Description of a Zostera marina catalase gene involved in responses to temperature stress
- Published
- Accepted
- Received
- Academic Editor
- Bruno Marino
- Subject Areas
- Ecology, Marine Biology, Molecular Biology
- Keywords
- CAT, Temperature stress, mRNA expression, Enzyme activity
- Copyright
- © 2018 Zang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2018. Description of a Zostera marina catalase gene involved in responses to temperature stress. PeerJ 6:e4532 https://doi.org/10.7717/peerj.4532
Abstract
Catalase (CAT) is an antioxidant enzyme that plays a significant role in cellular protection against oxidative damage by degradation of hydrogen peroxide to oxygen and water. In the present study, the complete CAT cDNA sequence of Zostera marina was identified through expressed sequence tags (EST) analysis and the rapid amplification of cDNA ends (RACE) technique. The nucleotide sequence of ZmCAT cDNA consisted of 1,816 bp with a 1,434 bp open reading frame (ORF), encoding a polypeptide of 477 amino acid residues, which possessed significant homology to other known plant CATs. The molecular mass of the predicted protein was 55.3 kDa with an estimated isoelectric point of 6.40. Phylogenetic analysis showed that ZmCAT was closely related to CAT from gramineous species. In response to temperature stress, H2O2 and MDA contents in Z. marina increased significantly with cold stress (<10 °C) and heat stress (>25 °C). ZmCAT expression was significantly upregulated at temperatures from 5 to 10 °C and then gradually downregulated, reaching its lowest expression at 30 °C. Recombinant ZmCAT protein exhibited strong antioxidant activity over a wide temperature range, with the highest rZmCAT activity observed at 25 °C and a higher relative activity retained even with heat stress. All these results indicated that ZmCAT was a member of the plant CAT family and involved in minimizing oxidative damage effects in Z. marina under temperature stress.
Introduction
Seagrass meadows, covering ∼0.1–0.2% of the global ocean floor, are highly productive ecosystems that provide key ecological services to the marine environment (Erftemeijer & Robin Lewis III, 2006; Orth et al., 2006). Zostera marina (or eelgrass), one of the most widespread seagrass species throughout the temperate northern hemisphere, plays an important ecological engineering role by influencing sediment stabilization, carbon cycling, and food web structure (Fourqurean et al., 2012; Hemminga & Duarte, 2000). In recent decades, Z. marina, as well as other species of seagrass is declining as a result of aquatic environmental changes, which will become more noticeable with sustained global climate change (Orth et al., 2010; Short et al., 2011). Temperature has long been recognized as a major environmental factor affecting the biogeographical distribution and health of seagrass meadows (Bulthuis, 1987; Ralph, 1998). Furthermore, recent large-scale seagrass populations have been precipitously decimated worldwide from historical abundances, caused by extreme summer heat waves (Moore, Shields & Parrish, 2014; Thomson et al., 2015), suggesting the question regarding how global climate change might aggravate this decline.
Temperature stress is known to accelerate the generation of reactive oxygen species (ROS), such as singlet oxygen (1O2), hydrogen peroxide (H2O2), superoxide radical (O2⋅), and hydroxyl radicals (⋅OH), thereby leading to oxidative damage (Potters et al., 2007; Jena et al., 2013; Mittler, 2002). ROS production is a necessary process that can occur naturally and is related to both respiratory and photosynthetic metabolism, representing normal aerobic metabolism of plants. However, under abiotic stressful situations, homeostasis between the production and scavenging of ROS is altered and causes ROS accumulation (Wang et al., 2010). Excessive accumulation of ROS can cause oxidative damage through increasing lipid peroxidation (LPO), protein oxidation and degradation, and double-strand DNA breakage (Matés, 2000). To maintain homeostasis and protect against oxidative stress, plant cells have evolved antioxidant defense mechanisms, such as antioxidant enzymes, including catalase (CAT), superoxide dismutase, ascorbate peroxidase, glutathione peroxidase, glutathione reductase, and glutathione peroxidase (Hasanuzzaman, Nahar & Fujita, 2013). Among these enzymes, CAT plays a critical role in antioxidant defense pathways by efficiently catalyzing, in peroxisomes and glyoxysomes, the conversion of two molecules of H2O2 to two molecules of H2O and an O2(2H2O2 → 2H2O +O2) through the transfer of two electrons, thus counteracting H2O2 toxicity (Kashiwagi et al., 1997; Srivastava et al., 2012). Regulation mechanisms of CAT have been investigated at the transcriptional and enzymatic levels, which demonstrated that mechanisms of plant adaptions to heating and chilling stress might be positively correlated with CAT responses in maize (Scandalios, Acevedo & Ruzsa, 2000) cucumber (Gao, Qin & Yu, 2009) olive (Cansev, Gulen & Eris, 2011), broccoli (Lin, Huang & Lin, 2010) and banana (Figueroa-Yáñez et al., 2012).
The recent development of molecular resources for different seagrass species has provided important new insights into the knowledge of the transcriptional control of temperature stress responses (Franssen et al., 2014; Gu et al., 2012; Marin-Guirao et al., 2017). Our first attempts have already been carried out in the MnSOD gene of Z. marina and found that it was beneficial in minimizing oxidative damage effects of temperature stress (Liu et al., 2016). For seagrass, CAT activity has been detected in many species, such as Zostera japonica (Lin et al., 2016) and Posidonia oceanica (Sureda et al., 2008), but there have been no reports regarding detailed analysis and characterization of genes encoding CAT from any species of seagrass except for a partial CDS deposited in GenBank from Cymodocea nodosa. Study of the structural characteristics and antioxidant responses of ZmCAT will help us to determine the functional roles of ZmCAT in response to temperature stress. Thus, the purposes of this study were to: (1) clone the full-length cDNA sequence of ZmCAT and analyze the evolution of ZmCAT among other species (2) investigate the effects of temperature stress on H2O2 and malondialdehyde (MDA) contents in Z. marina (3) validate mRNA expression and recombinant protein activity of ZmCAT in response to temperature treatments.
Materials and Methods
Plant material
Eelgrass vegetative shoots with attached roots were collected from the subtidal zone in Huiquan Bay (36°05′19.4″N, 120°34′46.2″E; Qingdao City, China) The research permit was issued to the College of Marine Life Science, Ocean University of China (Permit number: 2013OUCOLS0618). Plants were maintained in plastic boxes and transported within 2 h to the laboratory. Plant surface epiphytes and the older leaves were removed and individual shoots planted in plastic hydroponic planting baskets (100 mm inner diameter), which were filled with washed and sterilized silt (sediment particle size <63 µm) from a collection site in Huiquan Bay. The baskets were placed at aquaria bottoms and provided aerated filtered natural seawater at a constant temperature of 15 °C and allowed to acclimatize for 7 d. Fluorescent lamps provided an overhead light source with an applied light intensity of 150 µmol m−2 s−1 and light/dark regime of 12/12 h. Healthy shoots of Z.marina were used for the following experiments.
Temperature treatments
A total of 30 eelgrass plants were employed for the temperature stress stimulation treatment, with plants were randomly divided into 6 groups. Five eelgrass plants were evenly spaced and kept upright in each of 6 glass aquariums (195 L). Temperature treatments were at 5, 10, 15, 20, 25, and 30 °C for 96 h, maintained by an automatic water temperature regulator (RESUN, 800W/S2TS2500) in each aquarium. During the experiment, aquaria seawater was adequately aerated and circulated using multifunctional submersible pumps to ensure mixing and homogeneous temperature within each aquarium. All experimental seawater was completely changed daily to avoid nutrient limitations, and the salinity and water temperature was checked daily. After treatment, leaf samples (n = 5) were cut into 100 mg (fresh weight, FW) sections after 5 cm segment from the leaf base, flash frozen in liquid nitrogen and rapidly stored at −80 °C until further use.
Total RNA extraction and cDNA synthesis of ZmCAT
Total RNA was extracted from fresh leaf samples (50 mg) of eelgrass with RNA Extract Kit (Aidlab Biotechnologies Ltd., Beijing, China) according to the manufacturer’s instructions. The quality and integrity of total RNA were assessed by a NanoDrop (Thermo Fisher Scientific, Delaware, USA) and 1% agarose gel electrophoresis. The first-strand synthesis was carried out using the RQ1 RNase-free DNase I (Promega Corp., Madison, WI, USA) treated raw RNA (4 µg) as template and adaptor primer-oligo (dT) as primer (Table S1). The reaction were performed at 55 °C for 1 h and terminated by heating at 70 °C for 5 min. Products from this reaction were subsequently stored at −80 °C.
Expressed sequence tags analysis and cloning of full-length ZmCAT cDNA
BLAST analysis of all ESTs sequences from the Z. marina cDNA library in National Center for Biotechnology Information (NCBI) showed that an EST of 487 bps sequence (Accession No. AM768877) showed high similarity to identified CATs from Musa acuminata subsp. Malaccensis (accession number XM009402721); thus, this EST sequence was selected for further cloning the full-length cDNA sequence of ZmCAT. Two gene specific primers, ZmCAT-Race-F1 and ZmCAT- Race-F2 (Table S1), were designed to amplify the sequence of ZmCAT cDNA by the rapid amplification of cDNA ends (RACE) technique. PCR products were gel-purified and cloned into the T-A cloning vector pMD19-T simple (Takara Bio Inc. Shiga, Japan). After transformation into competent cells of E. coli DH5 α0, positive recombinants were recognized by blue-white selection on ampicillin-containing Luria-Bertani plates and white colonies subsequently screened by PCR with M13-47 and RV-M primers (Table S1). Positive clones were sequenced on an ABI Prism™ 3730 automated DNA sequencer (Thermo Fisher Scientific Inc., Pittsburgh, PA, USA).
Bioinformatical analysis of nucleotide and amino acid sequences
The nucleotide and deduced amino acid sequences of ZmCAT cDNA were analyzed using the BLAST software of NCBI and signal peptides predicted with the SignalP 4.1 Server. Besides, functional domains and signature motifs were identified using the normal mode of the Simple Modular Architecture Research Tool (Letunic, Doerks & Bork, 2012) and PROSITE database. Cellular localization of the inferred amino acid sequence was carried out with the PSORT Prediction tool of the PSORT WWW Server. A multiple sequence alignment was generated using the Clustal Version 2.0 program (Larkin et al., 2007). A phylogenetic tree was then produced by the neighbor-joining (NJ) approach of the MEGA 6.0 program (Tamura et al., 2013), using 5,000 bootstrap replications.
Analysis of the H2O2 and MDA contents
Leaf samples were ground under liquid nitrogen and homogenized in 0.9 mL of precooled extraction solution, containing 0.05 mol/L phosphate buffer solution (pH 7.8) and 1% polyvinylpyrrolidone. Then, tissue homogenates were chilled to 4 °C until used for further processes. H2O2 concentrations were measured using the Hydrogen Peroxide Assay Kit (Beyotime Biotechnology Inc, Shanghai, China) following manufacturer’s instructions. Absorbance was read at 560 nm using a microplate reader (PerkinElmer Inc., Waltham, USA). H2O2 concentrations were calculated according to a standard concentration curve and expressed as nmol/ml. MDA contents were measured using a Plant Malondialdehyde assay kit (Nanjing Jiancheng Bioengineering Inst., Nanjing, China), which was based on the reaction of thiobarbituric acid (TBA) with MDA. The absorbance of red TBA-MDA complex was read at 532 nm using a microplate reader and the MDA content (nmol/g) was calculated according to manufacturer’s instructions.
Analysis of ZmCAT mRNA expression by quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from leaf samples and assessed as described above. Raw RNA (0.5 µg) was digested using RNase-free DNase I and then reverse-transcribed to cDNA using an AMV First Strand cDNA Synthesis Kit (Sangon Biotech Inc., Shanghai, China). The resulting cDNA samples were analyzed and quantitative PCR was performed in a Bio-Rad iQ5™ Multicolor Real-time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA), according to manufacturer’s instructions. The qPCR analysis was performed in a total liquid volume of 25.0 µL, containing 1 × SYBR Green Master Mix, 4.0 µL of the diluted cDNA templates (n = 5), 0.4 mM of each primer and 7.5 µL of DEPC-treated RNA-free water. A 140 bps product was amplified with Zm-CAT-qRT F and ZmCAT-qRT-R (Table S1) and sequenced to verify PCR specificity. The housekeeping gene eukaryotic initiation factor 4A (eIF4A) was used as an internal control to check the RT-qPCR reaction (Ransbotyn & Reusch, 2006). Primers of the internal control gene used for RT-qPCR were listed in Table S1 and produced a fragment of 125 bps. Conditions for the reactions were 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, and then 60 °C for 40 s. A melting curve was generated for each sample at the end of the RT-qPCR reaction to confirm the purity of the amplified products. Data from RT-qPCR were analyzed using the 2−ΔΔCt method of the ABI 7500 software 2.0.6 (Applied Biosystems, Inc., Foster City, USA).
Recombinant overexpression and purification of ZmCAT
The pEASY-E1 vector was used for high level, inducible expression of ZmCAT recombinant proteins, as has been reported by Liu et al. (2016). In brief, the cDNA fragment encoding the mature peptide of ZmCAT was obtained after PCR amplification with the primers, ZmCAT-recombinant-F and ZmCAT-recombinant-R (Table S1). The plasmid pEASY-E1/ZmCAT was isolated by E.Z.N.A.® Plasmid Mini Kit (Omega Bio-Tek Inc., Norcross, GA, USA) and then transformed into E. coli strain BL21 (DE3) to produce recombinant protein. The resulting strain was grown at 37 °C in liquid SOB medium coupled with ampicillin (100 mg mL−1) until the OD600 reached 0.4–0.6. The bacteria were induced for protein expression by IPTG addition to 1 mM. After incubation for 4 h at 30 °C, the bacteria were harvested by centrifugation and the recombinant protein purified by MagExtractorTM His Tag Kit (Toyobo Co Ltd., Osaka, Japan) under denaturing conditions (8 mol L−1 urea). The purified protein was next refolded in a gradient urea-TBS glycerol buffer and the resultant protein mixed 1/1 (wt/wt) with 2× Coomassie blue loading dye. Each sample was loaded on a 12% SDS-PAGE gel and Precision Plus Protein™ standards (Bio-Rad Laboratories Inc., Hercules, CA, USA) added to provide molecular mass markers.
Analysis of enzymatic activity of recombinant ZmCAT
Enzymatic activities of recombinant ZmCAT (rZmCAT) were examined using a Catalase Assay Kit (Beyotime Biotechnology Inc., Jiangsu, China) as previously developed methods (Liu et al., 2016). The purified rZmCAT protein was quantified using an Enhanced BCA Protein Assay Kit (Beyotime Biotechnology Inc, Shanghai, China). The enzymatic activities of rZmCAT protein was characterized by treating protein samples at 5 °C intervals between 0 and 40 °C for 24 h.
Statistical analysis
The data of mRNA expression and enzymatic activities were normalized to 20 °C. All data were presented as the mean ±SD (n = 5). One-way analysis of variance (ANOVA) followed by a Student-Newman-Keuls (S-N-K) multiple comparison method was used to analyze the significance of differences in the SPSS 22.0 software and p-values of <0.05 considered statistically significant differences.
Results
Cloning and sequence analysis of full-length ZmCAT cDNA
The full-length cDNA of ZmCAT was obtained by the RACE technique and deposited into GenBank (accession number KJ766310). The complete cDNA of ZmCAT consisted 1,816 bp, including a 1,434 bp open reading frame (ORF), 73 bp 5′-untranslated region (UTR), and a 309 bp 3′-UTR (Fig. 1). The 3′-UTR contained a 16 bp poly(A)-tail but no putative polyadenylation signal (AATAAA). The ORF encoded a polypeptide of 477 amino acid residues with a calculated molecular mass of 55.3 kDa, and a theoretical pI point of 6.40. No signal peptide in the deduced amino acid sequence was revealed by SignalP program analysis. Localization prediction software suggested that ZmCAT would be localized to the chloroplast. PROSITE analysis showed that the ZmCAT amino acid sequence contained highly conserved motifs, which included the proximal active site signature (54FDRERIPERVVHARGAS70) and proximal heme-ligand signature sequences (344RIFSYADTQ352). In addition, the His65 catalytic residue was found in the active site responsible for proper binding and reduction of peroxide (Fig. 1). The putative ZmCAT contained two potential N-glycosylation sites at N-28 (NNSS) and N-247 (NHSH). A CAT core domain at 18–398 (Thr18 to Rrg401) and CAT immune-responsive domain (Pro420 to Lys475) were also detected.
Figure 1: Nucleotide sequence and deduced amino acid sequences of ZmCAT catalase.
The number of nucleotides and deduced amino acid residues are shown in the left margin. A catalase domain (Thr18 to Rrg401) and a catalase-rel domain (Pro420 to Lys475) are shaded. The asterisk indicated the stop codon. The lowercase letters represent nucleotide sequences, and the capital letters indicate amino acid sequences.Multiple sequence alignment and phylogenetic analysis of ZmCAT
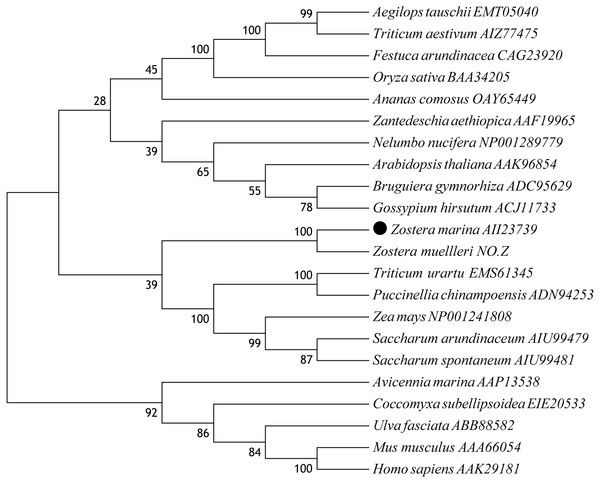
A multiple alignment of ZmCAT was performed by selecting CAT sequences of nine representative plant species (Fig. 2). The deduced amino acid sequence of ZmCAT exhibited high similarity with other previously identified CAT, such as an 88% identity with CAT from Ananas comosus, 85% identity with Zea mays, 84% identity with CATs from Zantedeschia aethiopica, Bruguiera gymnorhiza, Oryza sativa, Triticum aestivum, Saccharum arundinaceum and Festuca arundinacea and 83% identity with Arabidopsis thaliana. The molecular evolutionary relationship of ZmCAT was examined by aligning sequences of known CAT members from different taxa and constructing a phylogenetic tree from the conserved regions by the NJ method. The NJ phylogenetic tree was positioned separately into three main branches and ZmCAT found to be clustered with CAT from Zostera muelleri and located in the angiospermous cluster sub-branch (Fig. 3).
Figure 2: Multiple alignment of deduced amino acid sequences of ZmCAT with selected sequences from other plant species.
Ananas comosus (OAY65449), Zea mays (NP_001241808), Zantedeschia aethiopica (AAF19965), Bruguiera gymnorhiza (ADC95629), Oryza sativa (BAA34205) Triticum aestivum (AIZ77475), Saccharum arundinaceum (AIU99479), Festuca arundinacea (CAG23920), Arabidopsis thaliana (AAK96854). The black shaded regions represent identical amino acids, while the gray regions represent similar amino acids.Figure 3: Phylogenetic analysis based on amino acid sequences of ZmCAT and other known CATs.
Numbers above or below branches at tree nodes indicate percent bootstrap values after 5,000 replicates. The GenBank accession numbers are displayed next to each sequence. Accession NO.Z: The protein sequences of CAT in Zostera mueller was download from http://appliedbioinformatics.com.au/index.php/Seagrass_Zmu_Genome.Effects of temperature stress on the content of H2O2 and MDA
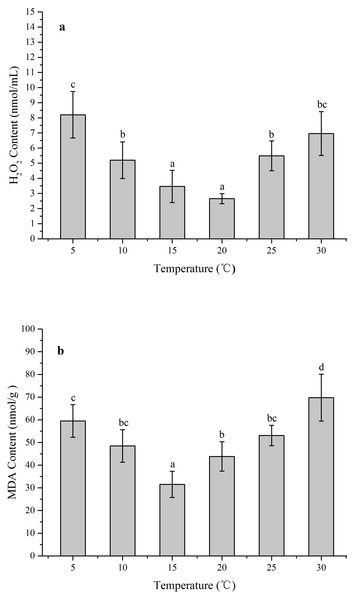
The ROS levels and membrane injury in Z. marina under different temperature treatments (5, 10, 15, 20, 25, and 30 °C) after 96 h was investigated in an experiment with different H2O2 and MDA contents When the temperature decreased from 15 to 5 °C and increased from 20 to 30 °C, the H2O2 content increased significantly (Fig. 4A). The greatest H2O2 content increases occurred in the 5 treatment (8.20 ± 1.54 nmol/mL) with 3.1-fold higher than 20 °C (p < 0.05), while the H2O2 content exhibited no significant change and kept at the temperature ranged from 15 to 20 °C. In response to temperature treatments, there were significantly increased MDA contents at the temperature decreased from 15 to 5 °C and increased from 15 °C to 30 °C (Fig. 4B). The highest MDA content occurred in the 30 °C treatments (69.74 ± 10.32 nmol/g) with 2.2-fold higher than 15 °C (p < 0.05).
Figure 4: The effects of temperature stress on H2O2 and MDA contents in Z. marina during a 96 h experiment.
(A) H2O2 content. (B) MDA content. Values shown as means ± SD (n = 5), bars marked with dissimilar letters represent significant differences (p < 0.05) from each other according to S-N-K multiple comparisons tests.Effects of temperature stress on ZmCAT mRNA expression
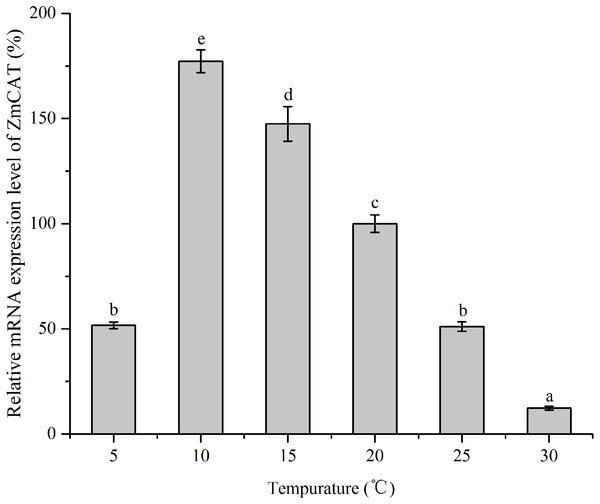
RNA levels of CAT gene were quantified using RT-qPCR technique of reverse transcripts of RNA from Z. marina that had been subjected to various temperature treatments for 96 h (Fig. 5). The data were then standardized relative to the RNA expression levels of eIF4A, a housekeeping gene consistently expressed in plants. Although relative mRNA expression levels were detected in all treatments, the CAT gene was significantly upregulated from 5 to 15 °C, with the highest expression at 10 °C. In contrast, this gene revealed rather low expression in heat-stressed conditions (25 and 30 °C); and ZmCAT expression at 10 °C was 13-fold higher than at 30 °C (p < 0.05). Hence, temperature stress suppressed ZmCAT expression, but ZmCAT was expressed at lower values with elevated heat stress than with cold stress.
Figure 5: The effects of temperature stress on the relative mRNA expression levels of ZmCAT.
Amplification of eIF4A gene was used as an internal control. The ZmCAT expression levels at 20°C was set to 100%. Values shown as means ± SD (n = 5), bars marked with dissimilar letters represent significant differences (p < 0.05) from each other according to S-N-K multiple comparisons tests.Effects of temperature on recombinant ZmCAT activity
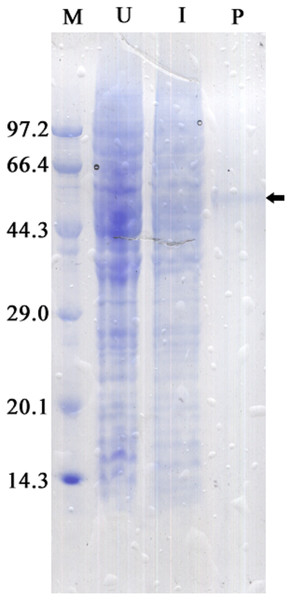
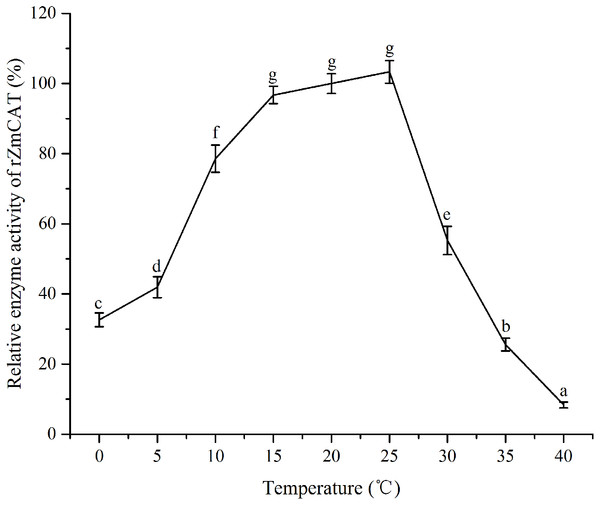
The recombinant protein activity of ZmCAT was determined to examine whether it was functionally active in the antioxidant response under temperature stress. Highly purified ZmCAT proteins for biochemical characterization under temperature stress were obtained using E. coli BL21 (DE3) as a heterologous expression system to express ZmCAT protein and demonstrate ZmCAT’s antioxidant activity, based on H2O2 consumption. Recombinant ZmCAT (rZmCAT) protein was analyzed by SDS-PAGE, appearing as a distinct band at a molecular weight of ∼55 kDa, which was close to the calculated molecular mass of ZmCAT (55.3 kDa, Fig. 6). The enzymatic activity of recombinant protein was 19,310.5 ± 83.5 U mg−1 at 20 °C. The stability of rZmCAT was investigated by measuring the enzymatic activities of its recombinant protein under different temperature treatments (0–40 °C). The enzymatic activity increased rapidly from 0 to 15 °C and remained at a high level from 15 to 25 °C, with >95% of the relative enzymatic activity retained in the temperature range of 15–25 °C. After reaching peak activity at 25 °C the rZmCAT gradually lost enzymatic activity and almost devitalized at 40 °C. It was noteworthy that, although the relative activity was clearly reduced from 25 to 40 °C, >20% of its activity was retained after incubation at 35 °C for 96 h (Fig. 7).
Figure 6: SDS-PAGE analysis of recombinant Zm. CAT protein expressed in E. coli BL21 (DE3).
Samples analyzed included low molecular weight protein markers (M, lane 1), un-induced expression of pEASY-E1/ZmCAT in total BL21 (DE3) cell lysates (U, lane 2), 1 mM IPTG induced expression of pEASY-E1/ZmCAT in BL21 (DE3) (I, lane 3), and the purified recombinant protein of ZmCAT (P, lane 4).Figure 7: The effects of temperature stress on the enzymatic activity of recombinant ZmCAT protein.
The enzymatic activity at 20°C was set to 100%. Each values is shown as mean ± S.D. (n = 5), and bars with different characters were significantly different (p < 0.05).Discussion
Catalase plays an important role in the antioxidant defense system by degrading the H2O2 to H2O and O2 ( Unwin, 1975). In recent years, several CAT genes have been identified in different studies (Du, Zhou & Huang, 2013; Lin, Huang & Lin, 2010), but the present study was the first to deal with CAT cloned and characterized from eelgrass. ZmCAT was found to possess most of the main characteristic amino acid residues, motifs, and elements of the CAT protein family. The sequence and multiple alignment results indicated that the active site signature (54FDRERIPERVVHARGAS70) and heme-ligand signature motifs (344RIFSYADTQ352) were conserved in ZmCAT (Fig. 1). Multiple alignments revealed high sequence identity with CATs from other known plant species, ranging from 83 to 88% (Fig. 2). Phylogenetic relationship collectively suggested that ZmCAT was grouped within the angiosperms and might have similar functions to CATs from other plant species (Fig. 3). All present results showed that ZmCAT was a representative member of the plant CAT family.
Temperature stress usually causes injury such as excessive accumulation of ROS in plant cells, which seriously damage cell membranes, protein, and nucleic acids (Gill & Tuteja, 2010; Vighi et al., 2016). Low temperature stress can induce destructive oxidative processes leading to an increase of MDA content in tissue, which could reflect the degree of LPO and structural integrity of plant membranes (Li et al., 2013). Therefore, redox states and membrane integrity responses of Z. marina exposed to temperature stress were identified by investigating the effects of such stress on H2O2 and MDA contents. In response to temperature stress, H2O2 and MDA contents in Z. marina significantly increased at cold stress (<10 °C) and heat stress (>25 °C) and the highest H2O2 and MDA content respectively occurred at 5 and 30 °C (Fig. 4). The observed increased H2O2 and MDA contents were indications that Z. marina was unable to suppress oxidative stress under extreme temperature stress, which might influenced its physiological functions and survival.
The expression profile of CAT was a key part of the plant defense system against oxidative stress (Bi et al., 2017). In the present study, ZmCAT appeared to be regulated at the transcriptional level for detoxifying increased H2O2 during cold stress conditions. The optimal temperature for Z. marina growth has been reported to be 15.3 ±1.6 °C (Lee, Sang & Kim, 2007), while the expression concentration increased rapidly from 5 to 10 °C and the highest ZmCAT gene expression observed at 10 °C (Fig. 4). Cold stress can lead to excess H2O2 accumulation and increased H2O2 content might act as signals for inducing antioxidant gene expressions. Therefore, it was deduced that the ZmCAT gene was induced and upregulated to reduce oxidative damage under lower temperature stress, but sustained extreme low temperature stress (5 °C) damages tissue and leads to decreased ZmCAT expression and increased H2O2 content and membrane injury Similar results have been obtained by Baek & Skinner (2003), who found that CAT gene expression was significantly higher in winter wheat after cold-stress exposure. However, ZmCAT gene expression was suppressed when the temperature increased from 25 to 30 °C, which showed the inhibiting effects of heat stress conditions on ZmCAT gene expression that, in turn, might have resulted in intracellular H2O2 accumulation.
Information on the recombinant enzyme activity of the ZmCAT after temperature treatments was helpful for a better understanding of its physiological function. The relative enzymatic activity retained with >95% by the rZmCAT at 15–25 °C. Field surveys and experiments have confirmed that high water temperatures >25 °C in summer heat waves increasingly threatens seagrass performance and >30 °C can be lethal (Ehlers, Worm & Reusch, 2008; Kaldy, 2014; Nejrup & Pedersen, 2008). However, the highest H2O2 removal activity by rZmCAT was observed at 25 °C. Moreover, a relative activity of 55% was retained by rZmCAT at 30 °C (Fig. 6). On the basis of its strong antioxidant activity at high temperatures, ZmCAT was suggested here to play a role against heat stress, as a potent ROS-detoxifying enzyme in eelgrass. Several CATs have been demonstrated to have high thermal stability, including Pyropia yezoensis CAT (Py CAT), Oryza sativa CAT-A (Os CatA), and CAT-C (Os CatC), with reported optimal temperatures of ∼30 °C (Li et al., 2012; Vighi et al., 2016). In addition, rZmCAT exhibited higher heat sensitivity than that reported for Py CAT. The rZmCAT activity declined rapidly as temperature exceeded 25 °C and <10% activity was retained at 40 °C, similar to that reported for OsCatC. A potential reason for this difference might have been because CAT in Z. marina and P. yezoensis must cope with different living environments, with P. yezoensis living in the upper intertidal zone with high and variable temperature, while Z. marina is a subtidal population and in a more stable environment. Thus, a more stable enzyme is generated in P. yezoensis as an adaptation to harsh living conditions. It should be noted that the increased H2O2 and MDA content at extreme temperature stress (5 °C and 30 °C) might have resulted from both ZmCAT gene expression and enzymatic activity, which were significantly inhibited. Interestingly, ZmCAT expression characterization was different from enzyme activity under heat stress. ZmCAT mRNA expression was downregulated after 25 and 30 °C temperature treatments for 96 h, while the enzymatic activity first increased and then declined. ZmCAT appeared to respond more sensitively at the transcriptional level than at the enzymatic level. These results indicated that ZmCAT in eelgrass survived under heat stress through elevated enzymatic activity rather than upregulated gene transcription to protect cellular components against ROS effects produced as a consequence of oxidative stress. This information might be valuable for further analysis of the profile and functional diversity of CAT.
Conclusions
In this study, a novel CAT gene, named as ZmCAT, was identified in Z. marina and analysis of its deduced amino acid sequence showed typical CAT functional domains, including the CAT proximal heme-ligand signature and CAT proximal activity site. Multiple sequence alignment as well as phylogenetic analysis provided evidence for the evolutionary conservation of ZmCAT, which was a typical member of the plant CAT family. Temperature stress increased oxidative stress and membrane injury in Z. marina. To counter this stress, ZmCAT presented an antioxidant defense and protection by high enzymatic activity under heat stress and by upregulated gene transcription under cold stress. However, both ZmCAT gene expression and enzymatic activity were significantly suppressed at extreme temperatures (5 and 30 °C) which indicated that the ability of ZmCAT to scavenge H2O2 was weakened and led to changes in the cellular redox state and influenced its physiological function in eelgrass. Gene expression and enzymatic activity of ZmCAT varied under different temperature stresses, which suggested that ZmCAT might have played a central role in mitigating oxidative damage under temperature stress.