DNA-barcoding of forensically important blow flies (Diptera: Calliphoridae) in the Caribbean Region
- Published
- Accepted
- Received
- Academic Editor
- Dezene Huber
- Subject Areas
- Biodiversity, Entomology, Molecular Biology, Taxonomy, Veterinary Medicine
- Keywords
- Calliphoridae, Caribbean, DNA-barcoding, Forensic entomology, Diptera
- Copyright
- © 2017 Yusseff-Vanegas and Agnarsson
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2017. DNA-barcoding of forensically important blow flies (Diptera: Calliphoridae) in the Caribbean Region. PeerJ 5:e3516 https://doi.org/10.7717/peerj.3516
Abstract
Correct identification of forensically important insects, such as flies in the family Calliphoridae, is a crucial step for them to be used as evidence in legal investigations. Traditional identification based on morphology has been effective, but has some limitations when it comes to identifying immature stages of certain species. DNA-barcoding, using COI, has demonstrated potential for rapid and accurate identification of Calliphoridae, however, this gene does not reliably distinguish among some recently diverged species, raising questions about its use for delimitation of species of forensic importance. To facilitate DNA based identification of Calliphoridae in the Caribbean we developed a vouchered reference collection from across the region, and a DNA sequence database, and further added the nuclear ITS2 as a second marker to increase accuracy of identification through barcoding. We morphologically identified freshly collected specimens, did phylogenetic analyses and employed several species delimitation methods for a total of 468 individuals representing 19 described species. Our results show that combination of COI + ITS2 genes yields more accurate identification and diagnoses, and better agreement with morphological data, than the mitochondrial barcodes alone. All of our results from independent and concatenated trees and most of the species delimitation methods yield considerably higher diversity estimates than the distance based approach and morphology. Molecular data support at least 24 distinct clades within Calliphoridae in this study, recovering substantial geographic variation for Lucilia eximia, Lucilia retroversa, Lucilia rica and Chloroprocta idioidea, probably indicating several cryptic species. In sum, our study demonstrates the importance of employing a second nuclear marker for barcoding analyses and species delimitation of calliphorids, and the power of molecular data in combination with a complete reference database to enable identification of taxonomically and geographically diverse insects of forensic importance.
Introduction
Forensic entomology is the application of the study of insects in legal investigations. Although several groups of insects, mainly of the orders Diptera and Coleoptera, are associated with cadaveric decomposition, blow flies (Diptera: Calliphoridae) are among the most dominant and conspicuous insects in the decomposition process (Catts, 1992). They are useful to determine time of death and, in particular situations, cause of death (Goff, 2000) or relocation of a body (Matuszewski, Szafalowicz & Jarmusz, 2013). During the last five decades of intensive studies in forensic entomology (Byrd & Castner, 2010; Catts & Haskell, 1990; Goff, 2000; Smith, 1986; Tomberlin & Benbow, 2015), the acceptance of insects as evidence in legal investigations has increased gradually and they are now included as standard operating procedures in crime scene investigations in many countries (Tomberlin & Benbow, 2015). Determining the post mortem interval (PMI) is one of the most important tasks during an investigation, and the use of immature stages of Calliphoridae is essential whenever time of death is difficult to establish based on other means (Catts & Haskell, 1990). Although the accurate determination of PMI and period of insect activity (PIA) depend of several factors that are discussed in detail by Catts (1992), the first one, and most important to resolve, is the correct identification of the specimens found at the crime scene. As each species has a specific developmental rate and range of distribution, the accurate identification of insects, mainly the larval stages, is critical because the incorrect determination will invalidate the estimated post mortem interval and impact other interpretations of the evidence (Goff, 2000; Wells & LaMotte, 2001).
Morphology is most commonly used to identify adult insects involved in cadaveric decomposition and taxonomic keys are available for most of the Calliphoridae species. In general, these taxonomic keys include the detailed description of the male and female genitalia, which is examined when external characteristics are not sufficient to establish identity (Tantawi, Whitworth & Sinclair, 2017; Whitworth, 2010; Whitworth, 2014; Whitworth & Rognes, 2012). Identification of immature stages (eggs, larvae and pupae) is more challenging, but possible when detailed taxonomic descriptions exist (Greenberg & Szyska, 1984; Sukontason et al., 2005; Szpila et al., 2013a; Szpila et al., 2014; Szpila & Villet, 2011; Wells, Byrd & Tantawi, 1999). However, in places like the Caribbean, where forensic entomology has not yet been developed, this approach is limited due to the lack of detailed descriptions of immature stages. For instance, from the 18 forensically important calliphorid species currently recognized in the Caribbean, plus the most important livestock pest parasite in the Americas, C. hominivorax (Whitworth, 2010), only eight have been documented well enough to be identified based on larvae, mainly using morphology of the third instar (Florez & Wolff, 2009; Wells, Byrd & Tantawi, 1999). For the other species, the identification of immature specimens would need to be done by rearing them to adulthood (Goff, 2000), which is time consuming, may delay legal investigations, and relies on the survival of larvae in the laboratory. Given local endemism, the scarce studies on this group in the Caribbean, and the lack of knowledge of immature stages for at least 11 species, developing alternative tools for identification is important.
With the advances in molecular methods, DNA barcoding has become a widely used technique for species delimitation and identification. This approach allows the identification of specimens during any development stage, including incomplete or damaged specimens, does not require taxonomic expertise, and it is also useful to recognize cryptic species that morphological approaches may not detect (Hebert et al., 2003; Hebert et al., 2004a; Hebert et al., 2004b). Worldwide many authors have used this method to identify species of the family Calliphoridae and these studies showed the potential of the ‘standard barcoding gene’ cytochrome c oxidase subunit I (COI) to distinguish between forensically significant species (Aly & Wen, 2013; Chen et al., 2009; Harvey et al., 2003; Liu et al., 2011; Nelson, Wallman & Dowton, 2007; Wells & Williams, 2007). However, COI does not reliably distinguish among certain closely related calliphorid species, specifically Chrysomya saffranea and Ch. megacephala (Harvey et al., 2008; Nelson, Wallman & Dowton, 2007), Ch. semimetalica and Ch. latifrons (Nelson, Wallman & Dowton, 2007), Calliphora stygia and C. albifrontalis, C. dubia and C. augur (Harvey et al., 2008; Wallman & Donnellan, 2001), C. aldrichia and C. montana (Tantawi, Whitworth & Sinclair, 2017), Cochliomyia macellaria and Co. aldrichi (Yusseff-Vanegas & Agnarsson, 2016), Lucilia mexicana and L. coeruleiviridis (DeBry et al., 2013; Whitworth, 2014), L. bazini and L. hainanenesis (Chen et al., 2014), L. illustris and L. caesar (Reibe, Schmitz & Madea, 2009; Sonet et al., 2012), L. cuprina and L. sericata (Williams & Villet, 2013). Given the serious implications of misidentification of forensic insects, an improved protocol for accurate identification is necessary. We propose using the nuclear internal transcribed spacer ITS2 as a second barcoding locus for taxonomic species determinations in calliphorids as suggested by GilArriortua et al. (2014). Although evaluations of ITS2 as unique identification marker have limitations for some taxa (Agnarsson, 2010), several studies have shown the potential application of ITS2 for blowfly species identification (Jordaens et al., 2013a; Nelson, Wallman & Dowton, 2007; Nelson, Wallman & Dowton, 2008, Song, Wang & Liang, 2008; Yusseff-Vanegas & Agnarsson, 2016). We expect a combination of barcodes from the nuclear and mitochondrial genomes to offer a general, simple and reliable way of identifying forensically important insects, even problematic sister species, as successfully done in certain other arthropod groups (Anslan & Tedersoo, 2015; Cao et al., 2016).
The success of DNA barcoding directly links to the quality of the underlying database (Candek & Kuntner, 2015; Coddington et al., 2016; DeBry et al., 2013; Harvey et al., 2003) not only in terms of the quality of identifications but also in terms of taxon sampling (species, geographic localities, populations). Existing efforts in this respect are lacking for Calliphoridae in the Caribbean, limiting the reliability of this technique for delimitation of species. Hitherto, three studies have included molecular data of a few Calliphoridae from the Caribbean (McDonagh, Garcia & Stevens, 2009; Whitworth, 2014; Yusseff-Vanegas & Agnarsson, 2016); they lack the geographic variation necessary to estimate the ratio between intraspecific variation and interspecific divergence from which barcoding accuracy depends (Meyer & Paulay, 2005). Our study provides the first thorough molecular study of Caribbean Calliphoridae.
Our aims are: (1) to establish COI barcode libraries for all Caribbean species and to test if barcodes offer reliable means of their identification, (2) to assess the usefulness of ITS2 as a second barcoding locus in species delimitation and identification, and, (3) to improve online databases with sequences from the Caribbean, including specimens from multiple localities in each island covering the geographic range for each species. To achieve these goals, we sampled 468 specimens of Calliphoridae representing 19 species.
| Genus | Species | Voucher ID | Country | Latitude | Longitude | COI | ITS2 |
|---|---|---|---|---|---|---|---|
| Calliphora | maestrica | DR084 | Hispaniola | N18.82138 | W70.67935 | MF097182 | MF097580 |
| Calliphora | maestrica | DR085 | Hispaniola | N18.82138 | W70.67935 | MF097183 | – |
| Calliphora | maestrica | DR086 | Hispaniola | N18.82138 | W70.67935 | MF097184 | – |
| Calliphora | maestrica | DR087 | Hispaniola | N18.82138 | W70.67935 | MF097185 | – |
| Calliphora | maestrica | DR088 | Hispaniola | N18.82138 | W70.67935 | MF097186 | MF097581 |
| Chloroprocta | idioidea | CU008 | Cuba | N20.054178 | W76.917603 | MF097187 | MF097582 |
| Chloroprocta | idioidea | CU047 | Cuba | N21.582414 | W77.783464 | MF097188 | MF097583 |
| Chloroprocta | idioidea | CU048 | Cuba | N21.582414 | W77.783464 | MF097189 | MF097584 |
| Chloroprocta | idioidea | CU049 | Cuba | N21.582414 | W77.783464 | MF097190 | – |
| Chloroprocta | idioidea | DR031 | Hispaniola | N18.316572 | W71.576447* | MF097191 | – |
| Chloroprocta | idioidea | DR044 | Hispaniola | N18.316572 | W71.576447* | MF097192 | MF097585 |
| Chloroprocta | idioidea | DR045 | Hispaniola | N18.316572 | W71.576447* | MF097193 | – |
| Chloroprocta | idioidea | DR051 | Hispaniola | N19.06753 | W69.46445 | MF097194 | – |
| Chloroprocta | idioidea | DR052 | Hispaniola | N19.06753 | W69.46445 | MF097195 | MF097586 |
| Chloroprocta | idioidea | ME001 | Mexico | N21.07645 | W89.501083 | – | MF097587 |
| Chloroprocta | idioidea | ME002 | Mexico | N21.07645 | W89.501083 | MF097196 | MF097588 |
| Chrysomya | albiceps | CO003 | Colombia | N5.900544 | W74.852897* | – | MF097589 |
| Chrysomya | albiceps | CO004 | Colombia | N5.900544 | W74.852897* | – | MF097590 |
| Chrysomya | albiceps | CO005 | Colombia | N5.900544 | W74.852897* | – | MF097591 |
| Chrysomya | albiceps | LA103 | Martinique | N14.47428 | W60.81463 | MF097199 | MF097592 |
| Chrysomya | albiceps | LA104 | Martinique | N14.47428 | W60.81463 | MF097200 | MF097593 |
| Chrysomya | albiceps | LA125 | Saint Lucia | N14.100031 | W60.92654 | MF097201 | MF097594 |
| Chrysomya | albiceps | LA135 | Barbados | N13.2051667 | W59.5295556 | MF097197 | – |
| Chrysomya | albiceps | LA136 | Barbados | N13.2051667 | W59.5295556 | MF097198 | – |
| Chrysomya | megacephala | CO006 | Colombia | N5.900544 | W74.852897* | MF097202 | MF097595 |
| Chrysomya | megacephala | CO007 | Colombia | N5.900544 | W74.852897* | – | MF097596 |
| Chrysomya | megacephala | CO008 | Colombia | N6.266242 | W77.374903* | MF097203 | MF097597 |
| Chrysomya | megacephala | CO009 | Colombia | N5.900544 | W74.852897* | – | MF097598 |
| Chrysomya | megacephala | DR017 | Hispaniola | N19.89155 | W071.65806 | MF097205 | – |
| Chrysomya | megacephala | DR018 | Hispaniola | N19.89155 | W071.65806 | MF097206 | – |
| Chrysomya | megacephala | DR068 | Hispaniola | N19.06710 | W69.46004 | MF097207 | – |
| Chrysomya | megacephala | DR069 | Hispaniola | N19.06710 | W69.46004 | MF097208 | – |
| Chrysomya | megacephala | DR101 | Hispaniola | N18.35698 | W68.61609 | MF097209 | – |
| Chrysomya | megacephala | DR102 | Hispaniola | N18.35698 | W68.61609 | MF097210 | – |
| Chrysomya | megacephala | DR103 | Hispaniola | N18.35698 | W68.61609 | MF097211 | – |
| Chrysomya | megacephala | DR104 | Hispaniola | N18.35698 | W68.61609 | MF097212 | – |
| Chrysomya | megacephala | DR116 | Hispaniola | N18.32902 | W68.80995 | MF097213 | MF097599 |
| Chrysomya | megacephala | DR117 | Hispaniola | N18.32902 | W68.80995 | MF097214 | MF097611 |
| Chrysomya | megacephala | DR118 | Hispaniola | N18.32902 | W68.80995 | MF097215 | – |
| Chrysomya | megacephala | DR119 | Hispaniola | N18.32902 | W68.80995 | MF097216 | – |
| Chrysomya | megacephala | FL003 | Florida, USA | N25.614383 | W80.584467 | KX529521 | KX529561 |
| Chrysomya | megacephala | FL004 | Florida, USA | N25.614383 | W80.584467 | MF097218 | – |
| Chrysomya | megacephala | FL011 | Florida, USA | N25.086633 | W80.452217 | MF097219 | – |
| Chrysomya | megacephala | JA004 | Jamaica | N18.0598056 | W77.5311944 | – | MF097600 |
| Chrysomya | megacephala | LA062 | Dominica | N15.34066 | W61.33351 | MF097220 | – |
| Chrysomya | megacephala | LA001 | Saint Eustatius | N17.47637 | W62.97470 | MF097225 | – |
| Chrysomya | megacephala | LA003 | Saint Eustatius | N17.47637 | W62.97470 | MF097217 | – |
| Chrysomya | megacephala | LA025 | Saint-Martin | N18.07779 | W63.05772 | MF097235 | – |
| Chrysomya | megacephala | LA055 | Saint Barthélemy | N17.91924 | W62.86366 | MF097234 | – |
| Chrysomya | megacephala | LA063 | Dominica | N15.34066 | W61.33351 | MF097204 | – |
| Chrysomya | megacephala | LA088 | Guadeloupe | N16.37752 | W61.47869 | MF097221 | – |
| Chrysomya | megacephala | LA089 | Guadeloupe | N16.37752 | W61.47869 | MF097222 | – |
| Chrysomya | megacephala | LA093 | Nevis | N17.14145 | W62.57784 | MF097226 | – |
| Chrysomya | megacephala | LA116 | Saint Kitts | N17.3404083 | W62.7410389 | MF097223 | – |
| Chrysomya | megacephala | LA117 | Saint Kitts | N17.3404083 | W62.7410389 | MF097224 | – |
| Chrysomya | megacephala | LA123 | Saint Lucia | N14.100031 | W60.92654 | – | MF097604 |
| Chrysomya | megacephala | ME013 | Mexico | N25.598592 | W103.441156 | – | MF097601 |
| Chrysomya | megacephala | ME014 | Mexico | N25.598592 | W103.441156 | – | MF097602 |
| Chrysomya | megacephala | PR038 | Puerto Rico | N18.412972 | W66.026619 | MF097227 | – |
| Chrysomya | megacephala | PR124 | Puerto Rico | N18.370953 | W66.026619 | MF097228 | – |
| Chrysomya | megacephala | PR125 | Puerto Rico | N18.370953 | W66.026619 | MF097229 | MF097603 |
| Chrysomya | megacephala | PR1251 | Puerto Rico | N18.370953 | W66.026619 | MF097230 | – |
| Chrysomya | megacephala | PR126 | Puerto Rico | N18.370953 | W66.026619 | MF097231 | – |
| Chrysomya | megacephala | PR138 | Puerto Rico | N18.447911 | W65.948617 | MF097232 | – |
| Chrysomya | megacephala | PR139 | Puerto Rico | N18.447911 | W65.948617 | MF097233 | – |
| Chrysomya | rufifacies | LA056 | Saint Barthélemy | N17.91924 | W62.86366 | MF097236 | – |
| Chrysomya | rufifacies | LA057 | Saint Barthélemy | N17.91924 | W62.86366 | MF097237 | – |
| Chrysomya | rufifacies | CU001 | Cuba | N20.054178 | W76.917603 | MF097238 | – |
| Chrysomya | rufifacies | CU003 | Cuba | N20.054178 | W76.917603 | MF097239 | – |
| Chrysomya | rufifacies | CU004 | Cuba | N20.054178 | W76.917603 | KX529555 | KX529562 |
| Chrysomya | rufifacies | CU005 | Cuba | N20.054178 | W76.917603 | MF097240 | – |
| Chrysomya | rufifacies | CU009 | Cuba | N20.054178 | W76.917603 | MF097241 | – |
| Chrysomya | rufifacies | CU034 | Cuba | N22.621386 | W83.725944 | MF097242 | – |
| Chrysomya | rufifacies | CU035 | Cuba | N22.621386 | W83.725944 | MF097243 | – |
| Chrysomya | rufifacies | CU036 | Cuba | N22.621386 | W83.725944 | MF097244 | – |
| Chrysomya | rufifacies | CU037 | Cuba | N22.621386 | W83.725944 | MF097245 | – |
| Chrysomya | rufifacies | DR001 | Hispaniola | N19.89155 | W71.65806 | MF097248 | – |
| Chrysomya | rufifacies | DR002 | Hispaniola | N19.89155 | W71.65806 | MF097249 | – |
| Chrysomya | rufifacies | DR003 | Hispaniola | N19.89155 | W71.65806 | MF097250 | – |
| Chrysomya | rufifacies | DR004 | Hispaniola | N19.89155 | W71.65806 | MF097251 | – |
| Chrysomya | rufifacies | DR006 | Hispaniola | N19.89155 | W71.65806 | MF097252 | – |
| Chrysomya | rufifacies | DR007 | Hispaniola | N19.89155 | W71.65806 | MF097253 | – |
| Chrysomya | rufifacies | DR008 | Hispaniola | N19.89155 | W71.65806 | MF097254 | – |
| Chrysomya | rufifacies | DR016 | Hispaniola | N19.89155 | W71.65806 | MF097255 | – |
| Chrysomya | rufifacies | DR036 | Hispaniola | N18.316572 | W71.576447* | MF097256 | – |
| Chrysomya | rufifacies | DR037 | Hispaniola | N18.316572 | W71.576447* | MF097257 | – |
| Chrysomya | rufifacies | DR038 | Hispaniola | N18.316572 | W71.576447* | MF097258 | – |
| Chrysomya | rufifacies | DR039 | Hispaniola | N18.316572 | W71.576447* | MF097259 | – |
| Chrysomya | rufifacies | DR070 | Hispaniola | N19.06710 | W69.46004 | MF097260 | – |
| Chrysomya | rufifacies | DR071 | Hispaniola | N19.06710 | W69.46004 | MF097261 | MF097605 |
| Chrysomya | rufifacies | DR0711 | Hispaniola | N19.06710 | W69.46004 | – | MF097606 |
| Chrysomya | rufifacies | DR093 | Hispaniola | N18.35698 | W68.61609 | MF097262 | – |
| Chrysomya | rufifacies | DR094 | Hispaniola | N18.35698 | W68.61609 | MF097263 | – |
| Chrysomya | rufifacies | DR095 | Hispaniola | N18.35698 | W68.61609 | MF097264 | – |
| Chrysomya | rufifacies | DR096 | Hispaniola | N18.35698 | W68.61609 | MF097265 | – |
| Chrysomya | rufifacies | DR097 | Hispaniola | N18.35698 | W68.61609 | MF097266 | – |
| Chrysomya | rufifacies | DR098 | Hispaniola | N18.35698 | W68.61609 | MF097267 | – |
| Chrysomya | rufifacies | DR099 | Hispaniola | N18.35698 | W68.61609 | MF097268 | – |
| Chrysomya | rufifacies | DR100 | Hispaniola | N18.35698 | W68.61609 | MF097269 | – |
| Chrysomya | rufifacies | DR132 | Hispaniola | N18.32902 | W68.80995 | MF097270 | – |
| Chrysomya | rufifacies | DR133 | Hispaniola | N18.32902 | W68.80995 | MF097271 | – |
| Chrysomya | rufifacies | DR135 | Hispaniola | N19.741319 | W70.654975* | MF097272 | – |
| Chrysomya | rufifacies | DR150 | Hispaniola | N19.34405 | W70.14824 | MF097273 | – |
| Chrysomya | rufifacies | DR151 | Hispaniola | N19.34405 | W70.14824 | MF097274 | – |
| Chrysomya | rufifacies | DR152 | Hispaniola | N19.34405 | W70.14824 | MF097275 | – |
| Chrysomya | rufifacies | DR155 | Hispaniola | N19.34405 | W70.14824 | MF097276 | – |
| Chrysomya | rufifacies | DR157 | Hispaniola | N18.32902 | W68.80995 | MF097277 | – |
| Chrysomya | rufifacies | DR158 | Hispaniola | N18.32902 | W68.80995 | MF097278 | – |
| Chrysomya | rufifacies | DR159 | Hispaniola | N18.32902 | W68.80995 | MF097279 | – |
| Chrysomya | rufifacies | DR160 | Hispaniola | N18.32902 | W68.80995 | MF097280 | – |
| Chrysomya | rufifacies | DR161 | Hispaniola | N18.32902 | W68.80995 | MF097281 | – |
| Chrysomya | rufifacies | DR162 | Hispaniola | N18.32902 | W68.80995 | MF097282 | – |
| Chrysomya | rufifacies | DR163 | Hispaniola | N18.32902 | W68.80995 | MF097283 | – |
| Chrysomya | rufifacies | FL001 | Florida, USA | N25.614383 | W80.584467 | MF097288 | – |
| Chrysomya | rufifacies | FL010 | Florida, USA | N25.086633 | W80.452217 | MF097289 | MF097607 |
| Chrysomya | rufifacies | JA003 | Jamaica | N18.0598056 | W77.5311944 | MF097293 | MF097608 |
| Chrysomya | rufifacies | LA002 | Saint Eustatius | N17.47637 | W62.97470 | MF097284 | – |
| Chrysomya | rufifacies | LA004 | Saint Eustatius | N17.47637 | W62.97470 | MF097285 | – |
| Chrysomya | rufifacies | LA005 | Saint Eustatius | N17.47637 | W62.97470 | MF097286 | – |
| Chrysomya | rufifacies | LA006 | Saint Eustatius | N17.47637 | W62.97470 | MF097287 | – |
| Chrysomya | rufifacies | LA041 | Saint-Martin | N18.11677 | W63.03902 | MF097316 | – |
| Chrysomya | rufifacies | LA042 | Saint-Martin | N18.11677 | W63.03902 | MF097317 | – |
| Chrysomya | rufifacies | LA043 | Saint-Martin | N18.11677 | W63.03902 | MF097318 | – |
| Chrysomya | rufifacies | LA044 | Saint-Martin | N18.11677 | W63.03902 | MF097319 | – |
| Chrysomya | rufifacies | LA069 | Dominica | N15.34066 | W61.33351 | MF097246 | – |
| Chrysomya | rufifacies | LA072 | Dominica | N15.34066 | W61.33351 | MF097247 | – |
| Chrysomya | rufifacies | LA090 | Guadeloupe | N16.37752 | W61.47869 | MF097290 | – |
| Chrysomya | rufifacies | LA091 | Guadeloupe | N16.37752 | W61.47869 | MF097291 | – |
| Chrysomya | rufifacies | LA092 | Guadeloupe | N16.37752 | W61.47869 | MF097292 | – |
| Chrysomya | rufifacies | LA101 | Martinique | N14.47428 | W60.81463 | MF097310 | – |
| Chrysomya | rufifacies | LA108 | Montserrat | N16.77608 | W62.30904 | MF097309 | – |
| Chrysomya | rufifacies | LA110 | Saint Kitts | N17.3404083 | W62.7410389 | MF097294 | MF097609 |
| Chrysomya | rufifacies | M074 | Mona, Puerto Rico | N18.086239 | W67.906339 | MF097295 | – |
| Chrysomya | rufifacies | M075 | Mona, Puerto Rico | N18.086239 | W67.906339 | MF097296 | – |
| Chrysomya | rufifacies | M082 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097297 | – |
| Chrysomya | rufifacies | M083 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097298 | – |
| Chrysomya | rufifacies | M089 | Mona, Puerto Rico | N18.06301 | W67.88728 | MF097299 | – |
| Chrysomya | rufifacies | M090 | Mona, Puerto Rico | N18.06301 | W67.88728 | MF097300 | – |
| Chrysomya | rufifacies | M091 | Mona, Puerto Rico | N18.06301 | W67.88728 | MF097301 | – |
| Chrysomya | rufifacies | M093 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097302 | – |
| Chrysomya | rufifacies | M094 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097303 | – |
| Chrysomya | rufifacies | M095 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097304 | – |
| Chrysomya | rufifacies | M096 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097305 | – |
| Chrysomya | rufifacies | M101 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097306 | – |
| Chrysomya | rufifacies | M108 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097307 | – |
| Chrysomya | rufifacies | M109 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097308 | – |
| Chrysomya | rufifacies | PR117 | Puerto Rico | N18.370953 | W66.026619 | MF097311 | – |
| Chrysomya | rufifacies | PR118 | Puerto Rico | N18.370953 | W66.026619 | MF097312 | – |
| Chrysomya | rufifacies | PR119 | Puerto Rico | N18.370953 | W66.026619 | MF097313 | – |
| Chrysomya | rufifacies | PR120 | Puerto Rico | N18.370953 | W66.026619 | MF097314 | – |
| Chrysomya | rufifacies | PR130 | Puerto Rico | N18.093306 | W65.556083 | MF097315 | MF097610 |
| Cochliomyia | aldrichi | M080 | Mona, Puerto Rico | N18.084222 | W65.939417 | KX529529 | KX529563 |
| Cochliomyia | aldrichi | M084 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097320 | – |
| Cochliomyia | aldrichi | M085 | Mona, Puerto Rico | N18.11125 | W67.933447 | KX529530 | KX529564 |
| Cochliomyia | aldrichi | M086 | Mona, Puerto Rico | N18.06301 | W67.88728 | KX529531 | KX529565 |
| Cochliomyia | aldrichi | M087 | Mona, Puerto Rico | N18.06301 | W67.88728 | MF097321 | – |
| Cochliomyia | aldrichi | M088 | Mona, Puerto Rico | N18.06301 | W67.88728 | MF097322 | – |
| Cochliomyia | aldrichi | M102 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097323 | – |
| Cochliomyia | aldrichi | M103 | Mona, Puerto Rico | N18.11125 | W67.933447 | KX529532 | KX529566 |
| Cochliomyia | aldrichi | M104 | Mona, Puerto Rico | N18.085972 | W67.933447 | MF097324 | – |
| Cochliomyia | aldrichi | M105 | Mona, Puerto Rico | N18.085972 | W67.933447 | KX529533 | KX529567 |
| Cochliomyia | aldrichi | M106 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097325 | – |
| Cochliomyia | aldrichi | M107 | Mona, Puerto Rico | N18.084222 | W67.939417 | KX529534 | KX529568 |
| Cochliomyia | hominivorax | CO001 | Colombia | N5.900544 | W74.852897* | – | MF097612 |
| Cochliomyia | hominivorax | CU020 | Cuba | N22.621386 | W83.725944 | – | MF097613 |
| Cochliomyia | hominivorax | CU033 | Cuba | N22.621386 | W83.725944 | KX529556 | KX529571 |
| Cochliomyia | hominivorax | DR042 | Hispaniola | N18.316572 | W71.576447* | KX529557 | KX529572 |
| Cochliomyia | hominivorax | DR105 | Hispaniola | N18.35698 | W68.61609 | KX529558 | KX529573 |
| Cochliomyia | macellaria | LA137 | Saint Barthélemy | N17.910299 | W62.847221 | MF097326 | – |
| Cochliomyia | macellaria | LA139 | Saint Barthélemy | N17.910299 | W62.847221 | MF097327 | – |
| Cochliomyia | macellaria | CO002 | Colombia | N5.900544 | W74.852897* | KX529522 | KX529574 |
| Cochliomyia | macellaria | CO010 | Colombia | N6.266242 | W77.374903* | KX529545 | KX529575 |
| Cochliomyia | macellaria | CU012 | Cuba | N22.621386 | W83.725944 | MF097330 | – |
| Cochliomyia | macellaria | CU013 | Cuba | N22.621386 | W83.725944 | MF097331 | – |
| Cochliomyia | macellaria | CU014 | Cuba | N22.621386 | W83.725944 | KX529541 | KX529577 |
| Cochliomyia | macellaria | CU015 | Cuba | N22.621386 | W83.725944 | MF097332 | – |
| Cochliomyia | macellaria | CU016 | Cuba | N22.621386 | W83.725944 | MF097333 | – |
| Cochliomyia | macellaria | CU017 | Cuba | N22.621386 | W83.725944 | MF097334 | – |
| Cochliomyia | macellaria | CU018 | Cuba | N22.621386 | W83.725944 | KX529526 | KX529578 |
| Cochliomyia | macellaria | CU019 | Cuba | N22.621386 | W83.725944 | MF097335 | MF097614 |
| Cochliomyia | macellaria | CU050 | Cuba | N21.582414 | W77.750131 | MF097336 | – |
| Cochliomyia | macellaria | CU051 | Cuba | N21.582414 | W77.750131 | MF097337 | – |
| Cochliomyia | macellaria | DR009 | Hispaniola | N19.89155 | W71.65806 | MF097341 | – |
| Cochliomyia | macellaria | DR010 | Hispaniola | N19.89155 | W71.65806 | KX529536 | KX529579 |
| Cochliomyia | macellaria | DR011 | Hispaniola | N19.89155 | W71.65806 | MF097342 | – |
| Cochliomyia | macellaria | DR012 | Hispaniola | N19.89155 | W71.65806 | MF097343 | – |
| Cochliomyia | macellaria | DR013 | Hispaniola | N19.89155 | W71.65806 | MF097344 | – |
| Cochliomyia | macellaria | DR014 | Hispaniola | N19.89155 | W71.65806 | MF097345 | – |
| Cochliomyia | macellaria | DR015 | Hispaniola | N19.89155 | W71.65806 | MF097346 | – |
| Cochliomyia | macellaria | DR043 | Hispaniola | N18.316572 | W71.576447* | MF097347 | – |
| Cochliomyia | macellaria | DR062 | Hispaniola | N19.06710 | W69.46004 | MF097348 | – |
| Cochliomyia | macellaria | DR063 | Hispaniola | N19.06710 | W69.46004 | MF097349 | – |
| Cochliomyia | macellaria | DR064 | Hispaniola | N19.06710 | W69.46004 | MF097350 | – |
| Cochliomyia | macellaria | DR065 | Hispaniola | N19.06710 | W69.46004 | MF097351 | – |
| Cochliomyia | macellaria | DR066 | Hispaniola | N19.06710 | W69.46004 | MF097352 | – |
| Cochliomyia | macellaria | DR106 | Hispaniola | N18.35698 | W68.61609 | MF097353 | – |
| Cochliomyia | macellaria | DR107 | Hispaniola | N18.35698 | W68.61609 | MF097354 | – |
| Cochliomyia | macellaria | DR108 | Hispaniola | N18.35698 | W68.61609 | MF097355 | – |
| Cochliomyia | macellaria | DR109 | Hispaniola | N18.35698 | W68.61609 | MF097356 | – |
| Cochliomyia | macellaria | DR1091 | Hispaniola | N18.35698 | W68.61609 | MF097357 | – |
| Cochliomyia | macellaria | DR120 | Hispaniola | N18.32902 | W68.80995 | MF097358 | – |
| Cochliomyia | macellaria | DR121 | Hispaniola | N18.32902 | W68.80995 | MF097359 | – |
| Cochliomyia | macellaria | DR134 | Hispaniola | N19.741319 | W70.654975* | KX529527 | KX529580 |
| Cochliomyia | macellaria | DR154 | Hispaniola | N19.34405 | W70.14824 | MF097360 | – |
| Cochliomyia | macellaria | FL006 | Florida, USA | N25.614383 | W80.584467 | – | MF097615 |
| Cochliomyia | macellaria | FL009 | Florida, USA | N25.457514 | W80.4863 | MF097361 | – |
| Cochliomyia | macellaria | JA002 | Jamaica | N18.0598056 | W77.5311944 | – | MF097616 |
| Cochliomyia | macellaria | LA022 | Saint-Martin | N18.07779 | W63.05772 | MF097384 | – |
| Cochliomyia | macellaria | LA023 | Saint-Martin | N18.07779 | W63.05772 | MF097385 | – |
| Cochliomyia | macellaria | LA024 | Saint-Martin | N18.07779 | W63.05772 | MF097386 | – |
| Cochliomyia | macellaria | LA032 | Saint-Martin | N18.11677 | W63.03902 | MF097387 | – |
| Cochliomyia | macellaria | LA033 | Saint-Martin | N18.11677 | W63.03902 | MF097388 | – |
| Cochliomyia | macellaria | LA034 | Saint-Martin | N18.11677 | W63.03902 | MF097389 | – |
| Cochliomyia | macellaria | LA035 | Saint-Martin | N18.11677 | W63.03902 | MF097390 | – |
| Cochliomyia | macellaria | LA036 | Saint-Martin | N18.11677 | W63.03902 | MF097391 | – |
| Cochliomyia | macellaria | LA049 | Saint Barthélemy | N17.91924 | W62.86366 | MF097371 | – |
| Cochliomyia | macellaria | LA0491 | Saint Barthélemy | N17.91924 | W62.86366 | MF097372 | – |
| Cochliomyia | macellaria | LA050 | Saint Barthélemy | N17.91924 | W62.86366 | MF097373 | – |
| Cochliomyia | macellaria | LA053 | Saint Barthélemy | N17.91924 | W62.86366 | MF097383 | – |
| Cochliomyia | macellaria | LA054 | Saint Barthélemy | N17.91924 | W62.86366 | MF097374 | – |
| Cochliomyia | macellaria | LA066 | Dominica | N15.34066 | W61.33351 | MF097338 | – |
| Cochliomyia | macellaria | LA067 | Dominica | N15.34066 | W61.33351 | MF097339 | – |
| Cochliomyia | macellaria | LA068 | Dominica | N15.34066 | W61.33351 | MF097340 | – |
| Cochliomyia | macellaria | LA071 | Dominica | N15.34066 | W61.33351 | KX529525 | KX529583 |
| Cochliomyia | macellaria | LA079 | Guadeloupe | N16.37752 | W61.47869 | MF097362 | – |
| Cochliomyia | macellaria | LA080 | Guadeloupe | N16.37752 | W61.47869 | MF097363 | – |
| Cochliomyia | macellaria | LA081 | Guadeloupe | N16.37752 | W61.47869 | MF097364 | – |
| Cochliomyia | macellaria | LA094 | Nevis | N17.14145 | W62.57784 | MF097368 | – |
| Cochliomyia | macellaria | LA096 | Martinique | N14.47428 | W60.81463 | KX529524 | KX529584 |
| Cochliomyia | macellaria | LA097 | Martinique | N14.47428 | W60.81463 | MF097367 | – |
| Cochliomyia | macellaria | LA115 | Saint Kitts | N17.3404083 | W62.7410389 | MF097365 | – |
| Cochliomyia | macellaria | LA118 | Saint Kitts | N17.3404083 | W62.7410389 | MF097392 | – |
| Cochliomyia | macellaria | LA131 | Barbuda | N17.6054722 | W61.8005833 | MF097328 | – |
| Cochliomyia | macellaria | LA132 | Barbuda | N17.6054722 | W61.8005833 | MF097329 | – |
| Cochliomyia | macellaria | LA138 | Saint Barthélemy | N17.897522 | W62.849694 | MF097375 | – |
| Cochliomyia | macellaria | LA140 | Saint Barthélemy | N17.897522 | W62.849694 | MF097376 | – |
| Cochliomyia | macellaria | LA141 | Saint Barthélemy | N17.897522 | W62.849694 | MF097377 | – |
| Cochliomyia | macellaria | LA142 | Saint Barthélemy | N17.897522 | W62.849694 | KX529523 | KX529592 |
| Cochliomyia | macellaria | LA143 | Saint Barthélemy | N17.897522 | W62.849694 | MF097378 | – |
| Cochliomyia | macellaria | LA144 | Saint Barthélemy | N17.897522 | W62.849694 | MF097379 | – |
| Cochliomyia | macellaria | LA145 | Saint Barthélemy | N17.897522 | W62.849694 | MF097380 | – |
| Cochliomyia | macellaria | LA146 | Saint Barthélemy | N17.897522 | W62.849694 | MF097381 | – |
| Cochliomyia | macellaria | LA147 | Saint Barthélemy | N17.897522 | W62.849694 | MF097382 | |
| Cochliomyia | macellaria | ME015 | Mexico | N25.598592 | W103.441156 | – | MF097617 |
| Cochliomyia | macellaria | M077 | Mona, Puerto Rico | N18.086239 | W67.906339 | KX529539 | KX529585 |
| Cochliomyia | macellaria | M081 | Mona, Puerto Rico | N18.11125 | W67.933447 | KX529537 | KX529586 |
| Cochliomyia | macellaria | M112 | Mona, Puerto Rico | N18.11125 | W67.933447 | KX529544 | KX529589 |
| Cochliomyia | macellaria | ME004 | Mexico | N21.07645 | W89.501083 | MF097366 | – |
| Cochliomyia | macellaria | PR029 | Puerto Rico | N17.961111 | W66.863806 | MF097369 | – |
| Cochliomyia | macellaria | PR047 | Puerto Rico | N18.178722 | W66.488111 | MF097370 | – |
| Cochliomyia | macellaria | PR121 | Puerto Rico | N18.370953 | W66.026619 | KX529544 | KX529589 |
| Cochliomyia | macellaria | PR128 | Puerto Rico | N18.093306 | W65.552111 | KX529540 | KX529590 |
| Cochliomyia | macellaria | PR129 | Puerto Rico | N18.093306 | W65.552111 | KX529542 | KX529591 |
| Cochliomyia | minima | CU010 | Cuba | N20.054178 | W76.917603 | MF097393 | – |
| Cochliomyia | minima | CU021 | Cuba | N22.621386 | W83.725944 | MF097394 | – |
| Cochliomyia | minima | CU022 | Cuba | N22.621386 | W83.725944 | KX529549 | KX529593 |
| Cochliomyia | minima | CU023 | Cuba | N22.621386 | W83.725944 | KX529550 | KX529594 |
| Cochliomyia | minima | CU024 | Cuba | N22.621386 | W83.725944 | MF097395 | – |
| Cochliomyia | minima | CU025 | Cuba | N22.621386 | W83.725944 | MF097396 | – |
| Cochliomyia | minima | CU026 | Cuba | N22.621386 | W83.725944 | MF097397 | – |
| Cochliomyia | minima | CU027 | Cuba | N22.621386 | W83.725944 | MF097398 | – |
| Cochliomyia | minima | CU043 | Cuba | N20.517817 | W74.65865 | MF097399 | – |
| Cochliomyia | minima | CU044 | Cuba | N20.517817 | W74.65865 | MF097400 | – |
| Cochliomyia | minima | CU045 | Cuba | N20.517817 | W74.65865 | MF097401 | – |
| Cochliomyia | minima | CU046 | Cuba | N20.517817 | W74.65865 | KX529547 | KX529595 |
| Cochliomyia | minima | DR026 | Hispaniola | N19.04995 | W70.89046 | MF097402 | – |
| Cochliomyia | minima | DR027 | Hispaniola | N19.04995 | W70.89046 | MF097403 | – |
| Cochliomyia | minima | DR028 | Hispaniola | N19.04995 | W70.89046 | MF097404 | – |
| Cochliomyia | minima | DR029 | Hispaniola | N19.04995 | W70.89046 | MF097405 | – |
| Cochliomyia | minima | PR013 | Hispaniola | N18.316572 | W71.576447* | MF097406 | – |
| Cochliomyia | minima | DR032 | Hispaniola | N18.316572 | W71.576447* | MF097407 | – |
| Cochliomyia | minima | DR033 | Hispaniola | N18.316572 | W71.576447* | MF097408 | – |
| Cochliomyia | minima | DR034 | Hispaniola | N18.316572 | W71.576447* | MF097409 | – |
| Cochliomyia | minima | DR035 | Hispaniola | N18.316572 | W71.576447* | MF097410 | – |
| Cochliomyia | minima | DR053 | Hispaniola | N19.06753 | W69.46445 | MF097411 | – |
| Cochliomyia | minima | DR054 | Hispaniola | N19.06753 | W69.46445 | MF097412 | – |
| Cochliomyia | minima | DR055 | Hispaniola | N19.06753 | W69.46445 | KX529552 | KX529596 |
| Cochliomyia | minima | DR056 | Hispaniola | N19.06753 | W69.46445 | MF097413 | – |
| Cochliomyia | minima | DR067 | Hispaniola | N19.06710 | W69.46004 | MF097414 | – |
| Cochliomyia | minima | DR072 | Hispaniola | N19.34864 | W70.14910 | MF097415 | – |
| Cochliomyia | minima | DR073 | Hispaniola | N19.34864 | W70.14910 | MF097416 | – |
| Cochliomyia | minima | DR074 | Hispaniola | N19.34864 | W70.14910 | MF097417 | – |
| Cochliomyia | minima | DR075 | Hispaniola | N19.34864 | W70.14910 | MF097418 | – |
| Cochliomyia | minima | DR076 | Hispaniola | N19.34864 | W70.14910 | MF097419 | – |
| Cochliomyia | minima | DR136 | Hispaniola | N19.741319 | W70.654975 | KX529548 | KX529597 |
| Cochliomyia | minima | DR137 | Hispaniola | N19.741319 | W70.654975 | MF097420 | – |
| Cochliomyia | minima | DR138 | Hispaniola | N19.741319 | W70.654975 | MF097421 | – |
| Cochliomyia | minima | DR139 | Hispaniola | N19.741319 | W70.654975 | MF097422 | – |
| Cochliomyia | minima | DR153 | Hispaniola | N19.34405 | W70.14824 | MF097423 | – |
| Cochliomyia | minima | DR164 | Hispaniola | N18.32902 | W68.80995 | MF097424 | – |
| Cochliomyia | minima | PR006 | Puerto Rico | N18.412972 | W66.727222 | MF097425 | – |
| Cochliomyia | minima | PR007 | Puerto Rico | N18.412972 | W66.727222 | MF097426 | – |
| Cochliomyia | minima | PR016 | Puerto Rico | N18.321333 | W65.818722 | MF097427 | – |
| Cochliomyia | minima | PR018 | Puerto Rico | N18.321333 | W65.818722 | MF097428 | – |
| Cochliomyia | minima | PR019 | Puerto Rico | N18.321333 | W65.818722 | MF097429 | – |
| Cochliomyia | minima | PR041 | Puerto Rico | N18.174722 | W66.491861 | MF097430 | – |
| Cochliomyia | minima | PR131 | Puerto Rico | N18.093306 | W65.552111 | MF097431 | – |
| Cochliomyia | minima | PR132 | Puerto Rico | N18.093306 | W65.552111 | KX529553 | KX529598 |
| Cochliomyia | minima | PR133 | Puerto Rico | N18.093306 | W65.552111 | KX529554 | KX529599 |
| Cochliomyia | minima | PR140 | Puerto Rico | N18.447911 | W65.948617 | MF097432 | MF097618 |
| Cochliomyia | minima | PR141 | Puerto Rico | N18.447911 | W65.948617 | KX529551 | KX529600 |
| Cochliomyia | minima | PR145 | Puerto Rico | N18.449889 | W65.595333 | MF097433 | – |
| Cochliomyia | minima | PR146 | Puerto Rico | N18.449889 | W65.595333 | MF097434 | – |
| Lucilia | cluvia | FL005 | Florida, USA | N25.614383 | W80.584467 | – | MF097619 |
| Lucilia | cluvia | FL017 | Florida, USA | N25.136917 | W80.94855 | MF097436 | MF097620 |
| Lucilia | cluvia | FL018 | Florida, USA | N25.136917 | W80.94855 | – | MF097621 |
| Lucilia | cluvia | FL019 | Florida, USA | N25.323331 | W80.833094 | MF097437 | – |
| Lucilia | cluvia | FL020 | Florida, USA | N25.323331 | W80.833094 | MF097438 | MF097622 |
| Lucilia | cluvia | FL025 | Florida, USA | N25.423053 | W80.679114 | MF097439 | MF097623 |
| Lucilia | cluvia | FL026 | Florida, USA | N25.423053 | W80.679114 | MF097440 | MF097624 |
| Lucilia | cluvia | PR147 | Puerto Rico | N18.429222 | W66.178022 | MF097441 | MF097625 |
| Lucilia | cluvia | PR148 | Puerto Rico | N18.429222 | W66.178022 | MF097442 | MF097626 |
| Lucilia | coeruleiviridis | FL007 | Florida, USA | N25.457514 | W80.4863 | – | MF097627 |
| Lucilia | coeruleiviridis | FL013 | Florida, USA | N25.136917 | W80.94885 | MF097443 | MF097628 |
| Lucilia | coeruleiviridis | FL014 | Florida, USA | N25.136917 | W80.94855 | – | MF097629 |
| Lucilia | coeruleiviridis | FL015 | Florida, USA | N25.136917 | W80.94885 | MF097444 | MF097630 |
| Lucilia | coeruleiviridis | FL016 | Florida, USA | N25.136917 | W80.94885 | MF097445 | MF097631 |
| Lucilia | coeruleiviridis | FL023 | Florida, USA | N25.457514 | W80.4863 | MF097446 | MF097632 |
| Lucilia | coeruleiviridis | FL024 | Florida, USA | N25.457514 | W80.4863 | MF097447 | MF097633 |
| Lucilia | cuprina | FL027 | Florida, USA | N25.457514 | W80.4863 | MF097448 | MF097634 |
| Lucilia | cuprina | FL028 | Florida, USA | N25.457514 | W80.4863 | MF097449 | MF097635 |
| Lucilia | cuprina | FL029 | Florida, USA | N25.457514 | W80.4863 | MF097450 | MF097636 |
| Lucilia | cuprina | FL030 | Florida, USA | N25.457514 | W80.4863 | MF097451 | MF097637 |
| Lucilia | cuprina | PR070 | Puerto Rico | N18.370953 | W66.026619 | MF097452 | – |
| Lucilia | cuprina | PR071 | Puerto Rico | N18.370953 | W66.026619 | MF097453 | – |
| Lucilia | cuprina | PR072 | Puerto Rico | N18.370953 | W66.026619 | MF097454 | – |
| Lucilia | cuprina | PR073 | Puerto Rico | N18.370953 | W66.026619 | KX529559 | KX529602 |
| Lucilia | cuprina | PR122 | Puerto Rico | N18.370953 | W66.026619 | MF097455 | MF097638 |
| Lucilia | cuprina | PR123 | Puerto Rico | N18.370953 | W66.026619 | MF097456 | – |
| Lucilia | cuprina | PR153 | Puerto Rico | N18.461053 | W66.729803 | MF097457 | – |
| Lucilia | cuprina | PR154 | Puerto Rico | N18.461053 | W66.729803 | MF097458 | MF097639 |
| Lucilia | eximia | CO011 | Colombia | N5.900544 | W74.852897* | MF097459 | – |
| Lucilia | eximia | CO012 | Colombia | N5.900544 | W74.852897* | MF097460 | MF097640 |
| Lucilia | eximia | CO013 | Colombia | N5.900544 | W74.852897* | MF097461 | MF097641 |
| Lucilia | eximia | CO015 | Colombia | N5.900544 | W74.852897* | MF097462 | MF097642 |
| Lucilia | eximia | CO016 | Colombia | N5.900544 | W74.852897* | – | MF097643 |
| Lucilia | eximia | CO022 | Colombia | N6.067217 | W73.645411 | MF097463 | MF097644 |
| Lucilia | eximia | CO023 | Colombia | N6.067217 | W73.645411 | MF097464 | MF097645 |
| Lucilia | eximia | CU002 | Cuba | N20.054178 | W76.917603 | – | MF097646 |
| Lucilia | eximia | CU006 | Cuba | N20.054178 | W76.917603 | – | MF097647 |
| Lucilia | eximia | DR019 | Hispaniola | N19.89155 | W071.65806 | MF097467 | MF097650 |
| Lucilia | eximia | DR049 | Hispaniola | N18.316572 | W71.576447* | MF097468 | – |
| Lucilia | eximia | DR050 | Hispaniola | N18.316572 | W71.576447 | – | MF097651 |
| Lucilia | eximia | DR129 | Hispaniola | N18.32902 | W68.80995 | MF097469 | – |
| Lucilia | eximia | FL021 | Florida, USA | N25.086633 | W80.452217 | MF097470 | MF097652 |
| Lucilia | eximia | FL022 | Florida, USA | N25.086633 | W80.452217 | MF097471 | MF097653 |
| Lucilia | eximia | LA064 | Dominica | N15.34066 | W61.33351 | MF097465 | MF097648 |
| Lucilia | eximia | LA065 | Dominica | N15.34066 | W61.33351 | MF097466 | MF097649 |
| Lucilia | eximia | LA124 | Saint Lucia | N14.100031 | W60.92654 | MF097483 | MF097665 |
| Lucilia | eximia | LA126 | Saint Lucia | N14.100031 | W60.92654 | – | MF097666 |
| Lucilia | eximia | LA127 | Saint Lucia | N14.100031 | W60.92654 | MF097484 | MF097667 |
| Lucilia | eximia | M076 | Mona, Puerto Rico | N18.086239 | W67.906339 | MF097472 | MF097654 |
| Lucilia | eximia | M099 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097473 | – |
| Lucilia | eximia | M100 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097474 | – |
| Lucilia | eximia | M110 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097475 | MF097655 |
| Lucilia | eximia | M111 | Mona, Puerto Rico | N18.11125 | W67.933447 | MF097476 | MF097656 |
| Lucilia | eximia | ME005 | Mexico | N21.07645 | W89.501083 | MF097477 | MF097657 |
| Lucilia | eximia | ME006 | Mexico | N21.07645 | W89.501083 | – | MF097658 |
| Lucilia | eximia | ME007 | Mexico | N21.07645 | W89.501083 | MF097478 | MF097659 |
| Lucilia | eximia | PR050 | Puerto Rico | N18.449889 | W66.595333 | MF097479 | MF097660 |
| Lucilia | eximia | PR060 | Puerto Rico | N17.971611 | W66.865361 | MF097480 | MF097661 |
| Lucilia | eximia | PR111 | Mona, Puerto Rico | N18.11125 | W67.933447 | – | MF097662 |
| Lucilia | eximia | PR114 | Puerto Rico | N18.370953 | W66.026619 | MF097481 | – |
| Lucilia | eximia | PR134 | Puerto Rico | N18.093306 | W65.552111 | – | MF097663 |
| Lucilia | eximia | PR135 | Puerto Rico | N18.093306 | W65.552111 | – | MF097664 |
| Lucilia | eximia | PR150 | Puerto Rico | N18.084222 | W67.939417 | MF097482 | – |
| Lucilia | fayeae | M079 | Mona, Puerto Rico | N18.084222 | W67.939417 | MF097485 | MF097668 |
| Lucilia | fayeae | PR008 | Puerto Rico | N18.412972 | W67.727222 | MF097486 | MF097669 |
| Lucilia | fayeae | PR012 | Puerto Rico | N18.412972 | W67.727222 | MF097487 | – |
| Lucilia | fayeae | PR020 | Puerto Rico | N18.321333 | W65.818722 | MF097488 | – |
| Lucilia | fayeae | PR022 | Puerto Rico | N18.321333 | W65.818722 | MF097489 | – |
| Lucilia | fayeae | PR023 | Puerto Rico | N18.293444 | W65.791917 | MF097490 | MF097670 |
| Lucilia | fayeae | PR045 | Puerto Rico | N18.174722 | W66.491861 | MF097491 | MF097671 |
| Lucilia | fayeae | PR053 | Puerto Rico | N18.449889 | W66.595333 | MF097492 | MF097672 |
| Lucilia | fayeae | PR116 | Puerto Rico | N18.370953 | W66.032175 | MF097493 | – |
| Lucilia | lucigerens | JA005 | Jamaica | N18.0598056 | W77.5311944 | MF097494 | MF097673 |
| Lucilia | lucigerens | JA006 | Jamaica | N18.0598056 | W77.5311944 | MF097495 | – |
| Lucilia | lucigerens | JA007 | Jamaica | N18.0598056 | W77.5311944 | MF097496 | MF097674 |
| Lucilia | mexicana | ME016 | Mexico | N25.598592 | W103.441156 | MF097497 | MF097675 |
| Lucilia | mexicana | ME020 | Mexico | N25.598592 | W103.441156 | MF097498 | MF097676 |
| Lucilia | mexicana | ME021 | Mexico | N25.598592 | W103.441156 | MF097499 | MF097677 |
| Lucilia | retroversa | CU007 | Cuba | N20.054178 | W76.917603 | MF097500 | MF097678 |
| Lucilia | retroversa | CU028 | Cuba | N22.621386 | W83.725944 | MF097501 | – |
| Lucilia | retroversa | CU029 | Cuba | N22.621386 | W83.725944 | MF097502 | – |
| Lucilia | retroversa | CU030 | Cuba | N22.621386 | W83.725944 | MF097503 | MF097679 |
| Lucilia | retroversa | CU031 | Cuba | N22.621386 | W83.725944 | MF097504 | – |
| Lucilia | retroversa | CU038 | Cuba | N20.517817 | W20.517817 | MF097505 | – |
| Lucilia | retroversa | CU039 | Cuba | N20.517817 | W20.517817 | MF097506 | – |
| Lucilia | retroversa | CU040 | Cuba | N20.517817 | W20.517817 | MF097507 | – |
| Lucilia | retroversa | CU041 | Cuba | N20.517817 | W20.517817 | MF097508 | MF097680 |
| Lucilia | retroversa | CU042 | Cuba | N20.517817 | W20.517817 | MF097509 | – |
| Lucilia | retroversa | DR020 | Hispaniola | N19.04871 | W70.88084 | MF097510 | – |
| Lucilia | retroversa | DR021 | Hispaniola | N19.04871 | W70.88084 | MF097511 | – |
| Lucilia | retroversa | DR022 | Hispaniola | N19.04871 | W70.88084 | MF097512 | – |
| Lucilia | retroversa | DR023 | Hispaniola | N19.04871 | W70.88084 | MF097513 | – |
| Lucilia | retroversa | DR024 | Hispaniola | N19.04871 | W70.88084 | MF097514 | MF097681 |
| Lucilia | retroversa | DR025 | Hispaniola | N19.04871 | W70.88084 | MF097515 | – |
| Lucilia | retroversa | DR030 | Hispaniola | N19.04871 | W70.88084 | MF097516 | – |
| Lucilia | retroversa | DR040 | Hispaniola | N18.316572 | W71.576447 | MF097517 | – |
| Lucilia | retroversa | DR046 | Hispaniola | N18.316572 | W71.576447 | MF097518 | – |
| Lucilia | retroversa | DR047 | Hispaniola | N18.316572 | W71.576447 | MF097519 | – |
| Lucilia | retroversa | DR048 | Hispaniola | N18.316572 | W71.576447 | MF097520 | – |
| Lucilia | retroversa | DR057 | Hispaniola | N19.06753 | W69.46445 | MF097521 | – |
| Lucilia | retroversa | DR058 | Hispaniola | N19.06753 | W69.46445 | MF097522 | – |
| Lucilia | retroversa | DR059 | Hispaniola | N19.06753 | W69.46445 | MF097523 | – |
| Lucilia | retroversa | DR060 | Hispaniola | N19.06753 | W69.46445 | MF097524 | – |
| Lucilia | retroversa | DR061 | Hispaniola | N19.06753 | W69.46445 | MF097525 | – |
| Lucilia | retroversa | DR079 | Hispaniola | N19.34864 | W70.14910 | MF097526 | – |
| Lucilia | retroversa | DR080 | Hispaniola | N19.34864 | W70.14910 | MF097527 | – |
| Lucilia | retroversa | DR081 | Hispaniola | N19.34864 | W70.14910 | MF097528 | – |
| Lucilia | retroversa | DR082 | Hispaniola | N19.34864 | W70.14910 | MF097529 | – |
| Lucilia | retroversa | DR083 | Hispaniola | N19.34864 | W70.14910 | MF097530 | – |
| Lucilia | retroversa | DR089 | Hispaniola | N19.34864 | W70.14910 | MF097531 | – |
| Lucilia | retroversa | DR090 | Hispaniola | N19.34864 | W70.14910 | MF097532 | – |
| Lucilia | retroversa | DR091 | Hispaniola | N19.34864 | W70.14910 | MF097533 | – |
| Lucilia | retroversa | DR092 | Hispaniola | N19.34864 | W70.14910 | MF097534 | – |
| Lucilia | retroversa | DR111 | Hispaniola | N18.35698 | W68.61609 | MF097535 | – |
| Lucilia | retroversa | DR110 | Hispaniola | N18.35698 | W68.61609 | MF097536 | – |
| Lucilia | retroversa | DR112 | Hispaniola | N18.35698 | W68.61609 | MF097537 | – |
| Lucilia | retroversa | DR122 | Hispaniola | N18.32902 | W68.80995 | MF097538 | – |
| Lucilia | retroversa | DR123 | Hispaniola | N18.32902 | W68.80995 | MF097539 | MF097682 |
| Lucilia | retroversa | DR124 | Hispaniola | N18.32902 | W68.80995 | MF097540 | MF097683 |
| Lucilia | retroversa | DR125 | Hispaniola | N18.32902 | W68.80995 | MF097541 | – |
| Lucilia | retroversa | DR126 | Hispaniola | N18.32902 | W68.80995 | MF097542 | – |
| Lucilia | retroversa | DR128 | Hispaniola | N18.32902 | W68.80995 | MF097543 | – |
| Lucilia | retroversa | DR140 | Hispaniola | N19.741319 | W70.654975* | MF097544 | – |
| Lucilia | retroversa | DR141 | Hispaniola | N19.741319 | W70.654975* | MF097545 | – |
| Lucilia | retroversa | DR142 | Hispaniola | N18.09786 | W71.18925 | MF097546 | – |
| Lucilia | retroversa | DR143 | Hispaniola | N18.09786 | W71.18925 | MF097547 | – |
| Lucilia | retroversa | DR144 | Hispaniola | N18.09786 | W71.18925 | MF097548 | |
| Lucilia | retroversa | DR145 | Hispaniola | N18.09786 | W71.18925 | MF097549 | – |
| Lucilia | retroversa | DR146 | Hispaniola | N18.09786 | W71.18925 | MF097550 | – |
| Lucilia | retroversa | DR147 | Hispaniola | N18.09786 | W71.18925 | MF097551 | – |
| Lucilia | retroversa | DR148 | Hispaniola | N18.09786 | W71.18925 | MF097552 | – |
| Lucilia | rica | LA007 | Saint Eustatius | N17.47637 | W62.97470 | MF097558 | – |
| Lucilia | rica | LA008 | Saint Eustatius | N17.47637 | W62.97470 | MF097559 | – |
| Lucilia | rica | LA009 | Saint Eustatius | N17.47637 | W62.97470 | – | MF097684 |
| Lucilia | rica | LA010 | Saint Eustatius | N17.47637 | W62.97470 | MF097560 | – |
| Lucilia | rica | LA016 | Saint-Martin | N18.07779 | W63.05772 | MF097572 | – |
| Lucilia | rica | LA017 | Saint-Martin | N18.07779 | W63.05772 | MF097573 | MF097697 |
| Lucilia | rica | LA026 | Saba | N17.63980 | W63.23373 | MF097435 | – |
| Lucilia | rica | LA027 | Saba | N17.63980 | W63.23373 | – | MF097692 |
| Lucilia | rica | LA028 | Saba | N18.07779 | W63.05772 | MF097569 | MF097693 |
| Lucilia | rica | LA037 | Saint-Martin | N18.11677 | W63.03902 | MF097574 | – |
| Lucilia | rica | LA045 | Saint Barthélemy | N17.91924 | W62.86366 | MF097570 | MF097694 |
| Lucilia | rica | LA061 | Saint Barthélemy | N17.91924 | W62.86366 | MF097571 | MF097696 |
| Lucilia | rica | LA073 | Nevis | N17.14145 | W62.57784 | MF097567 | MF097690 |
| Lucilia | rica | LA074 | Nevis | N17.14145 | W62.57784 | MF097568 | MF097691 |
| Lucilia | rica | LA098 | Martinique | N14.47428 | W60.81463 | MF097565 | MF097688 |
| Lucilia | rica | LA099 | Martinique | N14.47428 | W60.81463 | MF097566 | MF097689 |
| Lucilia | rica | LA106 | Montserrat | N16.77608 | W62.30904 | MF097564 | MF097687 |
| Lucilia | rica | LA114 | Saint Kitts | N17.3404083 | W62.7410389 | MF097563 | – |
| Lucilia | rica | LA128 | Antigua | N17.0358611 | W61.8246389 | MF097553 | – |
| Lucilia | rica | LA129 | Antigua | N17.0358611 | W61.8246389 | MF097554 | – |
| Lucilia | rica | LA130 | Antigua | N17.0358611 | W61.8246389 | MF097555 | – |
| Lucilia | rica | LA133 | Barbuda | N17.6054722 | W61.8005833 | MF097556 | – |
| Lucilia | rica | LA134 | Barbuda | N17.6054722 | W61.8005833 | MF097557 | – |
| Lucilia | rica | LA083 | Guadeloupe | N16.37752 | W61.47869 | MF097561 | MF097685 |
| Lucilia | rica | LA087 | Guadeloupe | N16.37752 | W61.47869 | MF097562 | MF097686 |
| Lucilia | rica | TLW042 | Antigua and Barbuda | As publisheda | BNNR042∧ | – | |
| Lucilia | rica | TLW043 | Antigua and Barbuda | As publisheda | BNNR043∧ | – | |
| Lucilia | rica | TLW044 | Antigua and Barbuda | As publisheda | BNNR044∧ | – | |
| Lucilia | rica | TLW046 | Antigua and Barbuda | As publisheda | BNNR046∧ | – | |
| Lucilia | sp. | CO027 | Colombia | N6.067217 | W73.645411 | MF097575 | MF097698 |
| Lucilia | vulgata | CO019 | Colombia | N6.067217 | W73.645411 | MF097576 | MF097699 |
| Lucilia | vulgata | CO025 | Colombia | N6.067217 | W73.645411 | MF097577 | MF097700 |
| Lucilia | vulgata | CO026 | Colombia | N6.067217 | W73.645411 | MF097578 | MF097701 |
| Lucilia | vulgata | CO028 | Colombia | N6.067217 | W73.645411 | MF097579 | MF097702 |
| Outgroups | |||||||
| Neobellieria | bullata | BG64 | As publishedb | JQ807156.1 | – | ||
| Ravinia | stimulans | AZ60 | As publishedb | JQ807112.1 | – | ||
| Sarcophaga | carnaria | NICC0410 | As publishedc | JQ582094.1 | – | ||
| Blaesoxipha | alcedo | AY09 | As publishedb | JQ806830.1 | – | ||
| Blaesoxipha | masculina | AW36 | As publishedb | JQ806832.1 | – |
Methods
Specimens and DNA extraction
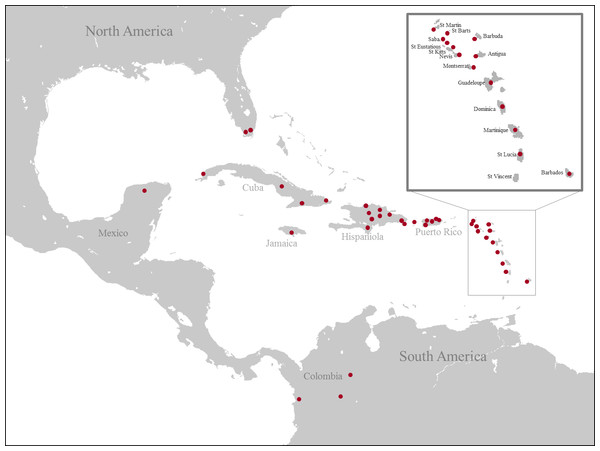
A total of 473 specimens were included in this study. Of these, 468 represented ingroup taxa and five represented outgroup taxa from the family Sarcophagidae (Sarcophaga Carnaria Linnaeus, 1758; Neobellieria bullata Parker, 1916; Ravinia stimulans Walker, 1849; Blaesoxipha masculina Aldrich, 1916 and Blaesoxipha alcedo Aldrich, 1916). We used a total of 600 DNA sequences and we obtained 521 (COI = 398, ITS2 = 123) while 79 (COI = 44, ITS2 = 35) were previously published (Table 1). The specimens were collected throughout the Caribbean (Fig. 1) from between 2011 and 2013 (see Table 1 for details). All specimens were collected under appropriate permits: USA, Florida, Everglades, United States Department of the Interior National Park Service EVER-2013-SCI-0028; Puerto Rico, DRNA: 2011-IC-035 (O-VS-PVS15-SJ-00474-08042011); Jamaica, NEPA, reference number #18/27; USA, USDI National Park Service, EVER-2013-SCI-0028; Costa Rica, SINAC, pasaporte científico no. 05933, resolución no. 019-2013-SINAC; Cuba, Departamento de Recursos Naturales, PE 2012/05, 2012003 and 2012001; Dominican Republic, Ministerio de Medio Ambiente y Recursos Naturales, no 0577; Colombia, Authoridad Nacional de Licencias Ambientales, 18.497.666 issued to Alexander Gómez Mejía; Saba, The Executive Council of the Public Entity Saba, no 112/2013; Martinique, Ministère de L’Écologie, du Développement Durable, et de L‘Énergie; Nevis, Nevis Historical & Conservation Society, no F001; Barbados, Ministry of Environment and Drainage, no 8434∕56∕1 Vol. II. Although L. vulgata, L. mexicana and L. coeruleiviridis are not present in the Caribbean islands, they are included as outgroups to the Calliphoridae from the West Indies. James (1970) reported L. coeruleiviridis from Cuba, however, this is likely an error as no specimens have been seen in collections from the region (Whitworth, 2010) and no specimens were collected during this study. All specimens, except the ones from Mexico, were collected using a novel trap designed for this study. We modified a standard butterfly trap by adding a conic form on the top with a vessel attached to the highest point like in the Malaise trap. Flies entered the trap attracted by the bait (chicken) and funneled into the collecting vessel containing 95% ethanol. Traps were hung 1m off the ground and were used to collect flies for 2–3 days at each locality. These traps proved efficient in collecting specimens for our molecular purposes, given that caught specimens were preserved in ethanol while the trap remained in the field. Collected specimens were transferred to Whirl-paks with 95% ethanol and stored at −20 °C. Adults were identified using the Whitworth (2010) taxonomic keys and the specimens with uncertain identity were sent to Dr. Whitworth at Washington State University for detailed examination and species confirmation. DNA was isolated from thoracic muscle or two legs of each individual with the QIAGEN DNeasy Tissue Kit (Qiagen, Inc., Valencia, CA). The remainder of the specimen was retained as a voucher currently held by the Agnarsson Lab; they will be placed in the Zadock Thompson Zoological Collections at the UVM Natural History Museum following completion of other studies currently being conducted using the material.
Figure 1: Map of collecting localities of all specimens used for the molecular analysis.
(Image credit: https://commons.wikimedia.org/wiki/File:Caribbean_map_blank.png#filelinks).PCR amplification and sequencing
A region of the mitochondrial genome encoding COI was amplified in a single fragment using the primers LCO1490 (Folmer et al., 1994), and C1-N-2776 (Hedin & Maddison, 2001). Those primers amplified successfully in all Calliphoridae except Lucilia Robineau-Desvoidy. From the eight Caribbean species of Lucilia, only Lucilia retroversa amplified successfully using these primers. For the remaining Lucilia species two different primer-pairs were used. The Primer 1 (Gibson et al., 2011) with C1-N-2191 (Simon et al., 1994) and the C1-J-1751 (Gibson et al., 2011) with C2-N-3014. For the second internal transcribed spacer ITS2 we used the primers ITS4 and ITS5.8 (White et al., 1990). The primer sequences and protocols are listed in Table 2. Amplified fragments were sequenced in both directions by University of Arizona Genetics Core. Sequences were interpreted from chromatograms using Phred and Phrap (Green, 1999; Green & Ewing, 2002) using the Chromaseq module (Maddison & Maddison, 2010a) in Mesquite 3.03 (Maddison & Maddison, 2010b) with default parameters. The sequences were then proofread by examining chromatograms by eye. Alignments were done using MAFFT (Katoh et al., 2002) through the online portal EMBL-EBI with default settings. The matrices were exported to Mesquite 3.03 (Maddison & Maddison, 2010b) and the translation of coding sequences to proteins for COI were checked for potential errors.
| Primer name | Sequence (5′–3′) | Protocol | Source protocol | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | CY | D | AN | E | FE | ||||
| LCO1490 | F | GGTCAACAAATCATAAAGATATTGG | 95 °C 2 min | 35 | 95 °C 30 s | 44 °C 45 s | 72 °C 45 s | 72 °C 10 min | Agnarsson, Maddison & Aviles (2007) |
| CI-N-2776 | R | GGATAATCAGAATATCGTCGAGG | |||||||
| Primer 1 | F | TACAATTTATCGCCTAAACTTCAGCC | 95 °C 3 min | 35 | 94 °C 15 s | 51 °C 15 s | 72 °C 30 s | 72 °C 5 min | DeBry et al. (2013) |
| C1-N-2191 | R | CCCGGTAAAATTAAAATATAAACTTC | |||||||
| C1-J-1751 | F | GGAGCTCCTGACATAGCATTCCC | 94 °C 90 s | 36 | 94 °C 22 s | 48 °C 30 s | 72 °C 80 s | 72 °C 60 s | Harvey et al. (2003) |
| C2-N-3014 | R | TCCATTGCACTAATCTGCCATATTA | |||||||
| ITS4 | F | TCCTCCGCTTATTGATATGC | 94 °C 2 min | 38 | 94 °C 30 s | 44 °C 35 s | 72 °C 30 s | 72 °C 3 min | Agnarsson (2010) |
| ITS5.8 | R | GGGACGATGAAGAACGCAGC | |||||||
Notes:
- F
-
Forward
- R
-
Reverse
- ID
-
Initial denaturation
- CY
-
cycles
- D
-
Denaturation
- AN
-
annealing
- E
-
Extension
- FE
-
Final extension
Phylogenetic analysis
The COI gene was partitioned by codon positions, each partition and ITS2 gene were exported from Mesquite for model choice. The appropriate models were chosen using jModeltest v2.1.4 (Posada & Crandall, 1998), and the AIC criterion (Posada & Buckley, 2004). The corresponding model of evolution was used for the Bayesian analysis: GTR + Γ + I for COI1st, F81+ I for COI2nd, GTR + Γ for COI3rd and HKY + Γ + I for ITS2. We ran the MC3(Metropolis Coupled Markov Chain Monte Carlo) chain in MrBayes v3.2.3 (Huelsenbeck & Ronquist, 2001) through the online portal Cipres Science Gateway v3.3 (Miller, Pfeiffer & Schwartz, 2010). The analysis was run for 20,000,000 generations, sampling every 1,000 generations, and the sample points of the first 5,000,000 generations were discarded as ‘burnin’, after which the chains had reached stationarity as determined by analysis in Tracer (Rambaut & Drummond, 2009). Maximum likelihood (ML) analysis of the concatenated matrix was done in Garli (Zwickl, 2006) using the same partitioning scheme and models. Sequences were submitted to GenBank and BOLD.
Species delimitation
We used MEGA6 to calculate genetic distances within and among species level clades suggested by the barcoding analysis of the COI data and by morphology. We used the species delimitation plugin in Geneious 8.1.5 (Kearse et al., 2012; Masters, Fan & Ross, 2011) to estimate species limits under Rosenberg’s reciprocal monophyly P(AB) (Rosenberg, 2007) and Rodrigo’s P(RD) method (Rodrigo et al., 2008). For this analysis we used a 317 taxa subset of our data, produced by reducing the most densely sampled species like Co. minima, Co. macellaria, Ch. rufifacies and L. retroversa to 38 exemplars since P(RD) probability cannot be computed when there are more than 40 exemplars per clade. We also estimated the probability of population identification of a hypothetical sample based on the groups being tested P ID (Strict) and P ID (Liberal). The genealogical sorting index (gsi) statistic (Cummings, Neel & Shaw, 2008) was calculated using the gsi webserver (http://genealogicalsorting.org) on the estimated tree. As genetic distances in MEGA6, gsi and species delimitation metrics from Geneious require a priory species designation, 26 putative species were assigned to the data based on combined analysis of phylogenetic topology from COI and morphological and geographic information. Finally, we used a single locus Bayesian implementation (bPTP) of the Poisson tree processes model (Zhang et al., 2013) to infer putative species boundaries on a given single locus phylogenetic input tree available on the webserver: http://species.h-its.org/ptp/. The analysis was run as a rooted tree from the MrBayes analysis, for 500,000 generations with 10% burnin removed. For gsi and bPTP analysis we reduced the data to 103 taxa representing the 26 putative species because of limitations of the server.
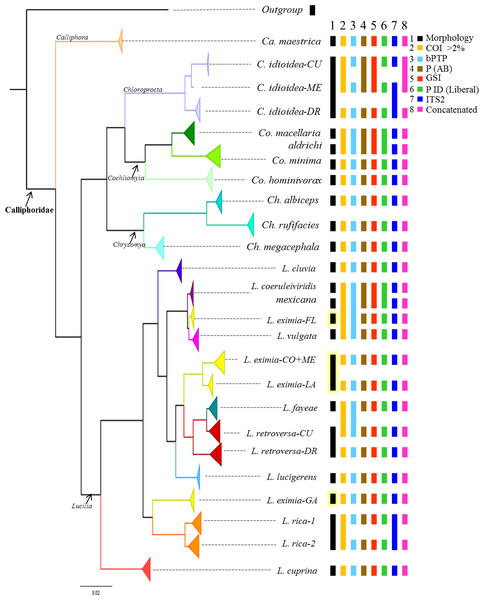
Figure 2: Summary of the Bayesian tree based on the COI dataset including 442 individuals, with the results of four different species delimitation approaches in addition to morphology, genetic distances of >2% mtDNA, ITS2 and the concatenated matrix.
See Fig. S1 for bootstrap support values.Results
We present by far the most extensive DNA barcoding dataset of Calliphoridae from the Caribbean. It includes a ∼1,200 bp fragment of the mitochondrial COI gene from 437 Calliphoridae specimens and ∼450 bp of the ITS2 gene from 158 specimens chosen to represent unique COI haplotypes of all putative species and all localities (20 different islands in the Caribbean plus Florida, Colombia and Mexico). Ninety nine of the sequences are from specimens collected in the mainland and the other 496 are from the Caribbean Islands. In total, we included 19 species of Calliphoridae identified morphologically (Whitworth, 2006; Whitworth, 2010), 16 of them reported from the Caribbean and three species, L. coeruleiviridis, L. mexicana and L. vulgata, from the mainland. The sequences from the Caribbean represent 16 of the 18 species of forensically important Calliphoridae that occur in the West Indies plus one of the most important livestock pest parasites in the Americas, C. hominivorax (Whitworth, 2010). The two species not included in this dataset are reported from Bahamas (Phormia regina) and Trinidad (Hemilucilia segmentaria), where we were not able to sample. For most species we included numerous exemplars, covering the geographic range of each species in the region.
Species delimitation using COI
Although based on traditional taxonomy we recognized 19 species of Calliphoridae in this study, COI gene analyses suggest that the diversity of Calliphoridae in the Caribbean is greater than morphology can detect. The phylogenetic analysis of COI recuperates 24 distinct clades (Fig. 2, Fig. S1), showing substantial geographic variation for L. eximia (four clades), C. idioidea (three clades), L. retroversa (two clades) and L. rica (two clades). However, COI did not distinguish between the pairs, Co. macellaria and Co. aldrichi from the Caribbean and L. coeruleiviridis and L. mexicana from the mainland. These four species are clearly identifiable based on morphological characteristics. Most putative species lineages showed genetic distances >2.7% (Table 3) and most of them are separated by a barcoding gap (Table 4). All species delimitation methods supported Ca. maestrica, C. idioidea-DR, Co. minima, Co. hominivorax, Ch. albiceps, Ch. rufifacies, Ch. megacephala, L. cluvia, L. cuprina, L. eximia-CO+ME, L. eximia-LA, Lucilia eximia-GA L. lucigerens, Lucilia retroversa-DR, and L. rica 1 and 2 (Fig. 2, Table 5); however, the other eight putative species were poorly supported in our analyses. Lower divergences, between 0.5 and 1.2% were found between clades, L. coeruleiviridis+L. mexicana, L. vulgata and L. eximia-FL, L. fayeae and L. retroversa CU, and between L. rica 1 and 2 (Table 3). All but bPTP methods of species determination supported L. eximia-FL clade, L. vulgata, L. fayeae, L. retroversa-CU (Table 5). Regarding C. idioidea, the Cuban and Mexico species-clades are only supported by bPTP and P ID (liberal). The bPTP analysis estimated between 21 and 29 species including the initial 26 putative species. Other species delimitation methods showed similar results, 22 putative species had P ID (liberal) higher of 89, 20 had significant Rosenberg values and 21 had GSI values of 100. All species determination methods fail in distinguishing between the pairs Co. macellaria and Co. aldrichi, and L. coeruleiviridis and L. mexicana as sequence divergences between species pairs are extremely low <0.08%. Given that no one method can distinguish between these species, the addition of ITS2 as a second barcoding locus was necessary to clarify the monophyly and validity of these species and increase the confidence of delimitation and identification of species with low genetic divergences.
| Putative species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ca. maestrica | ||||||||||||||||||||||||||
| 2 | C. idioidea-CU | 16.3 | |||||||||||||||||||||||||
| 3 | C. idioidea-DR | 15.3 | 2.8 | ||||||||||||||||||||||||
| 4 | C. idioidea-ME | 15.6 | 2.1 | 2.1 | |||||||||||||||||||||||
| 5 | Ch. albiceps | 15.5 | 13.8 | 13.1 | 13.1 | ||||||||||||||||||||||
| 6 | Ch. megacephala | 14.4 | 9.5 | 10.6 | 10.2 | 5.7 | |||||||||||||||||||||
| 7 | Ch. rufifacies | 15.5 | 14.5 | 14.1 | 14.1 | 2.8 | 6.7 | ||||||||||||||||||||
| 8 | Co. aldrichi | 14.4 | 10.6 | 9.5 | 9.9 | 10.2 | 9.2 | 12.0 | |||||||||||||||||||
| 9 | Co. hominivorax | 13.7 | 8.7 | 9.1 | 8.0 | 11.5 | 9.8 | 12.6 | 8.4 | ||||||||||||||||||
| 10 | Co. macellaria | 14.4 | 10.6 | 9.5 | 9.9 | 10.3 | 9.2 | 12.0 | 0.1 | 8.4 | |||||||||||||||||
| 11 | Co. minima | 15.7 | 10.5 | 10.5 | 10.2 | 9.8 | 8.8 | 11.0 | 4.2 | 9.7 | 4.2 | ||||||||||||||||
| 12 | L.. cluvia | 11.1 | 11.4 | 12.1 | 12.1 | 14.9 | 11.4 | 14.5 | 12.4 | 11.6 | 12.4 | 13.7 | |||||||||||||||
| 13 | L.. coeruleiviridis | 12.1 | 11.3 | 13.4 | 13.4 | 15.9 | 12.7 | 15.2 | 11.7 | 12.6 | 11.6 | 11.4 | 4.6 | ||||||||||||||
| 14 | L. cuprina | 11.6 | 9.2 | 9.5 | 10.2 | 13.1 | 9.5 | 13.8 | 10.6 | 11.9 | 10.6 | 11.2 | 8.2 | 8.5 | |||||||||||||
| 15 | L. eximia-CO-ME | 12.4 | 12.3 | 11.7 | 12.4 | 14.0 | 11.8 | 14.0 | 12.8 | 13.0 | 12.8 | 13.4 | 5.4 | 7.1 | 7.6 | ||||||||||||
| 16 | L. vulgata | 11.4 | 11.3 | 12.7 | 12.7 | 15.9 | 12.7 | 15.9 | 11.7 | 12.6 | 11.7 | 12.0 | 3.9 | 0.7 | 8.5 | 6.4 | |||||||||||
| 17 | L. eximia-FL | 11.6 | 11.0 | 12.4 | 12.4 | 15.5 | 12.4 | 14.8 | 12.0 | 12.4 | 11.9 | 11.1 | 4.8 | 1.2 | 8.7 | 6.9 | 1.2 | ||||||||||
| 18 | L. eximia-GA | 13.5 | 12.0 | 13.4 | 14.1 | 14.5 | 12.7 | 15.2 | 12.7 | 13.7 | 12.7 | 13.0 | 7.1 | 4.9 | 9.5 | 7.4 | 4.9 | 5.5 | |||||||||
| 19 | L. eximia-LA | 12.1 | 11.3 | 9.9 | 11.3 | 13.1 | 11.3 | 13.8 | 11.7 | 11.5 | 11.6 | 12.3 | 4.3 | 6.0 | 6.7 | 2.6 | 5.3 | 5.8 | 6.4 | ||||||||
| 20 | L. fayeae | 13.2 | 11.2 | 12.6 | 12.6 | 13.5 | 11.7 | 13.9 | 12.6 | 10.9 | 12.6 | 13.3 | 4.7 | 4.9 | 8.4 | 5.7 | 4.9 | 5.4 | 5.6 | 4.5 | |||||||
| 21 | L. lucigerens | 11.9 | 11.7 | 12.4 | 12.4 | 14.5 | 11.7 | 14.1 | 12.7 | 11.9 | 12.7 | 12.7 | 3.2 | 4.9 | 7.8 | 3.7 | 4.2 | 4.8 | 6.0 | 3.2 | 4.2 | ||||||
| 22 | L. mexicana | 12.1 | 11.3 | 13.4 | 13.4 | 15.9 | 12.7 | 15.2 | 11.7 | 12.6 | 11.6 | 11.4 | 4.6 | 0.0 | 8.5 | 7.1 | 0.7 | 1.2 | 4.9 | 6.0 | 4.9 | 4.9 | |||||
| 23 | L. retroversa-CU | 13.7 | 11.2 | 12.6 | 12.6 | 14.0 | 12.2 | 14.4 | 12.6 | 11.4 | 12.6 | 13.3 | 5.2 | 4.8 | 8.4 | 5.6 | 4.8 | 5.4 | 5.3 | 4.5 | 0.5 | 4.1 | 4.8 | ||||
| 24 | L. retroversa-DR | 13.5 | 12.4 | 13.1 | 13.1 | 14.8 | 13.4 | 14.5 | 12.7 | 13.3 | 12.7 | 13.4 | 4.0 | 5.0 | 9.2 | 5.4 | 4.3 | 4.8 | 5.7 | 4.6 | 2.8 | 3.6 | 5.0 | 2.7 | |||
| 25 | L. rica_1 | 13.9 | 12.1 | 12.1 | 11.6 | 14.8 | 11.9 | 14.7 | 13.0 | 11.1 | 13.0 | 13.6 | 6.1 | 6.8 | 8.2 | 6.6 | 6.1 | 6.6 | 7.5 | 5.4 | 5.0 | 5.4 | 6.8 | 4.7 | 5.0 | ||
| 26 | L. rica_2 | 13.4 | 11.9 | 11.9 | 11.2 | 14.7 | 11.6 | 14.8 | 12.6 | 10.7 | 12.6 | 13.3 | 5.6 | 6.3 | 8.0 | 6.4 | 5.6 | 6.1 | 7.3 | 5.6 | 5.6 | 5.2 | 6.3 | 5.3 | 5.3 | 1.0 | |
Notes:
- CO
-
Colombia
- CU
-
Cuba
- DR
-
Dominican Republic
- FL
-
Florida
- GA
-
Greater Antilles
- LA
-
Lesser Antilles
- ME
-
Mexico
| Putative species | % variation within species |
|---|---|
| Ca. maestrica | 0.14 |
| C. idioidea-CU | 0.00 |
| C. idioidea-DR | 0.00 |
| C. idioidea-ME | n/a |
| Ch. albiceps | 0.00 |
| Ch. megacephala | 0.00 |
| Ch. rufifacies | 0.01 |
| Co. aldrichi | 0.00 |
| Co. hominivorax | 0.24 |
| Co. macellaria | 0.15 |
| Co. minima | 0.29 |
| L.. cluvia | 0.10 |
| L.. coeruleiviridis | 0.00 |
| L. cuprina | 0.00 |
| L. eximia-CO-ME | 0.61 |
| L. vulgata | 0.00 |
| L. eximia-FL | 1.06 |
| L. eximia-GA | 0.00 |
| L. eximia-LA | 0.00 |
| L. fayeae | 0.14 |
| L. lucigerens | 0.00 |
| L. mexicana | 0.00 |
| L. retroversa-CU | 0.18 |
| L. retroversa-DR | 0.08 |
| L. rica_1 | 0.40 |
| L. rica_2 | 0.15 |
Notes:
- CO
-
Colombia
- CU
-
Cuba
- DR
-
Dominican Republic
- FL
-
Florida
- GA
-
Greater Antilles
- LA
-
Lesser Antilles
- ME
-
Mexico
| Putative species | Mono | D Intra | D Inter | Dtra/ Dter | P ID(Strict) | P ID(Liberal) | P(AB) | GSI | bPTP | Sp congru | Sp cons | Morph | Concat |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. C. maestrica | yes | 0.001 | 0.096 | 0.01 | 0.93 (0.80, 1.0) | 0.98 (0.88, 1.0) | NAN | 1 | Y | 1 | 1 | 1 | 1 |
| 2. C. idioidea-CU | yes | 0.0009 | 0.012 | 0.07 | 0.74 (0.57, 0.92) | 0.97 (0.82, 1.0) | 0.17 | 1 | Y | 2 | 2 | 2 | 2 |
| 3. C. idioidea-ME | yes | n/a | 0.012 | n/a | n/a | 0.96 (0.83, 1.0) | 0.17 | NA | Y | ||||
| 4. C. idioidea-DR | yes | 0.003 | 0.014 | 0.19 | 0.81 (0.68, 0.93) | 0.95 (0.85, 1.0) | 1.98E −03 | 1 | Y | 3 | 3 | 3 | |
| 5. Co. aldrichi | no | 0.0008 | 0.002 | 0.46 | 0.82 (0.75, 0.89) | 0.95 (0.91, 0.99) | NA | 0.39 | N | 4 | 4 | 3 | 4 |
| 6. Co. macellaria | no | 0.003 | 0.002 | 1.47 | 0.00 (0.00, 0.00) | 0.31 (0.28, 0.34) | NA | 0.61 | N | 4 | 5 | ||
| 7. Co. minima | yes | 0.002 | 0.030 | 0.07 | 0.97 (0.92, 1.0) | 0.99 (0.96, 1.0) | 6.30E −27 | 1 | Y | 5 | 5 | 5 | 6 |
| 8. Co. hominivorax | yes | 0.004 | 0.066 | 0.07 | 0.75 (0.57, 0.92) | 0.97 (0.83, 1.0) | 1.90E −07 | 1 | Y | 6 | 6 | 6 | 7 |
| 9. Ch. albiceps | yes | 0.002 | 0.033 | 0.05 | 0.90 (0.77, 1.0) | 0.97 (0.87, 1.0) | 4.90E −08 | 1 | Y | 7 | 7 | 7 | 8 |
| 10. Ch. rufifacies | yes | 0.0009 | 0.033 | 0.03 | 0.99 (0.93, 1.0) | 1.00 (0.97, 1.0) | 4.90E −08 | 1 | Y | 8 | 8 | 8 | 9 |
| 11. Ch. megacephala | yes | 0.001 | 0.054 | 0.02 | 0.99 (0.94, 1.0) | 1.00 (0.97, 1.0) | 1.40E −24 | 1 | Y | 9 | 9 | 9 | 10 |
| 12. L. cluvia | yes | 0.002 | 0.033 | 0.07 | 0.91 (0.81, 1.0) | 0.98 (0.92, 1.0) | 7.10E −12 | 1 | Y | 10 | 10 | 10 | 11 |
| 13. L. coeruleiviridis | no | 0.0008 | 0.0008 | 1.12 | 0.18 (0.05, 0.31) | 0.49 (0.38, 0.59) | NA | 0.59 | N | 11 | 11 | 11 | 12 |
| 14. L. mexicana | no | 0.0007 | 0.0008 | 0.88 | 0.20 (0.02, 0.39) | 0.51 (0.36, 0.66) | NA | 0.49 | N | 12 | 13 | ||
| 15. L. eximia-FL | yes | 0.002 | 0.005 | 0.40 | 0.39 (0.24, 0.54) | 0.74 (0.58, 0.89) | 0.03 | 1 | N | 13 | 14 | ||
| 16. L. vulgata | yes | 0.002 | 0.007 | 0.32 | 0.65 (0.51, 0.79) | 0.89 (0.78, 1.0) | 0.03 | 1 | N | 14 | 15 | ||
| 17. L. eximia-ME-CO | yes | 0.004 | 0.016 | 0.27 | 0.82 (0.71, 0.92) | 0.93 (0.87, 0.99) | 3.60E −04 | 1 | Y | 12 | 12 | 16 | |
| 18. L. eximia-LA | yes | 0.002 | 0.016 | 0.12 | 0.79 (0.64, 0.93) | 0.95 (0.84, 1.0) | 3.60E −04 | 1 | Y | 13 | 13 | ||
| 19. L. fayeae | yes | 0.002 | 0.008 | 0.31 | 0.82 (0.73, 0.91) | 0.94 (0.89, 0.99) | 2.40E −06 | 1 | N | 14 | 14 | 15 | 17 |
| 20. L. retroversa-CU | yes | 0.004 | 0.008 | 0.46 | 0.75 (0.67, 0.84) | 0.92 (0.87, 0.97) | 2.40E −06 | 1 | N | 16 | 18 | ||
| 21. L. retroversa-DR | yes | 0.002 | 0.024 | 0.09 | 0.96 (0.91, 1.0) | 0.99 (0.96, 1.0) | 2.60E −14 | 1 | Y | 15 | 15 | 19 | |
| 22. L. lucigerens | yes | 0.002 | 0.035 | 0.05 | 0.76 (0.58, 0.94) | 0.98 (0.84, 1.0) | 9.90E −07 | 1 | Y | 16 | 16 | 17 | 20 |
| 23. L. eximia-GA | yes | 0.001 | 0.048 | 0.03 | 0.98 (0.91, 1.0) | 1.00 (0.96, 1.0) | 1.30E −11 | 1 | Y | 17 | 17 | 21 | |
| 24. L. rica_1 | yes | 0.003 | 0.011 | 0.24 | 0.90 (0.83, 0.96) | 0.97 (0.92, 1.0) | 4.40E −09 | 1 | Y | 18 | 18 | 18 | 22 |
| 25. L. rica_2 | yes | 0.002 | 0.011 | 0.22 | 0.90 (0.83, 0.97) | 0.97 (0.92, 1.0) | 4.40E −09 | 1 | Y | 19 | 23 | ||
| 26. L. cuprina | yes | 0.002 | 0.076 | 0.03 | 0.98 (0.91, 1.0) | 1.00 (0.96, 1.0) | 4.30E −19 | 1 | Y | 20 | 19 | 19 | 24 |
Notes:
- CO
-
Colombia
- CU
-
Cuba
- DR
-
Dominican Republic
- FL
-
Florida
- GA
-
Greater Antilles
- LA
-
Lesser Antilles
- ME
-
Mexico
Phylogenetic inference
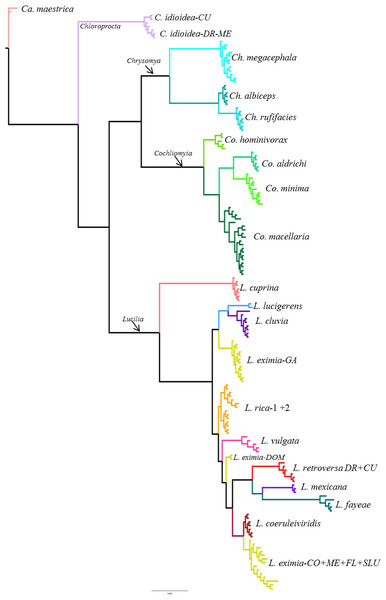
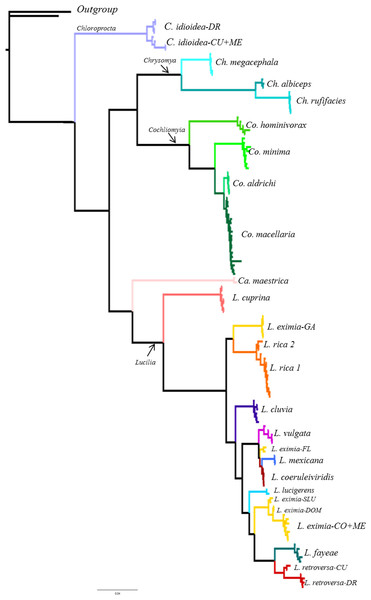
From the 26 putative species analyzed here, 25 were represented by multiple individuals and one by a single individual in the COI analysis. All phylogenetic analyses (COI, ITS2, COI+ITS2) yielded well resolved trees with strong posterior probability support for most of the branches and broadly agreed on species limits but with some differences in topology (Figs. 2–4, Figs. S1–S3). The Bayesian analysis of the ITS2 supported the monophyly of 21 of 26 putative species. It recovered the monophyly of Co. aldrichi, Co. macellaria, L. mexicana and L. coeruleiviridis, which failed with all other analysis. However it did not recover the geographic variation of C. idioidea from Mexico and Dominican Republic, L. retroversa from Cuba and Dominican Republic or L. rica 1 and 2, and it only recovers three of the four L. eximia clades indicated by COI analyses (Fig. 3, Fig. S2). The concatenated tree supports 24 of the 26 putative species including two clades within L. retroversa, L. rica, and C. idioidea, and three clades within L. eximia. The concatenated matrix did not support the monophyly of C. idioidea-CU that is nested within C. idioidea-ME and L. eximia-CO+ME nested within L. eximia-LA (Fig. 4, Fig. S3).
Discussion
Accurate identification of insects is a crucial step to using them as reliable evidence in legal investigations. Although morphology has been successfully used to identify immature specimens involved in cadaveric decomposition (Cardoso et al., 2014; Florez & Wolff, 2009; Szpila et al., 2013a; Szpila et al., 2013b; Szpila et al., 2014; Szpila & Villet, 2011; Wells, Byrd & Tantawi, 1999), this approach depends on the availability of taxonomic keys of the species present in the region. In the Caribbean, the immature stages of 11 species are unknown and other approaches are needed in order to identify them. Besides this, morphology may overlook potentially cryptic species and cannot be used on incomplete or destroyed specimens found on a crime scene. Here, we show DNA barcoding to be useful in overcoming these problems and provide tools to accelerate the identification and discovery of species. This is particularly important in areas like the Caribbean, where studies of insects involved in cadaveric decomposition are scarce (Whitworth, 2010; Yusseff-Vanegas & Agnarsson, 2016; Yusseff-Vanegas, 2007; Yusseff-Vanegas, 2014). One of the first steps required for this approach is creating a reliable DNA barcode database that can be used with confidence in order to identify unknown specimens found in death scenes investigation (DeBry et al., 2013; Harvey et al., 2003).
Figure 3: Bayesian tree based on ITS2 dataset including 158 specimens.
Individual terminal taxa have been replaced with species names, while full taxon clade structure is retained. Colors represent different species based on morphology. See Fig. S2 for bootstrap support values.Figure 4: Bayesian tree based on the concatenate dataset including 137 specimens.
Individual terminal taxa have been replaced with species names, while full taxon clade structure is retained. Colors represent different species based on morphology. See Fig. S3 for bootstrap support values.The success of DNA barcoding relies on the quality of the underlying database used to compare DNA sequences of new samples. A good database should contain DNA barcodes of expertly identified individuals, and preferably taxon sampling covering the distribution range of each species. Our study complies with both requirements and is the first thorough molecular study of Calliphoridae from the Caribbean. It includes a representative collection from all but two forensically relevant Calliphoridae from the region, and covers the whole geographic range of most of the investigated species (Table 1). All specimens in this study were carefully identified using traditional morphological taxonomy (Whitworth, 2006; Whitworth, 2010; Whitworth, 2014) and each individual was successfully allocated to one of the currently recognized calliphorid species, except for specimen CO027 that could only be identified to the genus level. Although morphological identification of specimens collected in this study corresponded to 19 previously reported species (Whitworth, 2010), our results based on molecular data indicate higher diversity. In all, 26 putative species lineages were identified, and in particular our results indicate that Lucilia and Chloroprocta are more diverse than suggested by current taxonomy. COI recovered substantial geographic variation for C. idioidea, L. eximia, L. retroversa and L. rica such that molecular data indicate up to eleven putative species lineages that cannot be, or at least have not been, recognized by morphology.
Lucilia eximia is considered a widespread species found from the southern United States through Central America to southern South America (Whitworth, 2014). Nevertheless, our molecular results show four distinct genetic clusters with an average inter-cluster divergence from 2.5 to 7.4% (Table 3). The clusters are geographically structured and three of them are widely separated (Fig. 2, Fig. S1). The first one is the Greater Antilles cluster (GA) that includes specimens from Puerto Rico, Mona Island and Dominican Republic, the second is a small cluster that includes specimens from Florida (FL), the third one contains specimens from Colombia and Mexico (CO-MEX), and the fourth contains specimens from the Lesser Antilles islands of Dominica and Saint Lucia (LA). Similar results were reported by Solano, Wolff & Castrol (2013) and Whitworth (2014) where widely separate clades of L. eximia were found using DNA barcodes. All species delimitation methods supported the uniqueness and genetic isolation of the four clades, each showing low intra-clade divergence (<1%, Table 4), and thus likely representing four distinct species. Although we found some morphological variation between L. eximia from the mainland and islands and among islands as previously reported (James, 1967; Whitworth, 2010; Woodley & Hilburn, 1994), detailed revision of those specimens by Dr. Whitworth from Washington State University concluded that there is not enough evidence to separate them as different morphological species, suggesting they may be morphologically cryptic species. Further studies on these populations will be necessary to establish their taxonomic status.
Lucilia rica was collected throughout the Lesser Antilles and is very abundant in most of the islands (personal observation). Although James (1970) listed this species from Puerto Rico, we did not find any specimens after very extensive collections on the island. Thus, we believe that L. rica is restricted to the Lesser Antilles and has not dispersed beyond Anguilla. Whitworth (2010) reported this species from Antigua, Bermuda, Guadalupe and St. Lucia; however, we found it in eight more islands (Table 1) and our data showed two geographic clusters (Figs. 2, 4; Figs. S1, S3). The first cluster (L. rica-1) contains specimens from St Martin, Saba, St. Eustatius, St. Kitts, Nevis and Martinique and the second one (L. rica 2) from Barbuda, Antigua, Montserrat and Guadeloupe. Although the genetic distance between clades is low (1%), it is much greater than the intra-clade divergences (<0.3%). While all species delimitation methods support the possibility of two different species (Tables 5 and 6), we did not find morphological evidence to support it. Nevertheless, given that this is the most abundant Lucilia species of the Lesser Antilles, additional studies on these populations are important to determine if the genetic difference is due to intraspecific variation or if they are cryptic species.
For Lucilia retroversa we find two geographic clusters, one from Cuba and one from the Dominican Republic with an average mtDNA distance of 2.5% (Table 3) and with low intra-clade divergence (<0.2%, Table 4). Whitworth (2010) reported some morphological differences between specimens from Bahamas (which share morphology with Cuban specimens) and the Dominican Republic, but after examination of male and female genitalia he concluded that those differences were intraspecific variation. However, he noticed that our L. retroversa specimens have a brown basicosta instead of white or yellow basicosta which is an important character used to separate L. retroversa from other species (see taxonomic key in Whitworth, 2010). Given that all of our species delimitation results support two possible cryptic species, we recommend further detailed molecular and morphological studies of these populations to determine if they merit the description of a separate species.
| Putative species | Closest species | Mono | D Intra | D Inter | Dtra/ Dter | P ID(Strict) | P ID(Liberal) | P(AB) |
|---|---|---|---|---|---|---|---|---|
| 1. Ca. maestrica | L. cuprina | yes | 0.005 | 0.19 | 0.03 | 0.58 (0.43, 0.73) | 0.97 (0.82, 1.0) | 1.00E−05 |
| 2. L. cluvia | L. coeruleiviridis | yes | 0.005 | 0.05 | 0.10 | 0.87 (0.74, 0.99) | 0.97 (0.87, 1.0) | 5.50E−09 |
| 3 L. coeruleiviridis | L. eximia-FL | yes | 0.001 | 0.01 | 0.14 | 0.84 (0.72, 0.97) | 0.96 (0.86, 1.0) | 0.01 |
| 4. L. mexicana | L. coeruleiviridis | yes | 0.0009 | 0.02 | 0.06 | 0.75 (0.58, 0.93) | 0.97 (0.83, 1.0) | 0.01 |
| 5. L. eximia-FL | L. coeruleiviridis | yes | 0.005 | 0.01 | 0.48 | 0.34 (0.19, 0.50) | 0.69 (0.53, 0.84) | 4.94E−03 |
| 6. L. vulgata | L. coeruleiviridis | yes | 0.003 | 0.02 | 0.18 | 0.75 (0.60, 0.89) | 0.94 (0.83, 1.0) | 0.1 |
| 7. L. eximiaCO-ME | L. eximia-LA | yes | 0.006 | 0.02 | 0.32 | 0.79 (0.69, 0.90) | 0.92 (0.86, 0.99) | 0.01 |
| 8. L. fayeae | L. retroversa-CU | yes | 0.006 | 0.04 | 0.15 | 0.84 (0.71, 0.96) | 0.96 (0.86, 1.0) | 4.30E−04 |
| 9. L. retroversa-CU | L. retroversa-DR | yes | 0.004 | 0.02 | 0.18 | 0.50 (0.35, 0.65) | 0.87 (0.72, 1.0) | 0.03 |
| 10. L. lucigerens | L. eximia-LA | yes | 0.003 | 0.04 | 0.08 | 0.55 (0.40, 0.70) | 0.93 (0.78, 1.0) | 3.10E−04 |
| 11 L. eximia-GA | L. rica 2 | yes | 0.002 | 0.06 | 0.04 | 0.91 (0.78, 1.0) | 0.98 (0.87, 1.0) | 2.70E−06 |
| 12. L. rica 1 | L. rica 2 | yes | 0.005 | 0.02 | 0.33 | 0.81 (0.72, 0.90) | 0.94 (0.88, 0.99) | 4.20E−04 |
| 13. L. rica 2 | L. rica 1 | yes | 0.004 | 0.02 | 0.30 | 0.59 (0.42, 0.77) | 0.84 (0.69, 0.98) | 4.20E−04 |
| 14. L. cuprina | L. cluvia | yes | 0.003 | 0.15 | 0.02 | 0.94 (0.83, 1.0) | 1.00 (0.94, 1.0) | 1.90E−11 |
| 15. Ch. albiceps | Ch. rufifacies | yes | 0.003 | 0.04 | 0.06 | 0.75 (0.57, 0.93) | 0.97 (0.83, 1.0) | 2.98E−03 |
| 16. Ch. rufifacies | Ch. albiceps | yes | 0.002 | 0.04 | 0.05 | 0.90 (0.78, 1.0) | 0.97 (0.87, 1.0) | 2.98E−03 |
| 17. Ch. megacephala | Ch. albiceps | yes | 0.003 | 0.11 | 0.02 | 0.92 (0.79, 1.0) | 0.98 (0.87, 1.0) | 2.80E−05 |
| 18. Co. aldrichi | Co. macellaria | yes | 0.002 | 0.01 | 0.13 | 0.85 (0.72, 0.97) | 0.96 (0.86, 1.0) | 4.70E−07 |
| 19. Co. macellaria | Co. aldrichi | yes | 0.007 | 0.01 | 0.52 | 0.84 (0.78, 0.89) | 0.96 (0.93, 0.99) | 4.70E−07 |
| 20. Co. minima | Co. aldrichi | yes | 0.007 | 0.05 | 0.14 | 0.88 (0.77, 0.99) | 0.96 (0.90, 1.0) | 4.50E−09 |
| 21. Co. hominivorax | Co. aldrichi | yes | 0.007 | 0.09 | 0.08 | 0.88 (0.76, 1.0) | 0.97 (0.87, 1.0) | 1.00E−07 |
| 22. C. idioidea-DR | C. idioidea-ME | yes | 0.003 | 0.02 | 0.19 | 0.74 (0.60, 0.88) | 0.94 (0.83, 1.0) | 4.08E−03 |
| 23 C. idioidea-CU | C. idioidea-ME | yes | 0.001 | 0.01 | 0.06 | 0.56 (0.41, 0.71) | 0.94 (0.79, 1.0) | 0.33 |
| 24: L. retroversa-DR | L. retroversa-CU | yes | 0.004 | 0.02 | 0.16 | 0.76 (0.62, 0.90) | 0.94 (0.83, 1.0) | 0.03 |
| 25: C. idioidea-ME | C. idioidea-CU | no | 0.005 | 0.01 | 0.38 | 0.40 (0.24, 0.55) | 0.75 (0.59, 0.90) | NA |
| 26. L. eximia-LA | L. eximiaCO-ME | no | 0.007 | 0.02 | 0.36 | 0.63 (0.48, 0.77) | 0.88 (0.77, 0.99) | NA |
Notes:
- CO
-
Colombia
- CU
-
Cuba
- DR
-
Dominican Republic
- FL
-
Florida
- GA
-
Greater Antilles
- LA
-
Lesser Antilles
- ME
-
Mexico
Chloroprocta idioidea, the only species of Chloroprocta, is a widespread species found from southern North America to southern South America (Dear, 1985; Whitworth, 2010). Our results show that C. idioidea is also geographically structured into three clades: one from Dominican Republic, one from Cuba and one from Mexico (Fig. 2, Fig. S1). Analysis of the genetic divergence between clades show more than 2% divergence between the Cuba and Dominican Republic clades but less than 2% divergence between the Mexico and Cuba clades (Table 3). Some authors (Hall, 1948; Shannon, 1926) believed there were two species of the genus in the Americas, however (Dear, 1985) concluded that there was only one single widespread species that exhibits some color variations which is dependent upon geographic distribution. Our molecular results indicate at least two, and perhaps three, separate species of Chloroprocta. All species delimitation methods (Table 5) and the concatenated matrix (Fig. 4, Fig. S3) suggest that the Dominican Republic versus the Cuba and Mexico clade are separate species, but were ambiguous about the status of C. idioidea-CU that is nested within C. idioidea-ME. Cuban and Mexican specimens are morphologically similar, dark-bluish in color with brownish to orange legs, however, as reported by Dear (1985) the Cuban females have brownish, instead of yellow-white calypters. Our specimens from Dominican Republic are similar to the southern USA specimens described by Dear (1985) but have darker post spiracles and clear wings with only the costa faintly tinted. Although we could see morphological differences between populations, those differences were based on a limited number of specimens (e.g., five specimens from Dominican Republic and three from Mexico). Further studies with larger number of specimens of C. idioidea, including detailed morphological descriptions and expanded molecular analysis, are necessary to further test species limits within this genus.
Our focus here is not to fully resolve calliphorid taxonomy. However, it is important to highlight the consequences of our findings for forensic entomology studies. Currently L. eximia is one of the most widespread and abundant Lucilia in the Neotropics (Whitworth, 2014). However, our results suggest that, in fact, this is not one widely distributed species, but potentially several species that differ in geographic range and possibly in biological traits (rates of development, diapause, habitat preference, feeding habits etc.). The same is true for L. retroversa and C. idioidea, both have genetically distinct clades in the Dominican Republic and in Cuba (Figs. 2 and 4). This finding will have direct consequences for the use of these species in legal investigations, if that variation reflects differences in behavior and biology, that can affect post mortem interval estimations (Tarone, Singh & Picard, 2015). Previous studies of Phormia regina (Byrd & Allen, 2001), C. macellaria and C. rufifacies (Yusseff-Vanegas, 2007) have shown that their developmental rate differ from different populations. Picard & Wells (2009) suggested that that variation is in part due to differences in population genetic structure, and for that reason, ecological data obtained from one population should not be generalized or extrapolated to other populations (Byrne et al., 1995). This is important at least for specimens collected in Cuba where both populations are present, probably as the result of recent dispersal of L. retroversa and C. idoidea from the Dominican Republic to Cuba. Our results (S1) show that two of the southeast Cuban specimens, CU007 (L. retroversa) and CU008 (C. idioidea), collected in Turquino National Park in Cuba (Table 1), cluster tightly with Dominican Republic specimens (S1). To confirm the genetic affinity of these specimens we added three more nuclear genes for a limited number of individuals from both populations and re-ran the analysis. The multi-gene analysis again strongly clustered CU007 and CU008 with the Dominican Republic specimens for each species. Thus, both the Dominican Republic and Cuban populations are clearly present in Cuba.
COI recuperated substantial geographic variation with high COI sequences divergence between populations of Lucilia eximia, L. retroversa, L. rica and C. idioidea (Fig. 2, Fig. S1), suggesting the possibility of different species (Hebert et al., 2003; Hebert, Ratnasingham & deWaard, 2003). However, genetic variation is not always indicative of species differentiation. For instance, studies including Phormia regina have found that the genetic distance between N American and W European populations is higher than 4% (Boehme, Amendt & Zehner, 2012; Desmyter & Gosselin, 2009). But after detailed molecular and morphological analysis of both populations, Jordaens et al. (2013a) concluded that the high differentiation at COI, COII and cytb, but low (16S, nDNA) and lack of morphological differentiation, was indicative of substantial intraspecific mtDNA sequence divergence, rather than a species level differentiation. In light of those results, definite conclusions cannot yet be drawn regarding the taxonomy of these species. Further population level studies of the four species in question are therefore necessary. A comprehensive molecular analysis including several mitochondrial and nuclear genes in combination with morphological examination and detailed description of the genitalia, are required to determine if they are in fact different species, or if the genetic difference between populations is the product of intraspecific variation. Meanwhile the use of these species for forensic purposes should be evaluated carefully and with reference to genetic and behavioral differences among its populations.
Regarding the other Calliphoridae species, Ca. maestrica, Co. minima, Co hominivorax, Ch. albiceps, Ch. rufifacies, Ch. megacephala, L. cluvia, L. cuprina and L. lucigerens, all showed reciprocal monophyly with strong posterior probability support and all can be successfully identified using the DNA barcoding approach. All species delimitation methods, phylogenetic analysis of ITS2, and the concatenated tree support their monophyly and species status, and the results are congruent with morphology. Calliphora maestrica is the only Calliphora species reported for the Caribbean and is endemic from the region. This species was originally described from Sierra Maestra region in Cuba (Peris et al., 1998) and later reported also from Jamaica and Dominican Republic (Whitworth, 2010). Although we collected on all three islands, we only found C. maestrica in Villa Pajon, Dominican Republic, a cold region at altitudes >2,140 m. We did not find it in Cuba or Jamaica, likely due to lack of sampling at altitudes above 1,200 m on both islands.
The three species of Chrysomya were recently introduced to the New World (Baumgartner & Greenberg, 1984). Although Whitworth (2010) reported Ch. megacephala and Ch. rufifacies from Dominica, Dominican Republic, Jamaica and Puerto Rico, they are abundantly present in most of the islands being found from Cuba to Martinique (Table 1). In contrast, Chrysomya albiceps has more restricted distribution being found in islands closer to South America (Table 1, Whitworth, 2010). Although Dear (1985) reported this species from Puerto Rico, we did not find it after extensive collections on the island. That report was based on a single larva found in a goat, probably of Ch. albiceps but the species was not confirmed (Gagne, 1981). We believe that Ch. albiceps has not dispersed beyond Dominica and that the species reported by Dear (1985) was in fact Ch. rufifacies. Given the high dispersal abilities of the species of this genus (Baumgartner & Greenberg, 1984) and their invasive behavior (Aguiar-Coelho & Milward-De-Azevedo, 1998; De Andrade et al., 2002; Faria et al., 1999; Wells & Greenberg, 1992), it is not surprising to find them widely distributed and very well established throughout the Caribbean. They do not show any geographic structure, suggesting their recent colonization from the mainland and the constant gene flow among populations.
Lucilia cluvia and L. cuprina, are widely distributed flies found in different parts of the world (Byrd & Castner, 2010). Lucilia cluvia is considered rare (Whitworth, 2010). Although it has been reported from several locations in Puerto Rico, Cuba, and Martinique, we have only found two specimens in a suburban area in Toa Baja, Puerto Rico. Lucilia cuprina is reported from several islands in the Caribbean, but we only found it in urban areas of Puerto Rico as our focus on other islands was in non-urban areas. Finally L. lucigerens is an endemic species from Jamaica and was collected abundantly throughout the island.
DNA barcoding in animals typically employs a single mitochondrial marker for identification and delimitation of species (Hebert et al., 2003; Hebert, Ratnasingham & DeWaard, 2003), and this approach has shown to be useful in Calliphoridae species identification. However it does not reliably distinguish among some recently diverged species (Harvey et al., 2003; Nelson, Wallman & Dowton, 2007), leading to doubt that COI alone is sufficient for identification of species (Nelson, Wallman & Dowton, 2007; Wells, Wall & Stevens, 2007). Rather, the use of multiple markers has been suggested as a means to increase the accuracy of species identification. Indeed, our results show that COI barcoding successfully identified most species, but did not distinguish between the pairs L. mexicana and L. coeruleiviridis as previously reported (DeBry et al., 2013; Whitworth, 2014; Williams, Lamb & Villet, 2016) and between Co. aldrichi and Co. macellaria (Tables 3 and 5, Fig. S1 ). The latter species is considered one of the most important Calliphoridae for forensic studies in the Americas (see discussion in Yusseff-Vanegas & Agnarsson, 2016). Additionally, COI showed very low genetic divergences (<0.7%, Table 3) between the putative species L. vulgata and L. coeruleiviridis, and L. fayeae and L. retroversa-CU; species that are clearly distinguished based on morphological characteristics. This low genetic divergence may reflect short histories of reproductive isolation (Hebert, Ratnasingham & DeWaard, 2003), or mitochondrial introgression. In either case the addition of the nuclear gene ITS2 resolved the monophyly of the four species that COI alone did not support, and added resolution for uncertain groups with mtDNA genetic distances lower than 2%. These findings agreed with previous studies where the analysis of ITS2 resolved complex species delimitation (GilArriortua et al., 2014; Song, Wang & Liang, 2008), however, not always addition of more genes resolved the monophyly of the sister species like the case of L. illustris and L. caesar, where, after analysis including six genes, the monophyly remain unresolved (Sonet et al., 2012).
In sum, our study demonstrates the importance employing a second nuclear marker for barcoding analyses and species delimitation of calliphorids and the power of molecular data in combination with a complete reference database to enable identification of taxonomically and geographically diverse insects of forensic importance. The combination of the two markers supported the higher diversity of Calliphoridae in the Caribbean recovering the monophyly of nine of the eleven possible cryptic species. However, definite conclusion about the taxonomy of these species will depend on further studies combining molecular and morphological approaches.
Conclusion
From almost a decade many studies have applied DNA-based methods for the identification of insects of forensic importance to enable identification of unknown insect specimens found in death scene investigations. However, this technique is not being implemented and the traditional time consuming methods of raising immature stages to adulthood is still in practice. The use of this approach has been unsuccessful because of lack of confidence due to sequence gaps and errors, unauthenticated reference DNA sequences in the database, and incomplete reference data set with partial taxon sampling. Thus, the base science foundation for application of DNA sequences analysis is unsolid for identification of evidentiary samples. Despite all studies of DNA based identification for insects involved in forensics, only a few of them include a complete reference data set. But even with a complete reference database, COI has failed in demonstrating reciprocal monophyly for several recently diverged species creating uncertainty about its use for identification. The addition of ITS2 as a second marker may be the key to increase certainty in identification and make this technique useful for forensic purposes. A great effort to build complete reference databases including extensive collections, accurate identification, geographical genetic variation for each targeted insect group and the addition of ITS2 as a second marker is needed. In general, COI barcodes are highly useful for species identification of the Caribbean calliphorids. ITS2 appears to be a good second marker that allows higher resolution and accurate identification of specimens that cannot be separated by COI alone. Our study provides, for the first time, a reliable dataset to accurately identify species of the family Calliphoridae from the Caribbean, and opens the door for future studies on biodiversity, biogeography, distribution and ecology of these forensically important flies.
Supplemental Information
Phylogenetic relationship within Calliphoridae based on a Bayesian analysis of nucleotide data from COI
Numbers indicate posterior probability support values. Specimen voucher codes referred to in Table 1 are shown following species names. For specimens from Lesser Antilles (LA), the three capital letters before the voucher code refers to the name of the islands abbreviated a follows: SBA, St. Barthelemy; SAB, Saba; BAR, Barbuda; NEV, Nevis; KIT, St. Kitts; MTQ, Martinique; ANT, Antigua; GUA, Guadeloupe; MON, Montserrat; EUS, St. Eustatius; SMA, St. Martin, SLU, St. Lucia; BBD Barbados.
Phylogenetic relationship within Calliphoridae based on a Bayesian analysis of nucleotide data from ITS
Numbers indicate posterior probability support values. Specimen voucher codes referred to in Table 1 are shown following species names. For specimens from Lesser Antilles (LA), the three capital letters before the voucher code refers to the name of the islands abbreviated a follows: SBA, St. Barthelemy; SAB, Saba; BAR, Barbuda; NEV, Nevis; KIT, St. Kitts; MTQ, Martinique; ANT, Antigua; GUA, Guadeloupe; MON, Montserrat; EUS, St. Eustatius; SMA, St. Martin, SLU, St. Lucia; BBD Barbados.
Phylogenetic relationship within Calliphoridae based on based on partitioned Bayesian analysis of the combined gene (COI and ITS2) data set
Numbers indicate posterior probability support values. Specimen voucher codes referred to in Table 1 are shown following species names. For specimens from Lesser Antilles (LA), the three capital letters before the voucher code refers to the name of the islands abbreviated a follows: SBA, St. Barthelemy; SAB, Saba; BAR, Barbuda; NEV, Nevis; KIT, St. Kitts; MTQ, Martinique; ANT, Antigua; GUA, Guadeloupe; MON, Montserrat; EUS, St. Eustatius; SMA, St. Martin, SLU, St. Lucia; BBD Barbados.