | MG913747 | MG913785 | Guatemala | UCFC |
| YMZ367_Leiophron_HON | MG926900 | MG913748 | – | Honduras | UCFC |
| YMZ368_Leiophron_VEN | MG926901 | MG913749 | – | Venezuela | UCFC |
| YMZ370_Euphoriella_PER | MG926902 | MG913750 | – | Peru | UCFC |
| YMZ371_Leiophron_PER | MG926903 | MG913751 | MG913786 | Peru | UCFC |
| YMZ372_Peristenus_PER | MG926904 | MG913752 | – | Peru | UCFC |
| YMZ373_Leiophron_THA | MG926905 | – | MG913787 | Thailand | UCFC |
| YMZ375_Leiophron_KEN | MG926906 | – | Kenya | UCFC | |
| YMZ376_Leiophron_THA | MG926907 | MG914753 | – | Thailand | UCFC |
| YMZ377_Leiophron_THA | MG926908 | MG914754 | MG913788 | Thailand | UCFC |
| YMZ378_Leiophron_THA | MG926909 | MG914755 | MG913789 | Thailand | UCFC |
| YMZ380_Leiophron_CON | MG926910 | MG914756 | MG913790 | Congo | UCFC |
| YMZ382_Leiophron_KOR | MG926911 | – | MG913791 | South Korea | UCFC |
| YMZ383_Leiophron_KOR | MG926912 | – | MG913792 | South Korea | UCFC |
| YMZ384_Leiophron_KOR | MG926913 | MG914757 | – | South Korea | UCFC |
| YMZ385_Leiophron_KOR | MG926914 | MG914758 | – | South Korea | UCFC |
| YMZ386_Leiophron_CON | MG926915 | MG914759 | – | Congo | UCFC |
| YMZ388_Leiophron_pallidistigma_KOR | MG926916 | MG914760 | – | South Korea | UCFC |
| AB102 Microctonus (Perilitini) | KJ591529 | KJ591329 | KJ591412 | USA | HIC |
| JS01000218 Townesilitus (Townesilitini) | KJ591440 | KJ591228 | KJ591353 | Sweden | HIC |
| JS115 Chrysopophthorus (Helorimorphini) | MG926854 | MG913702 | – | Colombia | HIC |
Figure 1: Geographical distribution of specimens used in this study.
Blue dots are published data from Stigenberg, Boring & Ronquist (2015), red dots are newly sampled taxa for this study.Terminology and image capture
Terminology used for most morphological characters follows Chen & Van Achterberg (1997) and Stigenberg, Boring & Ronquist (2015). However, wing venation terminology follows Sharkey & Wharton (1997). Specimens were photographed using a Canon 7D Mark II with a Mitutoyo M Plan Apo 10× objective mounted onto the Canon EF Telephoto 70–200 mm zoom lens, and the Canon MT–24EX Macro Twin Lite Flash (Tokyo, Japan) with custom-made diffusers to minimize hot spots.
DNA protocols
A total of 80 taxa were sampled out of which three species represented outgroups. The taxon sampling covers the entire range of Euphorini, which is found on all continents except in Antarctica and Australia outside of Papua New Guinea (Yu, Van Achterberg & Horstmann, 2012). This is the largest sampling of any of the euphorine tribes, and is comprised of all three Euphorini genera, as well as all four subgenera of Leiophron s.l. Specimens were extracted, amplified, and sequenced either at the Molecular Systematics Laboratory, Swedish Museum of Natural History following the protocols listed in (Stigenberg, Boring & Ronquist, 2015), or at the Insect Systematics Laboratory at the University of Central Florida following the DNeasy™ Tissue Kit protocol (Qiagen, Valencia, CA, USA). Petioles were separated from mesosomas to ensure buffer penetration during tissue lysis, and the two body parts were mounted onto the same point post-extraction for vouchering. Voucher specimens deposition are listed in Table 1. Three genes were amplified: partial 28S domain 2 and 3 (rDNA), partial CAD (Carbamoyl-Phosphate Synthetase 2, Aspartate Transcarbamylase, and Dihydroorotase) and the 5′ region of mitochondrial COI. New Euphorini-specific primers were designed for CAD based on sequences from Sharanowski, Dowling & Sharkey (2011) and Stigenberg, Boring & Ronquist (2015). The faster rate of evolution of the mitochondrial genes is ideal for separating closely related species (Zhang, Ridenbaugh & Sharanowski, 2017), while the ribosomal and nuclear genes have slower rates of evolution and are more suitable for higher level phylogenetic relationships (Sharanowski, Dowling & Sharkey, 2011). All three genes are commonly used in Braconidae phylogenetics, including Euphorinae (Sharanowski, Dowling & Sharkey, 2011; Stigenberg, Boring & Ronquist, 2015; Zhang, Ridenbaugh & Sharanowski, 2017).
All PCRs were performed on a Bio-Rad MyCyclerTM thermal cycler, using approximately 1µg DNA extract, 1 × Standard Taq Buffer (10 mm Tris-HCl, 50 mm KCl, 1.5 mm MgCl2, pH 8.3, New England Biolabs, Ipswich, Massachusetts, USA), 200 μM dNTP (Invitrogen, Carlsbad, CA, USA), 4 mM MgSO4, 400 nM of each primer, 1 unit of Taq DNA polymerase (New England Biolabs), and purified water to a final volume of 25 µl. Primer information and PCR conditions are listed in Table 2. Amplicons of reaction products were cleaned with Agencourt CleanSEQ magnetic beads and sequenced in both directions using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) and the Applied Biosystems 3730xl DNA Analyzer at the University of Kentucky, Advanced Genetic Technologies Center (UK-AGTC). Contigs were assembled and edited using Geneious version 8.18 (Kearse et al., 2012), and alignment was conducted using MAFFT server (Katoh et al., 2002) (https://mafft.cbrc.jp/alignment/server/). The protein coding genes were aligned using default MAFFT settings, and for 28S we used Q-INS-I strategy (Katoh & Toh, 2008) which takes secondary RNA structure into account. New sequences obtained from this study were deposited in GenBank (See Table 1).
| Gene region | Primers | Sequence (5′ to 3′) | Source | Annealing temperature |
|---|---|---|---|---|
| 28S | D2F (fwd) | AGTCGTGTTGCTTGATAGTGCAG | Campbell, Steffen-Campbell & Werren (1994) | 55 °C |
| D2R (rev) | TTGGTCCGTGTTTCAAGACGGG | Campbell, Steffen-Campbell & Werren (1994) | 55 °C | |
| COI | LCO1490 (fwd) | GGTCAACAAATCATAAAGATATTGG | Folmer et al. (1994) | 49 °C |
| HCO2198 (rev) | TAAACTTCAGGGTGACCAAAAAATCA | Folmer et al. (1994) | 49 °C | |
| CAD | CAD2F (fwd) | TAYGAGCTTACCAAAATWGAYC | New primer | 52 °C |
| CAD2R (rev) | CATAAATGTCCATCACAACTTC | New primer | 52 °C |
Phylogenetic analyses
The three genes were analyzed separately and concatenated using Bayesian inference (BI) analysis with MrBayes v3.2.6 (Ronquist et al., 2012) on the CIPRES Science Gateway (Miller et al., 2009). Each analysis had two independent searches with four chains and were run for 10,000,000 generations, sampling every 1,000, with a 10% burnin discarded. For the concatenated analysis, partitions were separated by gene and codon position for protein-coding genes for a total of six preselected partitions. Six partitions (28S; CAD_1; CAD_2 +CAD_3; COI_1; COI_2; and COI_3) were chosen using PartitionFinder 2.1.1 (Lanfear et al., 2016), based on the greedy algorithm and Bayesian Information Criterion (Table 3). The same partitions were also used for a maximum likelihood (ML) analysis using the relatively new IQ-Tree method (Nguyen et al., 2014), with 1,000 ultrafast bootstraps developed by Hoang et al. (2018). The concatenated dataset was also analyzed with RAxML v8.2.0 (Stamatakis, 2006), using the GTR+ Γ model of nucleotide substitution and 1,000 nonparametric bootstraps. All alignment files and scripts files can be accessed on Figshare (https://figshare.com/articles/Concatenated_File/6160247). All resulting trees were visualized using FigTree v1.4.2 (Rambaut, 2012). Intraspecific distances between Mama mariae Belokobylskij and Leiophron reclinator (Ruthe) was calculated using MEGA v7.0.21 (Kumar, Stecher & Tamura, 2016) using the Kimura-2-parameter (K2P) model (Kimura, 1980).
Results
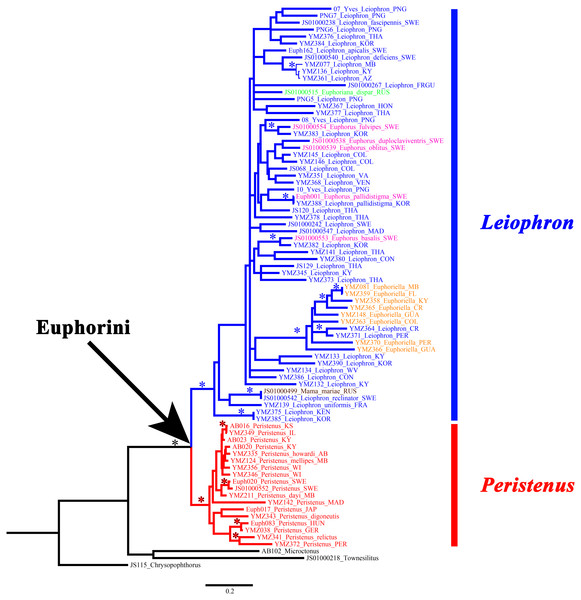
Here we present the most taxonomically comprehensive phylogeny of the euphorine braconid tribe Euphorini with all known genera and subgenera sampled. All genera and subgenera had multiple representatives except Euphoriana (only one exemplar included - E. dispar) and the monotypic Mama mariae. A total of 39 CAD, 71 28S, and 80 COI for a total of 190 sequences were used for the final analyses, 158 of which were newly generated for this study (Table 1). The summary statistics of all three genes can be found in Table 3. While we failed to amplify CAD sequences from some older specimens, the gene itself is informative (See Table 3) and should be used in other multilocus analyses of braconids. All the individual BI gene trees (Figs. S1–S3), as well as the concatenated BI and ML (Figs. S4–S6) analyses strongly supported the monophyly of the tribe Euphorini (1, 100, 100, for MrBayes posterior probability, RAxML bootstrap support, and IQ-Tree ultrafast bootstrap support, respectively) as well as the monophyly of the genera Peristenus (1, 95, 99) and Leiophron s.l. (1, 90, 90) (Fig. 2).
| Marker and partitions | #bp | #var | #par | CG% | Model | References |
|---|---|---|---|---|---|---|
| 28S nuclear DNA | ||||||
| 28S_123 | 592 | 263 | 179 | 43.3 | GTR+G | Tavaré (1986) |
| CAD nuclear DNA | 507 | 185 | 121 | 36.0 | ||
| CAD_1 | 169 | 133 | 96 | 26.5 | HKY+I+G | Hasegawa, Kishino & Yano (1985) |
| CAD_23 | 338 | 52 | 25 | 40.3 | HKY+I+G | Hasegawa, Kishino & Yano (1985) |
| COI mtDNA | 660 | 360 | 296 | 28.4 | ||
| COI_1 | 220 | 103 | 78 | 33.8 | GTR+I+G | Tavaré (1986) |
| COI_2 | 220 | 55 | 33 | 38.4 | GTR+G | Tavaré (1986) |
| COI_3 | 220 | 202 | 185 | 12.8 | GTR+I+G | Tavaré (1986) |
Figure 2: Concatenated gene tree for MrBayes, RAxML, and IQ-Tree.
Peristenus is colored red, and Leiophron is colored in blue, with subgenera within Leiophron shown in different colors (Leiophron sensu stricto in blue, Euphorus in purple, Euphoriana in green, Euphoriella in orange, and Mama in brown). Asterisks indicate strong nodal support for all three analyses (≥0.98 posterior probability support for MrBayes; ≥90 for bootstrap support for RAxML; and ≥90 for ultrafast bootstrap support for IQ-Tree).Mama mariae, the only species from the monotypic genus Mama, was not supported as a distinct genus and was instead recovered as a sister group of Leiophron reclinator (Fig. 2) with 0.8% divergence among COI intraspecific genetic distance.
The four subgenera of Leiophron s.l. (Leiophron s.s., Euphoriana, Euphoriella, Euphorus) were not supported as distinct clades within the monophyletic Leiophron s.l. in any of the phylogenetic analyses (Fig. 2, Figs. S1–S5).
Discussion
Generic concepts of Peristenus and Leiophron s.l.
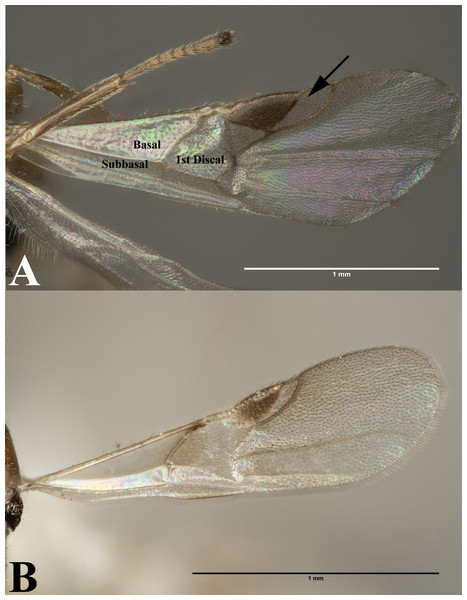
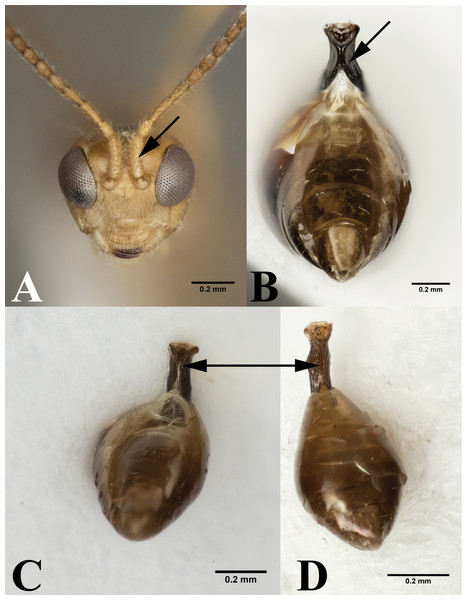
Our data supports the monophyly of Peristenus and Leiophron, corroborating the results of previous morphological studies (Loan, 1974a; Loan, 1974b; Shaw, 1985; Shaw, 1987; Chen & Van Achterberg, 1997). Additionally, with a much more focused taxon sampling we were able to delineate the finer relationships within the tribe Euphorini. Peristenus is largely uniform in morphology and exclusively attacks Miridae, while its sister taxon Leiophron is much more variable in both morphology and host breadth which likely has led to convergent morphology, and hence the subgeneric concepts and taxonomic confusion. Peristenus can be distinguished from Leiophron by the evenly setose 1st discal, basal, and subbasal cells in the forewing (Fig. 3A), and the 1st metasomal tergite, which is fused or touching basally (Fig. 4B).
Figure 3: Forewing.
(A) Forewing of Peristenus; arrowing pointing to marginal cell. (B) Forewing of Leiophron. Photos by JH Meyer.Figure 4: Leiophron vs Peristenus.
(A) Frontal view of Leiophron reclinator (Mama mariae syn. n), arrow pointing to spiny scape of antennae. (B) Ventral view of Peristenus metasoma, arrow pointing to the partially fused petiole. Ventral view of Leiophron metasoma: (C) arrowing pointing to ventral petiole showing unfused sclerite at midline. (D) arrowing pointing to completely fused sclerite at midline of petiole. Photos by JH Meyer.Representatives of all four subgenera of Leiophron s.l. defined by Stigenberg, Boring & Ronquist (2015): Euphoriana, Euphoriella, Euphorus, and Leiophron s.s., were included in this analysis. These subgeneric relationships were not supported in any of our analyses, as they failed to form monophyletic clades (Fig. 2). This is not surprising given the lack of consistent morphological characters as specimens often exhibit characteristics of different subgenera, leading to similar suggestions for synonymizations in the past (Muesebeck, 1936; Loan, 1974b; Shaw, 1985; Stigenberg, Boring & Ronquist, 2015). Morphological characters such as the presence or absence of the forewing vein (RS + M)a, hindwing vein cu-a, and complete occipital carina are all too variable to be used as defining characteristics (Chen & Van Achterberg, 1997; Stigenberg, Boring & Ronquist, 2015). These ambiguous distinguishing characters at the subgeneric level can easily lead to misidentification in ecological or applied studies. Euphoriella is morphologically unique based on the absence of radial cell, and the fusion of ventral side of petiole. However, this subgenus is rendered paraphyletic by Leiophron sp. YMZ364 and YMZ371. The type species of Euphoriella, Euphoriella incerta Ashmead, was originally collected from Florida (Muesebeck, 1936). However, the type specimen is too badly damaged to be compared with our equally damaged DNA voucher also collected from Florida to accurately confirm its identity. While there are no nomenclatural rules that requires subgenera to be monophyletic, we do not support paraphyly as these morphological similarities could be the result of convergent evolution and does not reflect evolutionary history. Therefore, based on our molecular evidence combined with the inconsistency of previously used morphological characters, we recommend treating Leiophron as a single genus without further subdivisions and synonymize the subgenera Euphoriana, Euphoriella, and Euphorus as junior synonyms of Leiophron. With this taxonomic update, Leiophron can be identified with the following combination of characters: 1st discal cell of the forewing is more setose than basal and subbasal cells (often glabrous) in Leiophron (Fig. 3B), but if not, then the ventral side of the 1st metasomal tergite (petiole) is not fused (Fig. 4C).
The exact age of the divergence between Peristenus and Leiophron is unknown, as the only known fossil record of Euphorini is a single specimen described as Euphorus indurescens Brues, found in Florissant, Colorado and dating back to Eocene at around 33.7–37 mya (Brues, 1910). In addition, both Leiophron and Peristenus have received little taxonomic attention outside of Europe, North America, and Asia (Loan, 1974a; Loan, 1974b; Chen & Van Achterberg, 1997; Belokobylskij, 2000b; Goulet & Mason, 2006; Stigenberg & Van Achterberg, 2016). We have included many undescribed species from Central and South America, Africa, and Papua New Guinea, which is unsurprising given the tremendous diversity of their major host Miridae. The first and second author are currently working on describing species from Papua New Guinea (J Stigenberg & YM Zhang, 2018, unpublished data), but a revision of the world Euphorini is needed.
Validity of the genus Mama
The validity of the enigmatic genus Mama, described based on a single species M. mariae from eastern Russia (Belokobylskij, 2000a), has been questioned before by Simbolotti, Villemant & Van Achterberg (2004). Both M. mariae and L. reclinator have long, compressed, and spiny scapes (Fig. 4A), but due to the poor condition of the L. reclinator lectotype no definitive conclusion was made (Simbolotti, Villemant & Van Achterberg, 2004). With the consistent placement of the two species as sister taxa with short branch lengths in all three genes (Figs. S1–S3) and concatenated dataset (Fig. 2), and identical in morphology (the second author has examined the holotypes of M. mariae and L. reclinator), we synonymize M. mariae syn. n. as a junior synonym of L. reclinator, thus effectively dissolving the monotypic genus Mama syn. n. as a junior synonym of Leiophron. The distribution of L. reclinator likely spans across Eurasia, as specimens are found from eastern Russia to Sweden and the United Kingdom (Stigenberg & Van Achterberg, 2016).
Updated generic concepts of Euphorini
Tribe Euphorini Foerster 1862
Diagnosis. Maxillary palp with five segments; labial palp with two to three segments; eye bare; first metasomal tergite petiolate; ovipositor slender and short, hardly protruding beyond metasoma; tarsal claws simple; forewing with marginal cell almost always equal or smaller than stigma (Fig. 3A); vein 3RSb (if present) strongly bent; vein r short or absent; vein 2M desclerotized; vein (RS+M)b absent; length of vein m-cu (if present) shorter than length of vein 2RS (Figs. 3A–3B).
Genus Peristenus Foerster 1862
Peristenus Foerster, 1862: 25; Shenefelt, 1969: 36 (as synonym of Leiophron Nees, 1818); (Shaw, 1985): 332. Type species (by original designation): Microctonus barbiger Wesmael, 1835 [= Leiophron pallipes Curtis, 1833].
Diagnosis. Antennal segments 16–33; labial palp with three segments; occipital carina complete or interrupted dorsally; notaulus well-defined, crenulate, posteriorly joining just before posterior margin of mesoscutum; forewing with marginal cell large, complete; basal, subbasal, and 1st discal cells of forewing similarly setose (Fig. 3A); veins (RS+M)a, 1m-cu, 2CUa, 2CUb of forewing fully developed (Fig. 3A); veins rs-m, 2-1A of forewing absent; vein M+CU of forewing unsclerotized; veins 1cu-a and 1-1A of hindwing fully present; first metasomal tergite widened apically, ventrally fused or touching basally (Fig. 4B); metasomal tergites behind first tergite smooth; second suture absent; second tergite with lateral fold; hypopygium medium-sized, densely setose; ovipositor sheath slender, short, and densely setose; ovipositor slender, distinctly curved downwards.
Biology. Koinobiont endoparasitoids of Miridae (Hemiptera). The early instar nymphs are parasitized and the mature parasite larva emerge from either the mature host nymphs or the adults.
Distribution. Cosmopolitan except for Antarctic, limited to Papua New Guinea in Australasia.
Genus Leiophron Nees, 1818
Leiophron Nees, 1818: 303; Shenefelt, 1969: 35; Shaw, 1985: 326. Type species (designated by Viereck, 1914): Leiophron apicalis Haliday, 1833.
Euphoriana Gahan, 1913: 433; Shenefelt, 1969: 33; Shaw, 1985: 326. Type species (by original designation): Euphoriana uniformis Gahan, 1913. Syn. by Loan, 1974b.
Euphoriella Ashmead, 1900: 116; Shenefelt, 1969: 34; Shaw, 1985: 323. Type species (by monotypy & original designation): Labeo incertus Ashmead, 1887. Syn. By Chen & Van Achterberg, 1997
Euphorus Nees, 1834: 360; Shenefelt, 1969: 35; Shaw, 1985: 326): Type species (by monotypy): Euphorus pallicornis Nees, 1834. Syn. by Muesebeck, 1958.
Mama Belokobylskij, 2000: 256; Stigenberg, Boring & Ronquist, 2015: 590. Type species (by monotypy & original desgination): Mama mariae Belokobylskij, 2000. Syn.n.
Diagnosis. Antennal segments 14–20; labial palp with two to three segments; occipital carina usually widely interrupted dorsally; notaulus usually absent; marginal cell of forewing small, incomplete, or absent; 1st discal cell of forewing often more setose than basal and subbasal cells (Fig. 3B); forewing vein 3SRb ending far beforewing apex; forewing vein (RS+M)a present or absent; forewing vein 2M present; forewing vein M+CU largely unsclerotized; forewing vein 1M usually thickened; forewing vein 1CUb sclerotized or unsclerotized; forewing veins 2CUa and 2CUb absent; hindwing vein cu-a partly present or absent; first metasomal tergite nearly parallel-sided or slightly widened apically, ventrally variable: largely open, separated by a split at the midline (Fig. 3C), largely touching, or entirely fused (Fig. 3D); second and third tergites without lateral fold and second metasomal suture absent; hypopygium small, straight ventrally and setose; ovipositor hardly visible, usually shorter than 0.25 times first tergite, slender and curved downwards.
Biology. Koinobiont endoparasitoids of nymphal Hemiptera (Miridae and Lygaeidae) and Psocodea (Psocidae). The early instar nymph of the host is parasitized and the mature larva emerges from the mature host nymph or adult.
Distribution. Cosmopolitan except for Antarctic, limited to Papua New Guinea in Australasia.
Conclusions
Using a multilocus phylogenetics approach and the most comprehensive taxon sampling of Euphorini to date, we were able to clarify the long standing taxonomic confusion within this tribe of economically important braconid wasps. The taxonomic uncertainty that has long impacted biological control studies of Euphorini is readily resolved with the revised generic concepts presented here, which reflects the strongly supported phylogenetic analyses, therefore providing clear distinguishing characters for the two genera Peristenus and Leiophron. With a phylogenetic framework to build upon, the next step should focus on the world revision of tribe Euphorini, with a strong alpha taxonomic component, as many of the species used in this study were undescribed or have an unknown biology.
Supplemental Information
Detailed collection locality information for all specimens used in this study
COI gene tree for MrBayes
Peristenus is colored red, and Leiophron is colored in blue, with subgenera within Leiophron shown in different colors (Leiophron sensu stricto in blue, Euphorus in purple, Euphoriana in green, Euphoriella in orange, and Mama in brown). Asterisks indicate strong nodal support ( ≥0.98 posterior probability).
28S gene tree for MrBayes
Peristenus is colored red, and Leiophron is colored in blue, with subgenera within Leiophron shown in different colors (Leiophron sensu stricto in blue, Euphorus in purple, Euphoriana in green, Euphoriella in orange, and Mama in brown). Asterisks indicate strong nodal support ( ≥0.98 posterior probability).
CAD gene tree for MrBayes
Peristenus is colored red, and Leiophron is colored in blue, with subgenera within Leiophron shown in different colors (Leiophron sensu stricto in blue, Euphorus in purple, Euphoriana in green, Euphoriella in orange, and Mama in brown). Asterisks indicate strong nodal support ( ≥0.98 posterior probability).
Concatenated gene trees for MrBayes
Peristenus is colored red, and Leiophron is colored in blue, with subgenera within Leiophron shown in different colors (Leiophron sensu stricto in blue, Euphorus in purple, Euphoriana in green, Euphoriella in orange, and Mama in brown). Asterisks indicate strong nodal support ( ≥0.98 posterior probability).
Concatenated gene trees for RAxML
Peristenus is colored red, and Leiophron is colored in blue, with subgenera within Leiophron shown in different colors (Leiophron sensu stricto in blue, Euphorus in purple, Euphoriana in green, Euphoriella in orange, and Mama in brown). Asterisks indicate strong nodal support ( ≥90 for bootstrap support).
Concatenated gene trees for IQ-Tree
Peristenus is colored red, and Leiophron is colored in blue, with subgenera within Leiophron shown in different colors (Leiophron sensu stricto in blue, Euphorus in purple, Euphoriana in green, Euphoriella in orange, and Mama in brown). Asterisks indicate strong nodal support ( ≥90 for ultrafast bootstrap support).