Chromosomal rearrangements and protein globularity changes in Mycobacterium tuberculosis isolates from cerebrospinal fluid
- Published
- Accepted
- Received
- Academic Editor
- Ugo Bastolla
- Subject Areas
- Bioinformatics, Genomics, Microbiology
- Keywords
- Meningitis, Comparative genomics, Mycobacterium tuberculosis
- Copyright
- © 2016 Saw et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2016. Chromosomal rearrangements and protein globularity changes in Mycobacterium tuberculosis isolates from cerebrospinal fluid. PeerJ 4:e2484 https://doi.org/10.7717/peerj.2484
Abstract
Background
Meningitis is a major cause of mortality in tuberculosis (TB). It is not clear what factors promote central nervous system invasion and pathology but it has been reported that certain strains of Mycobacterium tuberculosis (Mtb) might have genetic traits associated with neurotropism.
Methods
In this study, we generated whole genome sequences of eight clinical strains of Mtb that were isolated from the cerebrospinal fluid (CSF) of patients presenting with tuberculous meningitis (TBM) in Malaysia, and compared them to the genomes of H37Rv and other respiratory Mtb genomes either downloaded from public databases or extracted from local sputum isolates. We aimed to find genomic features that might be distinctly different between CSF-derived and respiratory Mtb.
Results
Genome-wide comparisons revealed rearrangements (translocations, inversions, insertions and deletions) and non-synonymous SNPs in our CSF-derived strains that were not observed in the respiratory Mtb genomes used for comparison. These rearranged segments were rich in genes for PE (proline-glutamate)/PPE (proline-proline-glutamate), transcriptional and membrane proteins. Similarly, most of the ns SNPs common in CSF strains were noted in genes encoding PE/PPE proteins. Protein globularity differences were observed among mycobacteria from CSF and respiratory sources and in proteins previously reported to be associated with TB meningitis. Transcription factors and other transcription regulators featured prominently in these proteins. Homologs of proteins associated with Streptococcus pneumoniae meningitis and Neisseria meningitidis virulence were identified in neuropathogenic as well as respiratory mycobacterial spp. examined in this study.
Discussion
The occurrence of in silico genetic differences in CSF-derived but not respiratory Mtb suggests their possible involvement in the pathogenesis of TBM. However, overall findings in this comparative analysis support the postulation that TB meningeal infection is more likely to be related to the expression of multiple virulence factors on interaction with host defences than to CNS tropism associated with specific genetic traits.
Background
Tuberculosis (TB) is an ancient communicable disease that has persisted throughout the ages to remain a major killer of the human race. The main portal of entry for the causative pathogen, Mycobacterium tuberculosis (Mtb) is the respiratory tract. Following inhalation into the pulmonary alveoli, these bacilli are phagocytosed by alveolar macrophages in which they survive to either cause local lesions or be disseminated to extra-pulmonary sites (Smith, 2003).
Tuberculous meningitis (TBM) is a severe form of extrapulmonary TB that is associated with high morbidity and mortality (Erhabor, Adewole & Ogunlade, 2006; Bidstrup et al., 2002). In parts of the world where the incidence of TB is high, TBM may occur in more than 10% of TB cases, especially among children and HIV infected individuals (Ige, Sogaolu & Ogunlade, 2005). The pathogenesis of this central nervous system (CNS) Mtb infection is still not clear. Both host susceptibility factors and specific mycobacterial genetic traits have been implicated. The former has been documented extensively by clinical reports on the greater risk of CNS infection in immunocompromised hosts (Keane et al., 2001; Vinnard & Macgregor, 2009; Elmas, Akinci & Bilir, 2011). Furthermore, various polymorphisms in human genes have been identified to be more strongly associated with susceptibility to meningeal than pulmonary TB (Hawn et al., 2006; Campo et al., 2015; Hoal-Van Helden et al., 1999). On the other hand, in vitro and animal model studies have provided evidence that some Mtb strains are better able to invade the CNS because of greater production of neurotropic factors (Huang & Jong, 2001). Mtb strains causing meningitis have been associated with distinct genotypes (Arvanitakis et al., 1998; de Viedma et al., 2006, Caws et al., 2008; Hesseling et al., 2010). At the Indian National Institute of Mental Health and Neurosciences, several unique DNA patterns were shown to be present in cerebrospinal fluid (CSF) Mtb isolates that were not present in the Mtb DNA pattern library made up of 23,000 strains from across the world (NIMHANS, 2010). Pando et al. (2010) reported that BALB/c mice infected via the intra-tracheal route by Mtb came down with meningitis when infected with isolates from CSF but not with isolates from sputum. Likewise, Be et al. (2008) identified five Mtb genes (Rv0311, Rv0805, Rv0931c, Rv0986, and MT3280) associated with invasion or survival in the CNS but not in lung tissues. In particular, the sensor domain of Mtb pknD (Rv0931c) was able to trigger the invasion of brain endothelia but not the lung epithelia (Be, Bishai & Jain, 2012).
The collective evidence from patient and microbial studies indicate that both host and microbial factors direct the pathogenesis of TBM. The importance of host-microbe interactions in this infection is illustrated in a number of studies. Caws et al. (2006) showed an association between the Beijing genotype and HIV status in Vietnamese patients with TBM and linked individuals with a TLR2 T597C mutation to a greater likelihood of infection with the East-Asian/Beijing genotype. Similarly, Reed et al. (2004) suggested that the virulence of the Beijing Mtb genotype is due to the presence of an intact polyketide synthase gene (pks 15/1) encoding a phenolic glycolipid (PGL) that is able to inhibit the release of the pro-inflammatory cytokines, tumour-necrosis factor-alpha and interleukins 6 and 12. It has been observed that Mtb of Euro-American lineage that do not have an intact pks15/1 gene are less capable of causing extrapulmonary TB including TBM (Caws et al., 2006). Hence, while host genotype determines susceptibility to infection by different Mtb strains, microbial virulence factors can influence disease manifestation by their specific interactions with host immune response.
In this study, we took advantage of whole genome sequencing technology and the accessibility to public Mtb genome databases to compare genomic features between Mtb isolates from CSF and respiratory specimens. We hoped to find polymorphisms that might extend existing knowledge on CNS tropism in clinical TB.
Materials and Methods
This study was approved by the Medical Ethics Committee, University Malaya Medical Centre, Kuala Lumpur, Malaysia (Reference no. 975.28).
Bacterial strains
The Mtb strains, UM-CSF01, UM-CSF04, UM-CSF05, UM-CSF06, UM-CSF08, UM-CSF09, UM-CSF15 and UM-CSF17 (hereafter referred to collectively as UM-CSF strains) were CSF isolates from patients treated for TBM in the University Malaya Medical Center, Kuala Lumpur, Malaysia. They were recovered from routine cultures in the BACTEC MGIT 960 liquid culture system (Becton Dickinson) and identified using a reverse line probe hybridisation assay (GenoType Mycobacterium CM/AS; Hain Lifescience GmbH, Germany). The corresponding sputum samples for these strains were culture negative for Mtb. The genotypes identified using the spoligotyping kit from Ocimum Biosolutions (Hyderabad, India) were Beijing ST1 (UM-CSF01, UM-CSF05, UM-CSF08, UM-CSF09 and UM-CSF17), EAI_IND (UM-CSF06), Unknown (UM-CSF04) and H3 ST50 (UM-CSF15). They were kept in Middlebrook 7H9 broth with 15% glycerol, at −80 °C, until required for further testing. For whole-genome sequencing, they were subcultured on Lowenstein-Jensen slants, heat inactivated at 80 °C for 2 h, and cooled down to room temperature before they were used for DNA extraction.
Extraction of genomic DNA
Bacterial genomic DNA was extracted with the Phenol/Chloroform/Isoamylalcohol (PCI) method (Sambrook, Fritsch & Maniatis, 1989) to obtain a high yield of DNA. Briefly, the inactivated Mtb culture was centrifuged to a maximum speed for 15 min. The pellet obtained was lysed by incubation in 10 mg/ml of lysozyme overnight at 37 °C, followed by the addition of 10% SDS and inactivation of DNases and RNases with 10 mg/ml Proteinase K. The mixture was vortex mixed and incubated at 55 °C for 5 h. Before the purification of nucleic acid, 5 M of sodium chloride was added, followed by the addition of phenol/chloroform/isoamyl alcohol (25:24:1) to remove all proteins. The mixture was then centrifuged at maximum speed. The aqueous supernatant containing DNA was transferred to a microcentrifuge tube and the purification step was repeated. Finally, nucleic acids were recovered from aqueous solution with ethanol precipitation using 3 M of sodium acetate and ice-cold isopropanol, and overnight incubation of the mixture at −20 °C. The pellet was washed with 80% ethanol and dried at room temperature. The required DNA precipitate was dissolved and diluted with autoclaved distilled water and its concentration and purity were measured in a spectrophotometer at OD260 (Nanophotometer, Implen USA).
Whole genome sequencing
Library preparation and sequencing
The DNA sequencing libraries were prepared using Nextera− DNA Sample Preparation kit (Illumina, San Diego, CA, USA). The quality of DNA library was validated by Bioanalyzer 2,100 using high sensitivity DNA kit (Agilent, USA) prior to sequencing. Upon sequencing, DNA (6 pM) was loaded into the sequencing cartridge and the sequencing was performed on the Illumina MiSeq platform.
Read quality assessment, assembly and annotation
The quality of raw sequences generated from MiSeq was checked using FastQC. Raw reads were trimmed at Phred probability score of 30 and were de novo assembled using CLC Genomic Workbench 5.1 (Qiagen Inc., Venlo, The Netherlands). Trimmed sequences were assembled with length fraction of 0.8 and similarity fraction of 0.8. All assemblies were evaluated based on statistical assessment, focusing on genome size, sequence continuity and number of contigs. The genomes were further screened for contamination against common contaminants databases and then used for downstream analyses.
To decrease the possibility of inaccurate assembly, we repeated the assembly and scaffolding of the genomes in IDBA-UD (Peng et al., 2012) and SSPACE (Boetzer et al., 2011) respectively.
The Whole Genome Shotgun project for UM-CSF01 and UM-CSF05 has been deposited at DDBJ/ENA/GenBank under the accessions LLXF00000000, LLXG00000000, respectively and that for UM-CSF04, UM-CSF06, UM-CSF08, UM-CSF09, UM-CSF15 and UM-CSF17 under the accessions LXGA00000000, LXGB00000000, LXGC00000000, LXGD00000000, LXGE00000000 and LXGF00000000, respectively.
Genomic analysis
To reduce the likelihood of observing traits peculiar to local strains, we included respiratory strains from Malaysian patients for comparison. Hence, in addition to the UM-CSF strains, we included for analysis, 13 other Mtb sputum isolates of patients managed in our medical centre over the same time period (2009–2011). All 13 strains were de novo sequenced (using Illumina Miseq technology) and annotated for another project currently in progress in our laboratory. For their use in this study, we reconstructed their genomes in IDBA-UD 1.0.9.
Genome rearrangements
We used Gepard (Krumsiek, Arnold & Rattei, 2007) and Mauve (Darling et al., 2004) to gain an overview of chromosomal structural variation between the UM-CSF strains, the H37Rv reference strain, the 16 genomes (Table S1) downloaded on 6th August 2015, from Genome Database (one representative from each group as defined in the database) (http://www.ncbi.nlm.nih.gov/genome/genomegroups/166?) and the 13 local sputum strains.
This was followed by pairwise-alignment and comparisons using Mugsy (Angiuoli & Salzberg, 2011), with default parameters, to identify the positions of rearranged regions in UM-CSF strains.
Identification of polymorphisms
For this analysis, we downloaded from NCBI’s SRA database, the sequencing reads for 56 Mtb genomes reported to be from sputum strains (Table S2). These reads and those from our UM-CSF and respiratory strains were mapped to H37Rv using Burrow-Wheeler Aligner. Gene variants were extracted using mpileup of samtools (Li et al., 2009) and annotated with snpEff (Cingolani et al., 2012). The variants identified were filtered based on the following criteria: minimum number of good quality read of three (DP4 ≥ 3); minimum mapping quality of 25 (MQ ≥ 25); SNP and Indel quality of 20 and 60 respectively.
Amino acid comparisons
The assembled genomes of UM-CSF strains were annotated using the self-training annotation algorithm in GeneMarkS (Besemer, Lomsadze & Borodovsky, 2001). Orthologous protein sequences were identified in the ProteinOrtho program, with e-value of 1 × 10−5 (Lechner et al., 2011). The effect of amino acid substitution was evaluated using the I-mutant webserver (Capriotti, Fariselli & Casadio, 2005) for change in protein stability and the GlobPlot standalone python script (Linding et al., 2003) for globularity.
Results
Genome overview
The genomes of UM-CSF strains were recovered from approximately 55–92X sequencing coverage respectively. The detailed statistical measurements of the genomes are shown in Table 1. Compared to H37Rv and the other 29 respiratory Mtb genomes (16 downloaded from NCBI and 13 extracted from Malaysian isolates), the UM-CSF strains appeared to have fewer gene duplications but a larger number of PE (proline-glutamate)/PPE(proline-proline-glutamate)/PGRS (polymorphic GC-rich sequence) proteins. There is no notable difference between the two groups in the structure of 16S rRNA and tmRNAs and in the number of tRNAs (20–30 for CSF strains and 19–33 for respiratory strains).
| Strain | N50 | No. contigs | No. scaffolds | Reads used (%) | Scaffolded genome size (bp) | No. protein CDS |
|---|---|---|---|---|---|---|
| UM-CSF01 | 36,496 | 315 | 234 | 96.67 | 4,282,569 | 4,271 |
| UM-CSF04 | 98,670 | 140 | 136 | 93.09 | 4,392,768 | 4,323 |
| UM-CSF05 | 74,553 | 182 | 138 | 97.56 | 4,338,921 | 4,190 |
| UM-CSF06 | 83,364 | 135 | 127 | 87.01 | 4,371,105 | 4,310 |
| UM-CSF08 | 91,928 | 174 | 156 | 93.77 | 4,352,163 | 4,353 |
| UM-CSF09 | 122,404 | 133 | 103 | 89.71 | 4,355,856 | 4,291 |
| UM-CSF15 | 91,928 | 129 | 126 | 94.05 | 4,374,121 | 4,313 |
| UM-CSF17 | 122,408 | 131 | 111 | 90.99 | 4,356,783 | 4,308 |
Genome rearrangements
The UM-CSF strains showed structural differences from H37Rv and the 29 respiratory Mtb genomes used for comparison. Rearrangement analysis by Gepard and Mauve gave identical results (Figs. S1–S6). Both analyses indicated sequence fragments that could have undergone rearrangement events in six of the eight UM-CSF strains (UM-CSF01, UM-CSF05, UM-CSF06, UM-CSF09, UM-CSF15 and UM-CSF17). The affected regions ranged from 223 to 500, 211 bp in one to eight contigs and involved one to 492 genes. Based on the annotation of H37Rv, many of the genes affected belonged to PE/PPE/PGRS gene families that are known to be associated with virulence in Mtb. Also affected were genes encoding mammalian cell entry (mce) proteins, transcriptional factors, metabolic enzymes and toxin-antitoxin (TA) proteins (Tables S3–S5).
In UM-CSF01 and UM-CSF05, the recombination sites for translocations and inversions were not within genes and hence, did not affect adjacent gene sequences. In the remainder four affected UM-CSF strains, the rearrangements caused deletions resulting in gene truncations. The affected genes are Rv3425 encoding PPE57 in UM-CSF06; Rv1141c encoding enoyl-CoA hydratase EchA11 in UM-CSF09; Rv1587c coding for a hypothetical protein in UM-CSF15, and Rv 3513c encoding a fatty-acid--CoA ligase FadD18 in UM-CSF17. The function of PPE57 is unknown but both EchA11 and FadD18 are suspected to be involved in lipid metabolism, the former in fatty acid oxidation and the latter in lipid degradation.
Micro-variants and protein globularity changes
Against the H37Rv genome, we identified 737–2,578 micro-variants from the UM-CSF strains. However, as we were interested in only micro-variants possibly associated with cerebrospinal invasion, we compared our UM-CSF strains with 69 other respiratory Mtb genomes (56 downloaded from the NCBI SRA database (Table S2) and 13 local respiratory Mtb genomes). We found 63–534 micro-variants specific to our eight UM-CSF strains within protein coding regions. None were in any of the regions of difference (RD1–RD16) where many Mtb strain-specific features and virulence factors are normally found. No variant was shared by all eight strains but 36 variants involving 10 genes (PE-PGRS10, PPE58, PE_PGRS49, lppD, PE_PGRS21, Rv0278c, embR, PE_PGRS19, PPE53 and PPE24) were found in at least four of the strains. The variants in eight of these genes led to amino acid changes but only two altered genes have known functions: PE_PGRS19, a putative outer membrane protein (Song et al., 2008) and embR which is involved in transcription, the biosynthesis of mycobacterial cell wall arabinan and resistance to ethambutol (Table S6).
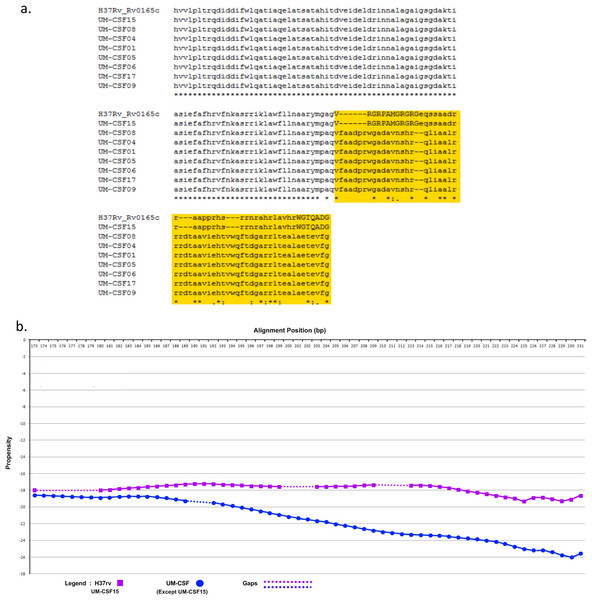
The amino acid sequence of a protein determines its folding into three-dimensional structures which may be ordered (globular with high hydrophobicity) or disordered (unstructured, typically with low hydrophobicity and a high proportion of polar and charged amino acids). There are many methods for predicting disordered proteins and various factors such as net charge, hydrophobicity and protein size can affect the accuracy of predictions. In UM-CSF strains, we noted 1,556 amino acid substitutions in 1,084 orthologs of proteins in H37Rv. With GlobPlot, 646 of the substitutions were predicted to be in disordered segments and 910 in globular segments. Figure 1 shows an example of the amino acid and globularity differences between UM-CSF strains and H37Rv. All proteins showing different globularity between UM-CSF strains and H37Rv were subjected to enrichment analysis using DAVID (Dennis et al., 2003) (Table S7). The single largest group of the enriched proteins involved transcription factors and transcription regulators. Four of the enriched proteins were found in three different CSF strains each. These were Rv0144 (probable transcriptional regulatory protein with Leu to Asp substitution), Rv3730c (hypothetical protein with Thr to Asp substitution), Rv0802c (possible succinyltransferase with Ser to Pro substitution) and Rv2034 (transcriptional regulatory protein with Ala to Thr substitution). All four showed higher propensity to disorder, and all, with the exception of Rv0802c, were accompanied by a decrease in stability. These changes are consistent with previous observations that amino acid substitutions leading to the acquisition of new protein functions are often accompanied by a loss of thermodynamic stability which may be compensated subsequently by the stabilizing effect of other unrelated mutations (Shoichet et al., 1995; Wang, Minasov & Shoichet, 2002; Tokuriki et al., 2008; Studer, Dessailly & Orengo, 2013). Furthermore, all four proteins were enriched (49–53%) in the disorder-promoting amino acids (A, R, G, Q, S, P, E, K) and depleted (28–34%) of order-promoting amino acids (W, C, F, I, Y, V, L, N) as described by Dunker et al. (2001). Hydrophobic amino acids form less than 50% of the amino acid sequence in each of these disordered proteins. As many disordered proteins or protein domains are functionally related to disease, it is possible that some of those identified in UM-CSF strains might contribute to an enhanced ability to cause TBM. However, a larger search for these protein structural and globularity changes found them to be present in many of our local respiratory strains as well.
Figure 1: (A) Alignment of Rv0165c sequences in eight UM-CSF strains and H37Rv, showing amino acid and globularity changes.
(B) Differences in propensity scores for amino acids in UM-CSF strains and H37Rv.Proteins reported to be associated with meningitis in M. tuberculosis
We looked for putative meningitis-associated factors that had been previously reported in scientific literature. Of 63 proteins reported for Mtb (Av-Gay & Everett, 2000; Pethe et al., 2001; Tsenova et al., 2005; Jain et al., 2006; Be et al., 2008; Be, Bishai & Jain, 2012) (Table S8), homologs of 56–60 were found in UM-CSF strains but only two (Rv0311 encoding a hypothetical protein and Rv0619 encoding a probable galactose-1-phosphate uridylyltransferase GalTb) were found in all eight CSF strains. Compared to H37Rv, both proteins showed amino acid and globularity differences. The Rv0311 protein in five of the eight UM-CSF strains had Glu (GAG) instead of Asp (GAT) at position 119 and was predicted to be disordered. Rv0619 in all eight UM-CSF strains had a substitution of Ala (GCC) for Thr (ACC) at position 174 and appeared as a globular segment with a change from polarity to hydrophobicity.
Additionally, we performed a similar search in five other mycobacteria that are associated with neuropathology. These comprised M. leprae and M. lepromatosis that cause different forms of leprosy, M. bovis that is usually linked with extrapulmonary TB, and two rapid-growers M. llatzerense and M. immunogenum that had been isolated from a case of brain abscess (Greninger et al., 2015). Fifty-six homologs of the 63 meningitis-related genes from Mtb were identified in M. bovis, followed by 16 in M. leprae, 15 in M. lepromatosis, 14 in M. llatzerense, and 11 in M. immunogenum. Our UM-CSF strains shared four of the 63 meningitis-related genes (Rv0014c, Rv1837c, Rv2176 and Rv0984) with all five of these mycobacterial species and five other genes (Rv1273c, Rv2318, Rv0983, Rv0966c and Rv0805) with the three slow-growing mycobacteria (Table 2). The Rv2947c (pks15/1) gene was found in the UM-CSF strains, M. leprae and M. ilatzerense. In the Beijing genotype of Mtb, an intact pks 15/1 is believed to be responsible for virulence and extrapulmonary disease (Reed et al., 2004). Consistent with their extrapulmonary (CNS) location in the host, five of our eight UM-CSF strains were genotyped as Beijing ST1 and each carried an intact pks 15/1 gene.
| Host | Rv number | Gene name | Product | Function |
|---|---|---|---|---|
| UM-CSF strains Mtb | Rv0311 | Rv0311 | Unknown protein | Unknown |
| UM-CSF strains Mtb | Rv0619 | galTB | Probable galactose-1-phosphate uridylyltransferase GalTb | Galactose metabolism (leloir pathway) catalytic activity |
| UM-CSF strains Mycobacteria, S pneumoniae | Rv1699 | pyrG | CTP synthase PyrG | Pyrimidine biosynthesis catalytic activity |
| UM-CSF strains Mycobacteria, S pneumoniae | Rv2606c | snzP | Pyridoxine biosynthesis protein SnzP | Biosynthesis of pyridoxine/pyridoxal 5-phosphate |
| UM-CSF strains Mycobacteria, S pneumoniae | Rv0357c | purA | Probable adenylosuccinate synthetase purA | AMP biosynthesis |
| UM-CSF strains Mycobacteria N meningitidis | Rv2457c | clpX | ATP-dependent CLP protease ATP-binding subunit ClpX | Directs the CLP protease to specific substrates; performs chaperone functions in the absence of CLP protease |
| UM-CSF strains N meningitidis | Rv2397c | cysA1 | Probable sulfate-transport ATP-binding protein ABC transporter cysA1 | Active transport of multiple sulfur-containing compounds across the cell membrane; energy coupling to the transport system |
| UM-CSF strains Mycobacteria | Rv1837c | glcB | Malate synthase G GlcB | glyoxylate bypass; alternative to TCA cycle |
| UM-CSF strains Mycobacteria | Rv0014c | pknB | Transmembrane serine/threonine-protein kinase B PknB | Signal transduction (via phosphorylation) |
| UM-CSF strains Mycobacteria | Rv2176 | pknL | Probable transmembrane serine/threonine-protein kinase L PknL | Signal transduction (via phosphorylation) |
| UM-CSF strains Mycobacteria | Rv0984 | moaB2 | Possible pterin-4-alpha-carbinolamine dehydratase MoaB2 | Molybdopterin biosynthesis |
| UM-CSF strains SG Mycobacteria | Rv1273c | Rv1273c | Probable drugs-transport transmembrane ATP-binding protein ABC transporter | Active transport of drugs across the membrane (export) |
| UM-CSF strains SG Mycobacteria | Rv2318* | uspC | Probable periplasmic sugar-binding lipoprotein UspC | Active transport of sugar across the membrane (import) |
| UM-CSF strains SG Mycobacteria | Rv0983 | pepD | Probable serine protease PepD (serine proteinase) | Unknown; possibly hydrolyzes peptides and/or proteins |
| UM-CSF strains SG Mycobacteria | Rv0966** | Rv0966 | Conserved protein | Unknown |
| UM-CSF strains SG Mycobacteria | Rv0805 | Rv0805 | Class III cyclic nucleotide phosphodiesterase | Hydrolyzes cyclic nucleotide monophosphate to nucleotide monophosphate |
Meningitis-associated genes from other bacterial pathogens
Streptococcus pneumoniae, Escherichia coli K-1 and Neisseria meningitidis are pathogens known to cause meningitis in humans. Of 141 proteins reported to be associated with S. pneumoniae meningitis (Orihuela et al., 2004; Molzen et al., 2011; Mahdi et al., 2012) (Table S9), three, Rv1699 (CTP synthase PyrG), Rv2606c (pyridoxine biosynthesis protein SnzP) and Rv0357c (adenylosuccinate synthetase PurA) were found in our UM-CSF strains. These genes showed 51–68% sequence similarity with their homologs in S. pneumoniae but were identical in all UM-CSF strains and H37Rv, in protein sequence as well as globularity (Fig. 2). When compared against 164 N. meningitidis virulence genes reported by Hao et al. (2011) (Table S10), UM-CSF strains shared two virulence homologs with this neuropathogen: Rv2457c, encoding ATP-dependent CLP protease ATP-binding subunit clpX and Rv2397c, encoding sulfate-transport ATP-binding protein ABC transporter CysA1. We failed to identify the genes for cell surface outer membrane Opa and Opc proteins that were previously reported to confer tissue tropism in N. meningitidis (Virji et al., 1993) and we did not find any homologs of previously reported E. coli K1 neurotropic genes such as IbeA, IbeB, AslA, YijP, and OmpA (Pouttu et al., 1999; Huang et al., 2001; Yao, Xie & Kim, 2006) (Table S11), in UM-CSF strains.
Figure 2: Alignment of meningitis-associated proteins from S. pneumoniae, H37Rv and eight UM-CSF strains.
Discussion
With the aid of whole genome sequencing, comparative genomics has become the mainstay analytical platform for the study of bacterial taxonomy, evolution and virulence. Even with impaired host immunity being recognized to be mainly responsible for extra-pulmonary dissemination in TB, it is still tempting to use the vast volume of bacterial sequence data amassed to look for mycobacterial factors that might direct the dissemination of Mtb to the CNS. Towards this end, we compared genomic features in CSF and respiratory Mtb isolates and searched the genomes for genes previously reported to be associated with CNS infection.
Although Mtb exhibits less genetic diversity than other bacteria, comparative genomic analyses have revealed SNPs, large sequence polymorphisms and variations in mobile elements among Mtb isolates from different sources (Fleischmann et al., 2002; Tsolaki et al., 2004). Large-scale chromosomal rearrangements have also been reported, such as the large inversions detected in the KwaZulu-Natal (KZN) strains from South Africa (Okumura et al., 2015) and the B0/W148 strains from Russia (Shitikov et al., 2014). In our UM-CSF strains, we found translocations, inversions, indels and SNPs not detected in 30–69 respiratory strains used for comparison. Many of the genes involved were putative virulence factors that included abundant PE and PPE proteins. The prominence of PE/PPE proteins here is not surprising as these hypervariable cell surface antigens form about 10% of the total coding sequences in the Mtb genome (Cole et al., 1998) and their association with large sequence polymorphisms (Talarico et al., 2007) has been reported. It is believed that these uniquely mycobacterial proteins are secreted by the ESX apparatus in the cell membrane (Abdallah et al., 2006) and are involved in many aspects of the infection process, such as promoting Mtb entry into macrophages and evasion of host immune responses, resulting in Mtb dissemination and pathology in different organs and tissues. Unfortunately, all the PE/PPE proteins we identified have no known function except for PE-PGRS30 which has been listed as a virulence factor. Also abundant in the rearranged fragments are TA and mce proteins. The TA systems in Mtb are up-regulated following bacterial entry into macrophages (Ramage, Connolly & Cox, 2009). The VapBC family which made up most of the TA proteins in our UM-CSF strains, was shown to control the persistence of uropathogenic E. coli within host tissues in cases of sepsis, meningitis and urinary tract infections (Norton & Mulvey, 2012). It is possible that these proteins also contribute to the persistence of Mtb in TBM. Similarly, mce proteins may enhance cellular invasion and persistence of Mtb in the CNS by facilitating the uptake and utilisation of cholesterol from host cells during infection (Pieters & Gatfield, 2002). We looked for the 11 putative serine-threonine protein kinases (pknA-pknL) in Mtb as Be, Bishai & Jain (2012) identified Mtb pknD (Rv0931c) as a key microbial factor required for CNS tropism. We found the pkn genes in our UM-CSF strains but only pknG (Rv0410c) was in a rearranged fragment in UM-CSF05.
We looked for globularity changes as the folding of proteins affects their function. In our DAVID analysis of proteins showing different globularity between UM-CSF strains and H37Rv, transcription factors and other transcription regulators predominate. It is expected that changes in transcription-related proteins would have profound effects on gene expression resulting in wide-ranging changes in Mtb behavior including altered tropism and virulence. An interesting finding is the enrichment of proteins involved in androgen and estrogen metabolism. This could be related to the role of sex hormones in the modulation of bacterial-host interactions. It is known that, while sex hormones from the host can affect the metabolism and virulence of bacteria, they are also degraded by bacteria to be used as carbon and energy sources (García-Gómez, González-Pedrajo & Camacho-Arroyo, 2013; Neyrolles & Quintana-Murci, 2009). It is plausible that a change in protein domain globularity in the Mtb proteins involved in sex hormone metabolism could upset the usual bacterial-host interactions to facilitate CNS invasion in TBM. Among other enriched proteins of interest are ABC transporters, glycerophospholipids and proteins involved in DNA repair. ABC transporters transfer molecules across cell membranes and are implicated in Mtb traversal across the blood-brain barrier (Jain et al., 2006). Glycerophospholipids are found in neural membranes (Farooqui, Horrocks & Farooqui, 2000). A functional change in these lipids could increase membrane permeability to Mtb. Bacterial DNA can be damaged by reactive oxygen species and reactive nitrogen intermediates generated from cellular metabolism and the host immune response (Davidsen et al., 2007). DNA repair mechanisms include nucleotide excision repair (NER) controlled by the UvrABCD endonuclease enzyme complex, base excision repair and DNA mismatch repair. Darwin & Nathan (2005) showed that the uvrB gene is required for Mtb to resist reactive oxygen and nitrogen molecules in vivo. In UM-CSF strains, we found the uvrC gene in a rearranged fragment in UM-CSF01 and identified homologs of mismatch repair genes among the enriched proteins with globularity changes.
In our search for common neurotropic genes, we compared our UM-CSF strains with five mycobacterial species associated with neuropathology (M. leprae, M. lepromatosis, M. bovis, M. ilatzerense and M. immunogenum) and three other established neuropathogens, S. pneumoniae, N. meningitidis and E. coli K1. We didn’t find any evidence of horizontal gene transfer of previously reported neurotropic factors, between UM-CSF strains and the other eight pathogens. We did find, however, homologs of genes associated with CNS disease in all the pathogens we examined, with the exception of E. coli K1 (Table 2). The number of Mtb gene homologs was, expectedly, highest in M. bovis, a member of the Mtb complex (56 of 63, 89%), followed by M. leprae (16 of 63, 25%) and M. lepromatosis (15 of 63, 24%). To our surprise, the number of homologs in M. llatzerense (14 of 63, 22%) and M. immunogenum (11 of 63, 17%), the two environmental rapid growers isolated from a brain abscess, was almost as high as in the established neuropathogens M. leprae and M. lepromatosis.
The nine common genes we identified in UM-CSF strains and other mycobacterial spp. are mostly involved in cell membrane transport, signal transduction, nucleotide biosynthesis, and energy metabolism (Table 2). Although these functions are common to all microbial cells, most of the genes have been previously associated with TB CNS pathology. For instance, Rv2318, Rv0983, Rv0984, and Rv0966c were found to be up-regulated in an in vitro model of TB infection of the human brain microvascular endothelium (Jain et al., 2006); Rv1837c was reported to be expressed in the early stages of TBM in children (Haldar et al., 2012); the transmembrane serine/threonine-protein kinase PknD (Rv0931c) was found by Be, Bishai & Jain (2012) to be required for the invasion of brain endothelia and Rv0805 was found to be important for the invasion and survival within brain tissue in a murine model (Be et al., 2008). The genes shared by UM-CSF strains and S. pneumoniae are involved in the biosynthesis of pyrimidine (pyrG, Rv1699), purine (purA, Rv0357c) and pyridoxine (Rv2606c). Both purA (adenylosuccinate synthetase) and pyrG (CTP synthase) were associated with attenuated S. pneumonia replication during experimental meningitis (Molzen et al., 2011) and pyridoxine is important for the functioning of nerves. One homolog of N. meningitidis (Rv2457c) is a chaperone of the ATP-dependent CLP protease that is required for in vitro and in vivo growth of Mtb (Raju et al., 2012). The other homolog (Rv2397c) is a part of the ABC transporter complex involved in sulfate/thiosulfate import (Szklarczyk et al., 2015).
In summary, in our UM-CSF strains, we found large-scale and smaller genomic rearrangements, indels, gene truncations and micro-variants that we did not detect in our comparison respiratory strains. All eight CSF strains shared common protein globularity differences against H37Rv and common homologs of meningitis-associated proteins in Mtb, S. pneumoniae and N. meningitidis. However, we failed to identify features more apparently related to TB neurotropism or neurovirulence such as proteins involved with changes in brain vascular endothelium or extracellular matrix composition that could result in increased permeability of the blood-brain-barrier. This could be partly due to the abundance of PE/PPE proteins and other proteins with unknown function, among the traits we observed. Future investigations could reveal their involvement in CNS tropism and pathology.
We were also disappointed to find that many of the globularity changes and virulence factors in our CSF strains (including those from S. pneumoniae and N. meningitidis) were also present in our comparison respiratory strains. However, this finding is consistent with the observation by other workers that many virulence genes are conserved in non-pathogenic bacteria. All four mce operons in the genome of Mtb (Kumar, Bose & Brahmachari, 2003) have been found in both pathogenic and non-pathogenic mycobacteria (Haile et al., 2002; Chitale et al., 2001). CLP proteases on the whole, are common in many bacterial spp. (De Mot et al., 1999). The ABC transporter complex involved in sulfate/thiosulfate import is found in pathogens as well as environmental bacteria (Szklarczyk et al., 2015). Many designated virulence genes in N. meningitidis were also found to be present in nonpathogenic species such as N. lactamica (Snyder & Saunders, 2006). All these observations suggest that pathogenic bacteria have adapted their genomes from a free-lifestyle to the intracellular environment with minimal acquisition of exclusive virulence genes (Forrellad et al., 2013). Hence, by analogy, we can also hypothesize that CNS tropism in TB is not driven by the presence of specific genetic traits but by the expression of multiple virulence factors, probably elicited in response to host immune defences.
Nonetheless, the detection of S. pneumoniae and N. meningitidis gene orthologs in our UM-CSF strains raises speculations on the existence of a pan-bacterial mechanism of CNS infection. Further investigations on our observations might lead to new understanding and new strategies in the management of tuberculous as well as other bacterial CNS infections.
Conclusion
Many genetic traits have been described for bacterial pathogens causing CNS infection. We detected large-scale rearrangements, short translocations, inversions, indels and nsSNPs in our CSF-derived strains, as well as protein globularity changes and orthologs of meningitis-associated genes previously reported in other neuropathogenic bacteria. Many of these features are, however, not CSF-specific or consistently present in all CSF strains. Hence, our findings suggest that neurotropism and neurovirulence in TBM is directed by the expression of multiple virulence factors selected by the interaction between pathogen and host immune responses rather than the presence of specific genetic traits.